The kinetochore is a macromolecular protein complex that acts as an adapter between chromosomes and spindle microtubules. The kinetochore is typically divided into two regions: the inner kinetochore, which binds to the centromeric DNA, and the outer kinetochore, which interacts with microtubules (Musacchio and Desai, 2017; Ariyoshi and Fukagawa, 2023). Proper kinetochore attachment to the microtubules is monitored by the Spindle Assembly Checkpoint (SAC), which delays anaphase by inhibiting the anaphase-promoting complex/cyclosome (APC/C) until proper attachment and alignment are achieved, thereby ensuring accurate chromosome distribution to daughter cells (Komaki and Schnittger, 2016; McAinsh and Kops, 2023).
Centromere identity is typically defined by the presence of an epigenetic marker: a centromere-specific variant of histone 3, known as CENH3 or CENP-A (CENPA). Research on plant centromeres has gained significant traction in recent years, due to its practical relevance for plant breeding, including haploid induction via centromere-mediated genome elimination (Marimuthu et al., 2021; Quiroz et al., 2024) and the design of stably inherited synthetic chromosomes (Puchta and Houben, 2024; Wang et al., 2024). Several reviews highlighting advances in plant centromere research have been recently published (Chandra et al., 2024; Naish and Henderson, 2024); therefore we do not focus on centromeres or centromeric histone in this manuscript. Here, we summarize recent discoveries in plant kinetochore research across various species, compare them with animal and fungal kinetochores, and discuss unanswered questions in the field.
1.1 Unusual composition of the inner kinetochore in plantsThe inner kinetochore, also known as the Constitutive Centromere-Associated Network (CCAN), comprises 16 subunits in human cells. Recent studies in yeast and human cells have shown that CENP-LN serves as the primary DNA-binding module, with other CCAN subunits enhancing DNA binding. CENP-C acts as a critical linker, connecting CCAN to CENP-A and the centromere (Musacchio and Desai, 2017; Yan et al., 2019; Pesenti et al., 2022). In some species, the number of CCAN proteins is highly reduced; for example, the inner kinetochores of C. elegans and D. melanogaster consist only of CENP-A and CENP-C, with the latter directly linking the centromere to the outer kinetochore (Drinnenberg et al., 2016).
In plants, homologues of the CCAN subunits CENP-C, CENP-O, CENP-S (MHF1), CENP-X (MHF2), and CENP-U (BIN4) have been identified (Table 1). In Arabidopsis CENP-C was shown to co-localize with the 180 bp centromeric regions of chromosomes throughout the cell cycle (Ogura et al., 2004). Another study combining biochemistry and in vivo analysis demonstrated that CENP-C binds DNA through a specific 122-amino-acid region in maize, with binding affinity enhanced in the presence of small single-stranded centromeric RNAs (Du et al., 2010), suggesting centromeric ssRNA may have a potential role in augmenting kinetochore formation. Another study has also demonstrated that the γ-tubulin complex protein 3-interacting proteins (GIPs), GIP1 and GIP2, are involved into the recruitment of both CENH3 and CENP-C to the centromeres (Batzenschlager et al., 2015).
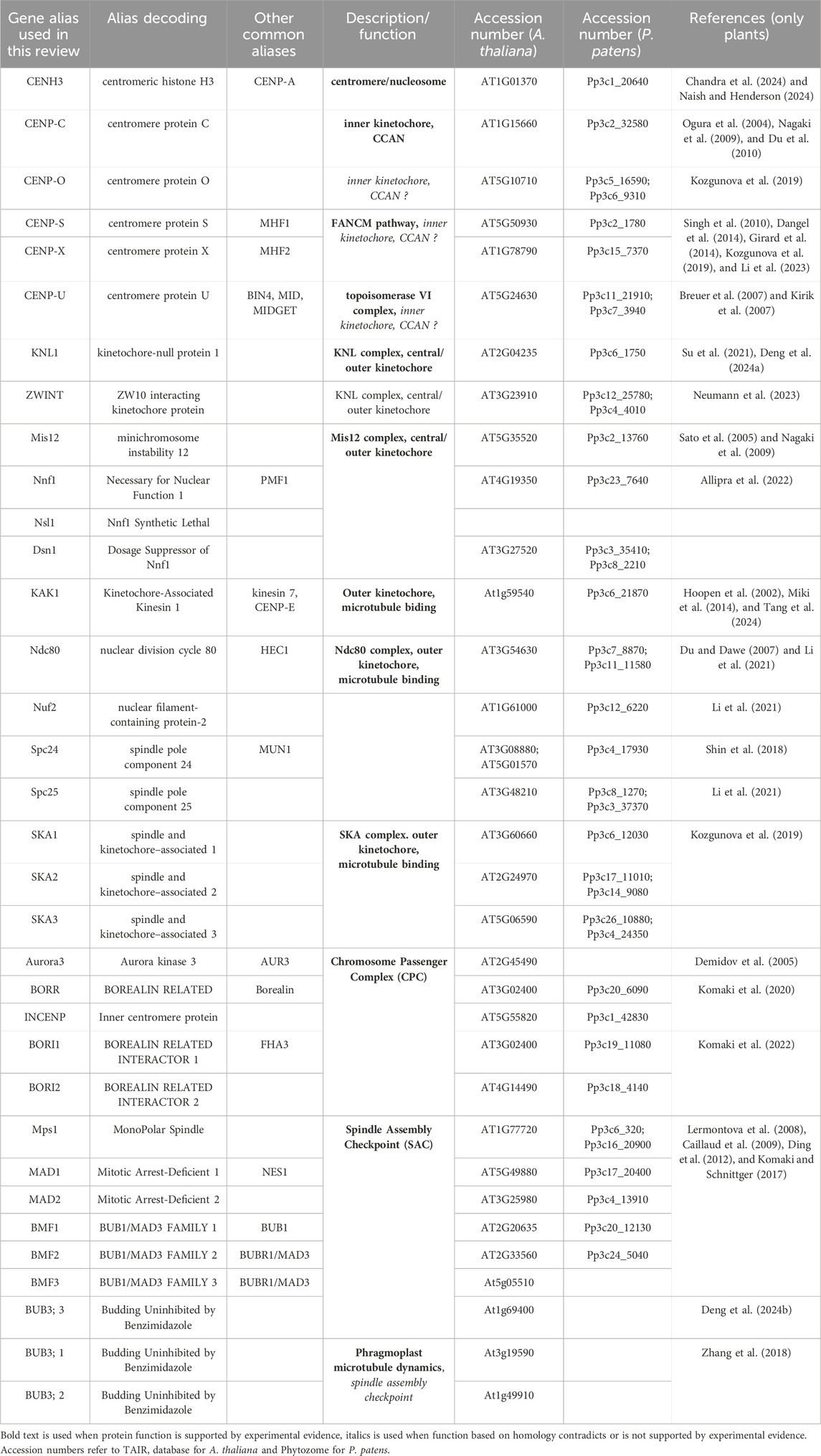
Table 1. Kinetochore and kinetochore-related proteins in plants.
Information about whether other CCAN proteins beside CENP-C play a role in plants’ kinetochore formation and cell division remains limited. CENP-U homologue is known as BIN4 or MIDGET, a part of the topoisomerase VI complex in plants. Mutations in BIN4 affect endoreduplication and produce brassinosteroid-insensitive dwarves with no known implications for kinetochore assembly or cell division (Breuer et al., 2007; Kirik et al., 2007). In human cells, CENP-U, as a part of CENP-OPQUR complex, recruits Polo-like kinase 1 (PLK1) to the kinetochore (Chen et al., 2021; Nguyen et al., 2021; Singh et al., 2021); the divergent role of the plant CENP-U is consistent with the absence of PLK1 in plant genomes.
CENP-S (MHF1) and CENP-X (MHF2) have been identified through genetic screening in Arabidopsis as factors that limit crossovers during meiosis (Girard et al., 2014), with similar results later shown in rice (Li et al., 2023). Another study has shown that MHF1 also works in the interstrand cross-link repair and is necessary for efficient homologous recombination (HR) in somatic cells (Dangel et al., 2014). A screen of kinetochore proteins in the bryophyte Physcomitrium patens (Physcomitrella) discovered that in moss cells, CENP-O, CENP-S, and CENP-X do not localize to kinetochores. Surprisingly, despite CENP-X’s lack of kinetochore localization, its knockdown via inducible RNA interference leads to chromosome missegregation defects during mitosis, resembling the phenotypes seen after knockdown of other kinetochore proteins (Kozgunova et al., 2019). Overall, the function of CENP-S and CENP-X in DNA repair and restricting meiotic crossovers appears to be highly conserved among eukaryotes (Singh et al., 2010), while their role in the inner kinetochore in plant cells remains ambiguous and calls for further investigation.
1.2 Outer kinetochore proteins in plantsThe outer kinetochore connects the inner kinetochore to microtubules, transmitting forces from microtubule depolymerization to move chromosomes. Known as the KMN protein assembly, it includes the KNL1, Mis12, and Ndc80 complexes, along with other important proteins like the SKA complex. Unlike the reduced inner kinetochore, outer kinetochore components are mostly conserved in plants (Figure 1; Table 1).
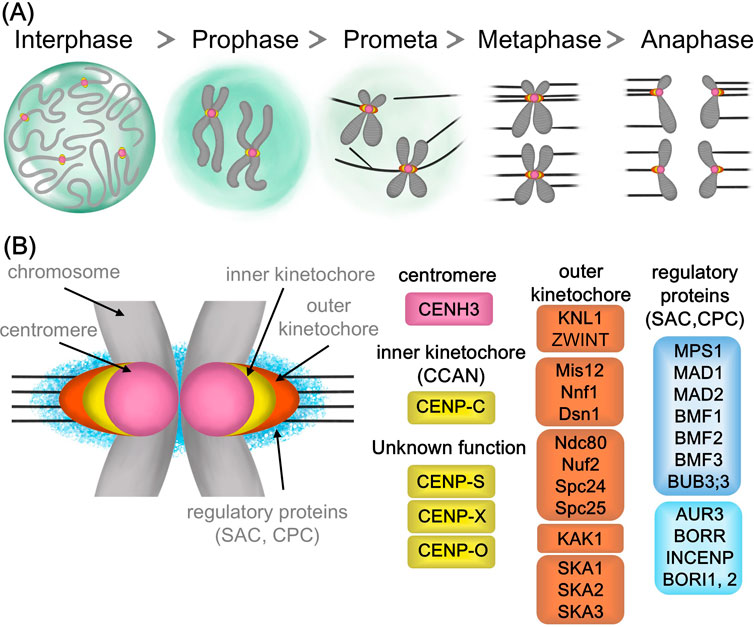
Figure 1. Cell division and kinetochore proteins in plant cells (A) Schematic of cell division stages and chromosome attachment to spindle microtubules by kinetochore (B) A color-coded model of kinetochore organization, showing centromere, inner kinetochore, outer kinetochore and regulatory proteins.
The Mis12 complex (MIND complex in S. cerevisiae), comprised of Mis12, Dsn1, Nuf2, and Nsl1 proteins, connects the inner and outer kinetochores by binding to CENP-C through Mis12 subunit, and the Ndc80 complex through Nsl1 and Dsn1 subunits, respectively (Musacchio and Desai, 2017). In Arabidopsis, a homologue of Mis12 co-localize with centromeric repeats throughout the cell cycle (Sato et al., 2005). Knockout of Nnf1 leads to embryonic lethality, indicating that it is essential for cell division. Interestingly, ectopic expression of GFP-Nnf1 rescues this lethality in homozygous Nnf1−/− mutants but results in dwarf plants, which is associated with decreased levels of endogenous polyamines (Allipra et al., 2022). The same study also suggested that the leucine zipper-like binding motif of Nnf1 may play a role in gibberellic acid (GA) metabolism. These findings highlight the intriguing possibility that kinetochore proteins can have additional functions beyond their roles in chromosome segregation.
The KNL1-ZWINT complex is known to recruits mitotic regulators, including SAC proteins BUBR1 and BUB1, to ensure accurate chromosome segregation. KNL1 features MELT-like motifs that, when phosphorylated, serve as docking sites for BUB1. However, in land plants, KNL1 lacks MELT repeats (Tromer et al., 2015), and the BUB1/MAD3 family has independently evolved distinct structures compared to animals and yeast (Komaki and Schnittger, 2016). This raises questions about how SAC proteins are recruited to the kinetochore and their binding partners. A recent study on maize identified KNL1 as a constitutive kinetochore component with an important role in chromosome segregation. A 145-amino acid region within maize KNL1 was found to interact with SAC proteins BMF1 and BMF2, but not BMF3 (Su et al., 2021). Interestingly, this BMF-interacting region is conserved in monocots but varies significantly in eudicots, suggesting different modes of SAC-to-kinetochore connections across plant lineages. This hypothesis was further confirmed in a study characterizing KNL1 homologue in Arabidopsis. The knl1 null mutant is viable but shows severe growth defects and chromosome missegregation phenotype. In the mutant, the kinetochore localization of several SAC components, including BUB3.3, BMF3, and MAD1, is lost, while BMF1 remains unaffected. The authors also identified a eudicot-specific domain responsible for recruiting BUB3.3 and BMF3 to the kinetochore. Cross-species rescue experiments confirmed that KNL1 from the dicot plant (tomato) could recover the phenotype in the Arabidopsis knl1 mutant, but KNL1 from monocots (rice) or bryophytes (P. patens) could not (Deng et al., 2024a). In animal cells, ZWINT recruits the RZZ complex, which facilitates the localization of dynein and dynactin to kinetochores. Neither dynein, dynactin, nor RZZ complex have been identified in plants. However, potential ZWINT homologues have been identified in Cuscuta and Arabidopsis, though their functions remain uncharacterized (Neumann et al., 2023).
Accurate chromosome segregation depends on stable end-on attachments between kinetochores and microtubules, with the microtubules’ plus-ends embedded in the kinetochore. The Ndc80 complex, a heterotetramer of Ndc80 (HEC1), Nuf2, Spc24, and Spc25, plays a crucial role in forming these attachments. The N-terminal regions of Ndc80 and Nuf2 mediate microtubule binding, while Spc24 and Spc25 connect to the Mis12 complex (Musacchio and Desai, 2017). In Arabidopsis, null mutants of Spc24 (meristem unstructured-1, MUN1) and Nuf2 are embryonically lethal, and all subunits of Ndc80 complex localize to centromeres/kinetochores throughout the cell cycle (Shin et al., 2018). A weak allele of MUN1 displays stunted growth, aneuploidy, and chromosome segregation defects and interaction of MUN1 with other Ndc80 complex components was confirmed through yeast two-hybrid assays and co-immunoprecipitation, establishing MUN1 as a functional homologue of Spc24 (Shin et al., 2018). Another study examined AtNuf2 role in partially complemented nuf2 mutant seedlings, revealing mitotic defects like aberrant spindle microtubules, chromosome bridges, and lagging chromosomes (Li et al., 2021). In maize, a homologue of Ndc80 is consistently present on kinetochores during the cell cycle and located outside the inner kinetochore protein CENP-C during meiosis (Du and Dawe, 2007).
In animal cells, CENP-E, also known as kinesin 7, is a motor protein that plays an important role in chromosome congression and SAC activation (Maiato et al., 2017). A study in P. patens identifying kinesin7-III as a potential CENP-E due to its co-localization with Mis12 (Miki et al., 2014), and another in barley showing antibodies against Cpel1 and Cpel2 label centromeres on mitotic chromosomes (Hoopen et al., 2002). This knowledge gap was bridged by a recent study in Arabidopsis which identified kinetochore-associated kinesin 1 (KAK1) from the kinesin 7 family as a downstream target of BUB3.3 driving chromosome congression (Tang et al., 2024).
The SKA complex is another microtubule-binding kinetochore subunit conserved in many eukaryotes. In the moss P. patens, SKA1 and SKA2 proteins localize around the nucleus during prophase and are recruited to kinetochores after nuclear envelope breakdown. Knockdown of SKA1 leads to various chromosome missegregation defects and mitotic delay, indicating its crucial role in moss kinetochores (Kozgunova et al., 2019). While the SKA complex is also present in angiosperms, no functional analyses have been conducted to date.
1.3 Regulatory proteins: spindle assembly checkpoint complex and chromosome passenger complexThe spindle assembly checkpoint (SAC), composed of BUB and MAD proteins, ensures that chromosomes are properly attached to the spindle microtubules. In fungi and animals, at the start of cell division, MAD1 and MAD2 are recruited to kinetochores, followed by BUBR1/MAD3, which associates with kinetochores through interaction with the WD40 repeat protein BUB3. Together, BUB3, BUBR1/MAD3, and MAD2 form the mitotic checkpoint complex (MCC) with CDC20, the activator of the APC/C (Lara-Gonzalez et al., 2021).
Although the core framework of SAC components is also found in plants (Caillaud et al., 2009; Komaki and Schnittger, 2016), the BUB1/MAD3 family has undergone several duplication events, evolving in a different way from Bub1 and BubR1/Mad3 in yeast and animal. In recognition of their differences, new gene nomenclature has been established: BUB1/MAD3 FAMILY 1 (BMF1) for BUB1, BMF2 for MAD3.1, and BMF3 for MAD3.2 (Komaki and Schnittger, 2017). Surprisingly, the SAC significance in plant development under normal conditions appears to be minor. This is supported by the fact that homozygous DNA insertion mutants of all putative core SAC genes could be isolated in Arabidopsis (Ding et al., 2012; Komaki and Schnittger, 2017; Zhang et al., 2018). Although the mutants are more sensitive to microtubule-depolymerizing drugs, they grow similarly to wild-type plants under normal conditions (Komaki and Schnittger, 2017). SAC proteins also exhibit localization patterns distinct from mammalian and yeast cells; for example, BMF3 and MAD2 show typical accumulation at kinetochores post-nuclear envelope breakdown, while BMF1 and MPS1 localize to centromeric regions throughout the cell cycle. BUB3 proteins display unique plant-specific localization: notably, BUB3; 1 and BUB3; 2 are observed in the phragmoplast midzone during cytokinesis (Komaki and Schnittger, 2017). Mutations in BUB3; 1 were originally reported to cause embryonic lethality (Lermontova et al., 2008); however a later study could isolate both bub3;1 mutant and bub3;1/bub3;2 double mutant (Zhang et al., 2018). BUB3; 1 and BUB3; 2 were shown to interact with MAP65-3 and play a role in regulating phragmoplast microtubule dynamics by enhancing the binding of MAP65-3 to microtubules (Zhang et al., 2018). In Arabidopsis, BUB3; 3 is detected at kinetochores throughout mitosis, and bub3;3 T-DNA insertion mutants often exhibit misaligned chromosomes and possess a non-functional SAC (Deng et al., 2024b). Another study discovered that chromosome misalignment in BUB3.3-depleted plants can be rescued by artificial tethering of KAK1 (kinesin-7) to kinetochores, suggesting KAK1 as a downstream target (Tang et al., 2024). Interestingly, the localization of SAC proteins, including MPS1, MAD1, BMF2, and BMF3, was found to be independent of BUB3; 3 recruitment to the kinetochore and vice versa. However, the interaction between BUB3; 3 and BMF3 enables the downstream recruitment of CDC20 to kinetochores (Deng et al., 2024b).
Another group of regulatory proteins which plays an important role in correcting kinetochore-microtubule attachments is the Chromosome Passenger Complex (CPC). The CPC resides mostly on inner centromere and consists of four proteins: the kinase Aurora B, INCENP, Borealin, and Survivin. In animal cells, Aurora B destabilizes incorrect kinetochore-microtubule attachments by selectively phosphorylating Ndc80 (Haase et al., 2017; Trivedi and Stukenberg, 2020). Plants also possess CPC, although with an altered composition. The homologue of Aurora B in plants is encoded by the AURORA 3 (AUR3) gene, which localizes to the centromeres and kinetochores (Demidov et al., 2005). A plant homologue of INCENP was identified through the study of the wyrd (wyr) mutant with abnormal ovule development (Kirioukhova et al., 2011). Further analysis revealed that both INCENP and homologue of Borealin, BOREALIN RELATED (BORR), localize to the centromere and kinetochore, similar to AUR3. Frameshift mutations in BORR are lethal, and knockdown of BORR compromises chromosome segregation and development (Komaki et al., 2020). Recently, two Survivin-like genes, BOREALIN RELATED INTERACTOR 1 and 2 (BORI1 and BORI2), were also identified in Arabidopsis. Loss of BORIs’ function is lethal, and reduced expression of BORIs leads to severe developmental defects. Similar to Survivin, BORIs are essential for targeting the CPC to chromatin and bind to phosphorylated histone H3 through their FHA domain (Komaki et al., 2022). Although CPC components have been identified in plants, their role in correcting kinetochore-microtubule attachments is still unclear, indicating a promising area for future research.
1.4 Non-canonical kinetochores in the Cuscuta parasitic plantsThe plant kingdom includes both monocentric species with a single centromere per chromosome and holocentric species with centromere activity along the chromosome length (Chandra et al., 2024; Naish and Henderson, 2024). The genus Cuscuta, consisting of around 200 species of parasitic plants, includes both monocentric and holocentric species. This diversity provides a unique opportunity to investigate the changes associated with the transition from monocentric to holocentric in a closely related species. In the holocentric species Cuscuta europaea, two variants of CENH3 are located in one to three discrete regions per chromosome, while the rest of the chromatin lack CENH3 signals. Despite this distribution, spindle microtubules attach uniformly along the entire length of the chromosomes, including the CENH3-free areas. This raises the question of whether CENH3 has lost its function or operates alongside an alternative CENH3-free mechanism for kinetochore positioning (Oliveira et al., 2020). The transition to holocentric chromosomes is accompanied by drastic changes in kinetochore composition, most notably the loss of KNL2 and several SAC genes, while CENP-C, KNL1, and ZWINT-1 homologues are truncated. Furthermore, in Cuscuta epithymum, no CENH3 signal is detected on the chromosomes; consequently, the centromeric localization of kinetochore proteins CENP-C, KNL1, Mis12, and Ndc80 is disrupted (Neumann et al., 2023). This suggests that some holocentric Cuscuta species do not form a conventional kinetochore and have either evolved unique kinetochore genes, reminiscent of kinetoplastids (Akiyoshi, 2016), or developed alternative kinetochore assembly mechanisms, like Lepidoptera species utilize divergent CENP-T, instead of CENH3-CENP-C, for kinetochore assembly (Cortes-Silva et al., 2020).
2 DiscussionAlthough the centromere/kinetochore tandem performs a highly conserved function in eukaryotes, there is considerable variability in centromeric repeats and kinetochore complex composition across species (Roach et al., 2012). For instance, the inner kinetochore in plants consists of only a few proteins compared to the 16 subunits of the human CCAN. CENP-C appears to be the most functionally conserved inner kinetochore protein characterized across different plant species (Ogura et al., 2004; Nagaki et al., 2009; Du et al., 2010). While it is possible that plants have lost many inner kinetochore components, like D. melanogaster or C. elegans (Drinnenberg et al., 2016), another possibility is that plants possess additional, yet-to-be-identified proteins that contribute to the inner kinetochore structure. A promising avenue for future studies would be to use co-immunoprecipitation or proximity labelling techniques to investigate the full composition of the inner kinetochore in plants.
Recent studies have shown that differences in kinetochore architecture and function exist even within the plant kingdom, such as the unique evolution of KNL1 and its interactions with spindle checkpoint proteins, as well as the distinctive kinetochores in Cuscuta species. In angiosperms, most outer kinetochore proteins and several SAC proteins are constitutively observed on centromeres throughout the cell cycle. In contrast, in the bryophyte P. patens, only CENH3 and CENP-C remain on the centromere for most of the cell cycle. The KNL1 and Mis12 complexes appear in prophase, while the Ndc80 and SKA complexes are recruited only after nuclear envelope breakdown, suggesting a time-dependent kinetochore assembly. However, it remains unclear whether this localization pattern is specific to P. patens or reflects a broader trend among non-vascular plants.
Many aspects of microtubule binding by plant kinetochores also remain uncertain. While key microtubule-binding proteins such as CENP-E (KAK1, kinesin-7), the Ndc80 complex, and the SKA complex appear to be conserved, functional analyses of their interaction with microtubules remain limited. Notably, a recent study found that Arabidopsis Spc25 has a higher affinity for microtubule binding compared to Nuf2 (Li et al., 2021), despite Nuf2 and Ndc80 forming a microtubule-binding module in animal kinetochores. Adding to the complexity, most SAC proteins, which regulate mitotic delay until proper microtubule-kinetochore attachments are established, are dispensable for plant development under normal conditions—a sharp contrast to their essential roles in animal cells. Could this dispensability of SAC be explained by an exceptionally robust microtubule binding by plant kinetochores?
While significant progress has been made in recent years, many questions remain regarding the complexities of kinetochore architecture and function in plants. The creation of hybrids and synthetic chromosomes, both closely linked to centromere-kinetochore function, are rapidly advancing fields that offer strong motivation to continue studying plant kinetochores.
Author contributionsEK: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Japan Society for the Promotion of Science (KAKENHI) 23K14210 and 22K20651 to EK.
AcknowledgmentsWe would like to thank Dr. Elsa Amelia Tungadi (ZWxzYV90dW5nYWRpQHlhaG9vLmNvbQ==) for the scientific illustration used in Figure 1 and Dr. Moe Yamada for advice and discussion on this manuscript.
Conflict of interestThe author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAllipra, S., Anirudhan, K., Shivanandan, S., Raghunathan, A., and Maruthachalam, R. (2022). The kinetochore protein NNF1 has a moonlighting role in the vegetative development of Arabidopsis thaliana. Plant J. 109, 1064–1085. doi:10.1111/tpj.15614
PubMed Abstract | CrossRef Full Text | Google Scholar
Batzenschlager, M., Lermontova, I., Schubert, V., Fuchs, J., Berr, A., Koini, M. A., et al. (2015). Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc. Natl. Acad. Sci. U.S.A. 112, 8656–8660. doi:10.1073/pnas.1506351112
PubMed Abstract | CrossRef Full Text | Google Scholar
Breuer, C., Stacey, N. J., West, C. E., Zhao, Y., Chory, J., Tsukaya, H., et al. (2007). BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell. 19, 3655–3668. doi:10.1105/tpc.107.054833
PubMed Abstract | CrossRef Full Text | Google Scholar
Caillaud, M.-C., Paganelli, L., Lecomte, P., Deslandes, L., Quentin, M., Pecrix, Y., et al. (2009). Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS One 4, e6757. doi:10.1371/journal.pone.0006757
PubMed Abstract | CrossRef Full Text | Google Scholar
Chandra, J. R., Kalidass, M., Demidov, D., Dabravolski, S. A., and Lermontova, I. (2024). The role of centromeric repeats and transcripts in kinetochore assembly and function. Plant J. 118, 982–996. doi:10.1111/tpj.16445
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, Q., Zhang, M., Pan, X., Yuan, X., Zhou, L., Yan, L., et al. (2021). Bub1 and CENP-U redundantly recruit Plk1 to stabilize kinetochore-microtubule attachments and ensure accurate chromosome segregation. Cell. Rep. 36, 109740. doi:10.1016/j.celrep.2021.109740
PubMed Abstract | CrossRef Full Text | Google Scholar
Cortes-Silva, N., Ulmer, J., Kiuchi, T., Hsieh, E., Cornilleau, G., Ladid, I., et al. (2020). CenH3-Independent kinetochore assembly in Lepidoptera requires CCAN, including CENP-T. Curr. Biol. 30, 561–572. doi:10.1016/j.cub.2019.12.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Dangel, N. J., Knoll, A., and Puchta, H. (2014). MHF 1 plays F anconi anaemia complementation group M protein (FANCM)-dependent and FANCM -independent roles in DNA repair and homologous recombination in plants. Plant J. 78, 822–833. doi:10.1111/tpj.12507
PubMed Abstract | CrossRef Full Text | Google Scholar
Demidov, D., Van Damme, D., Geelen, D., Blattner, F. R., and Houben, A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell. 17, 836–848. doi:10.1105/tpc.104.029710
PubMed Abstract | CrossRef Full Text | Google Scholar
Deng, X., He, Y., Tang, X., Liu, X., Lee, Y.-R. J., Liu, B., et al. (2024a). A coadapted KNL1 and spindle assembly checkpoint axis orchestrates precise mitosis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 121, e2316583121. doi:10.1073/pnas.2316583121
PubMed Abstract | CrossRef Full Text | Google Scholar
Deng, X., Peng, F. L., Tang, X., Lee, Y.-R. J., Lin, H.-H., and Liu, B. (2024b). The Arabidopsis BUB1/MAD3 family protein BMF3 requires BUB3.3 to recruit CDC20 to kinetochores in spindle assembly checkpoint signaling. Proc. Natl. Acad. Sci. U.S.A. 121, e2322677121. doi:10.1073/pnas.2322677121
PubMed Abstract | CrossRef Full Text | Google Scholar
Ding, D., Muthuswamy, S., and Meier, I. (2012). Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol. Biol. 79, 203–216. doi:10.1007/s11103-012-9903-4
PubMed Abstract | CrossRef Full Text | Google Scholar
Drinnenberg, I. A., Henikoff, S., and Malik, H. S. (2016). Evolutionary turnover of kinetochore proteins: a ship of theseus? Trends Cell. Biol. 26, 498–510. doi:10.1016/j.tcb.2016.01.005
PubMed Abstract | CrossRef Full Text | Google Scholar
Du, Y., Topp, C. N., and Dawe, R. K. (2010). DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6, e1000835. doi:10.1371/journal.pgen.1000835
PubMed Abstract | CrossRef Full Text | Google Scholar
Girard, C., Crismani, W., Froger, N., Mazel, J., Lemhemdi, A., Horlow, C., et al. (2014). FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42, 9087–9095. doi:10.1093/nar/gku614
PubMed Abstract | CrossRef Full Text | Google Scholar
Haase, J., Bonner, M. K., Halas, H., and Kelly, A. E. (2017). Distinct roles of the chromosomal passenger complex in the detection of and response to errors in kinetochore-microtubule attachment. Dev. Cell. 42, 640–654. doi:10.1016/j.devcel.2017.08.022
PubMed Abstract | CrossRef Full Text | Google Scholar
Kirik, V., Schrader, A., Uhrig, J. F., and Hulskamp, M. (2007). MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 19, 3100–3110. doi:10.1105/tpc.107.054361
PubMed Abstract | CrossRef Full Text | Google Scholar
Kirioukhova, O., Johnston, A. J., Kleen, D., Kägi, C., Baskar, R., Moore, J. M., et al. (2011). Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development 138, 3409–3420. doi:10.1242/dev.060384
PubMed Abstract | CrossRef Full Text | Google Scholar
Komaki, S., and Schnittger, A. (2016). The spindle checkpoint in plants — a green variation over a conserved theme? Curr. Opin. Plant Biol. 34, 84–91. doi:10.1016/j.pbi.2016.10.008
PubMed Abstract | CrossRef Full Text | Google Scholar
Komaki, S., and Schnittger, A. (2017). The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell. 43, 172–185. doi:10.1016/j.devcel.2017.09.017
PubMed Abstract | CrossRef Full Text | Google Scholar
Komaki, S., Takeuchi, H., Hamamura, Y., Heese, M., Hashimoto, T., and Schnittger, A. (2020). Functional analysis of the plant chromosomal passenger complex. Plant Physiol. 183, 1586–1599. doi:10.1104/pp.20.00344
PubMed Abstract | CrossRef Full Text | Google Scholar
Komaki, S., Tromer, E. C., De Jaeger, G., De Winne, N., Heese, M., and Schnittger, A. (2022). Molecular convergence by differential domain acquisition is a hallmark of chromosomal passenger complex evolution. Proc. Natl. Acad. Sci. U.S.A. 119, e2200108119. doi:10.1073/pnas.2200108119
PubMed Abstract | CrossRef Full Text | Google Scholar
Kozgunova, E., Nishina, M., and Goshima, G. (2019). Kinetochore protein depletion underlies cytokinesis failure and somatic polyploidization in the moss. eLife 43652. doi:10.7554/eLife.43652
PubMed Abstract | CrossRef Full Text | Google Scholar
Lara-Gonzalez, P., Pines, J., and Desai, A. (2021). Spindle assembly checkpoint activation and silencing at kinetochores. Seminars Cell. and Dev. Biol. 117, 86–98. doi:10.1016/j.semcdb.2021.06.009
PubMed Abstract | CrossRef Full Text | Google Scholar
Lermontova, I., Fuchs, J., and Schubert, I. (2008). The Arabidopsis checkpoint protein Bub3.1 is essential for gametophyte development. Front. Biosci. 13, 5202–5211. doi:10.2741/3076
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, J., Wang, Y., Zou, W., Jian, L., Fu, Y., and Zhao, J. (2021). AtNUF2 modulates spindle microtubule organization and chromosome segregation during mitosis. Plant J. 107, 801–816. doi:10.1111/tpj.15347
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y., Zhou, Y., Wang, B., Mu, N., Miao, Y., Tang, D., et al. (2023). FANCM interacts with the MHF1-MHF2 complex to limit crossover frequency during rice meiosis. Plant J. 116, 717–727. doi:10.1111/tpj.16399
PubMed Abstract | CrossRef Full Text | Google Scholar
Marimuthu, M. P. A., Maruthachalam, R., Bondada, R., Kuppu, S., Tan, E. H., Britt, A., et al. (2021). Epigenetically mismatched parental centromeres trigger genome elimination in hybrids. Sci. Adv. 7, eabk1151. doi:10.1126/sciadv.abk1151
PubMed Abstract | CrossRef Full Text | Google Scholar
McAinsh, A. D., and Kops, G. J. P. L. (2023). Principles and dynamics of spindle assembly checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 24, 543–559. doi:10.1038/s41580-023-00593-z
PubMed Abstract | CrossRef Full Text | Google Scholar
Miki, T., Naito, H., Nishina, M., and Goshima, G. (2014). Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. U.S.A. 111, E1053–E1061. doi:10.1073/pnas.1311243111
PubMed Abstract | CrossRef Full Text | Google Scholar
Nagaki, K., Kashihara, K., and Murata, M. (2009). Characterization of the two centromeric proteins CENP-C and MIS12 in Nicotiana species. Chromosome Res. 17, 719–726. doi:10.1007/s10577-009-9064-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Neumann, P., Oliveira, L., Jang, T.-S., Novák, P., Koblížková, A., Schubert, V., et al. (2023). Disruption of the standard kinetochore in holocentric Cuscuta species. Proc. Natl. Acad. Sci. U.S.A. 120, e2300877120. doi:10.1073/pnas.2300877120
PubMed Abstract | CrossRef Full Text | Google Scholar
Nguyen, A. L., Fadel, M. D., and Cheeseman, I. M. (2021). Differential requirements for the CENP-O complex reveal parallel PLK1 kinetochore recruitment pathways. MBoC 32, 712–721. doi:10.1091/mbc.E20-11-0751
PubMed Abstract | CrossRef Full Text | Google Scholar
Oliveira, L., Neumann, P., Jang, T.-S., Klemme, S., Schubert, V., Koblížková, A., et al. (2020). Mitotic spindle attachment to the holocentric chromosomes of Cuscuta europaea does not correlate with the distribution of CENH3 chromatin. Front. Plant Sci. 10, 1799. doi:10.3389/fpls.2019.01799
PubMed Abstract | CrossRef Full Text | Google Scholar
Pesenti, M. E., Raisch, T., Conti, D., Walstein, K., Hoffmann, I., Vogt, D., et al. (2022). Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization. Mol. Cell. 82, 2113–2131.e8. doi:10.1016/j.molcel.2022.04.027
PubMed Abstract | CrossRef Full Text | Google Scholar
Quiroz, L. F., Gondalia, N., Brychkova, G., McKeown, P. C., and Spillane, C. (2024). Haploid rhapsody: the molecular and cellular orchestra of in vivo haploid induction in plants. New Phytol. 241, 1936–1949. doi:10.1111/nph.19523
PubMed Abstract | CrossRef Full Text | Google Scholar
Roach, K. C., Ross, B. D., and Malik, H. S. (2012). “Rapid evolution of centromeres and centromeric/kinetochore proteins,” in Rapidly Evolving Genes and Genetic Systems. Editors R. S. Singh, J. Xu, and R. J. Kulathinal (Oxford University Press), 83–93. doi:10.1093/acprof:oso/9780199642274.003.0009
CrossRef Full Text | Google Scholar
Shin, J., Jeong, G., Park, J., Kim, H., and Lee, I. (2018). Mun (meristem unstructured), encoding a SPC24 homolog of NDC80 kinetochore complex, affects development through cell division in Arabidopsis thaliana. Plant J. 93, 977–991. doi:10.1111/tpj.13823
PubMed Abstract | CrossRef Full Text | Google Scholar
Singh, P., Pesenti, M. E., Maffini, S., Carmignani, S., Hedtfeld, M., Petrovic, A., et al. (2021). BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore receptors in mitosis. Mol. Cell. 81, 67–87.e9. doi:10.1016/j.molcel.2020.10.040
PubMed Abstract | CrossRef Full Text | Google Scholar
Singh, T. R., Saro, D., Ali, A. M., Zheng, X.-F., Du, C., Killen, M. W., et al. (2010). MHF1-MHF2, a histone-fold-containing protein complex, participates in the fanconi anemia pathway via FANCM. Mol. Cell. 37, 879–886. doi:10.1016/j.molcel.2010.01.036
PubMed Abstract | CrossRef Full Text | Google Scholar
Su, H., Liu, Y., Wang, C., Liu, Y., Feng, C., Sun, Y., et al. (2021). Knl1 participates in spindle assembly checkpoint signaling in maize. Proc. Natl. Acad. Sci. U.S.A. 118, e2022357118. doi:10.1073/pnas.2022357118
PubMed Abstract | CrossRef Full Text | Google Scholar
Tang, X., He, Y., Tang, Y., Chen, K., Lin, H., Liu, B., et al. (2024). A kinetochore-associated kinesin-7 motor cooperates with BUB3.3 to regulate mitotic chromosome congression in Arabidopsis thaliana. Nat. Plants 10, 1724–1736. doi:10.1038/s41477-024-01824-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Tromer, E., Snel, B., and Kops, G. J. P. L. (2015). Widespread recurrent patterns of rapid repeat evolution in the kinetochore scaffold KNL1. Genome Biol. Evol. 7, 2383–2393. doi:10.1093/gbe/evv140
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, M. L., Lin, X. J., Mo, B. X., and Kong, W. W. (2024). Plant artificial chromosomes: construction and transformation. ACS Synth. Biol. 13, 15–24. doi:10.1021/acssynbio.3c00555
PubMed Abstract | CrossRef Full Text | Google Scholar
Yan, K., Yang, J., Zhang, Z., McLaughlin, S. H., Chang, L., Fasci, D., et al. (2019). Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282. doi:10.1038/s41586-019-1609-1
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, H., Deng, X., Sun, B., Lee Van, S., Kang, Z., Lin, H., et al. (2018). Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nat. Plants 4, 485–494. doi:10.1038/s41477-018-0192-z
留言 (0)