Oral squamous cell carcinoma (OSCC) is the most common oral cancer and one of the deadliest malignancies with a 5-year survival rate of about 50% (Bray et al., 2024). Patients with OSCC are usually asymptomatic in the early stages and have insufficient understanding of risk factors including smoking, HPV infection, and alcohol (Ahmadi et al., 2019), which means they are typically diagnosed at an advanced stage, leading to poor prognosis and increased mortality. It is well known that HPV-positive (HPV+) OSCCs have a better prognosis than HPV-negative (HPV-) ones (Kumar et al., 2024). The prognostic difference between HPV+ and HPV- OSCCs is due to the impact of HPV infection on both microRNAs (miRNAs) and inflammatory cytokines. HPV + OSCCs exhibit distinctive miRNA profiles, which influence tumour progression and immune responses (Christianto et al., 2022). Additionally, HPV infection modulates the levels of inflammatory cytokines, possibly contributing to the unique tumour microenvironment observed in HPV + OSCCs (Castellano et al., 2022). Early diagnosis of OSCC is very important to improve its prognosis (Jeng et al., 2001; Guha et al., 2014; Cristaldi et al., 2019; Worthington et al., 2023).
The definitive approach for diagnosing OSCC involves clinical oral examination coupled with histological assessment of tissue samples from the affected area (Lopez-Escamez and Perez-Carpena, 2024). Currently, the emphasis in cancer diagnosis lies on identifying minimally invasive and cost-effective methods (Wang et al., 2017). Liquid biopsy (LB) represents a minimally invasive technique to obtain biological and dynamic information about cancer from a patient’s body fluid samples (Poulet et al., 2019). It facilitates the real-time monitoring of cancer progression. LB-related studies have been extended to a variety of physiological fluids, including urine and saliva, as well as to a variety of molecular entities such as DNA, RNA and extracellular vesicles (Alix-Panabières et al., 2023). Saliva biopsies have been proposed as an alternative method for cancer detection and prognosis due to their ease of collection (Patel et al., 2022).
Saliva contains cytokines, RNA and DNA molecules, tissue derivatives, and other components that contribute to disease diagnosis. The collection of saliva for identifying diagnostic markers represents a direct, non-invasive, convenient, and economical method. This potential has been thoroughly investigated over the past few decades. Recent studies have explored the feasibility of salivary biomarkers in diagnosing the onset and progression of OSCC (Kawahara et al., 2016; Chen et al., 2017; Sivadasan et al., 2020). Notably, salivary factors exhibit stable and disease-specific expression patterns in human peripheral blood and bodily fluids (Kunicki et al., 2024; Sharma et al., 2024; Tan et al., 2024), while miRNAs can serve as ideal biomarkers for this purpose (Chen et al., 2008; Ryu et al., 2023; Fattahi et al., 2024; Ko et al., 2024; Shaterabadi et al., 2024).
This review aims to emphasize advancements in salivary miRNAs and cytokines research over the last 3 years. The investigation of the possible effectiveness of salivary biomarkers (cytokines and miRNAs) in enhancing the diagnosis and prognosis of OSCC has been conducted over the past 3 years.
2 Salivary miRNAs associated with OSCC diagnosis and prognosismiRNAs consist of 20–24 nucleotides and were originally discovered in the roundworm Caenorhabditis elegans (Lee et al., 1993). miRNAs have the capacity to regulate gene expression post-transcriptionally. miRNAs can be isolated from cells, tissues and body fluids such as serum, plasma, tears or urine (Weber et al., 2010), and can also be selectively packaged in extracellular vesicles (Betel et al., 2008) (Table 1).
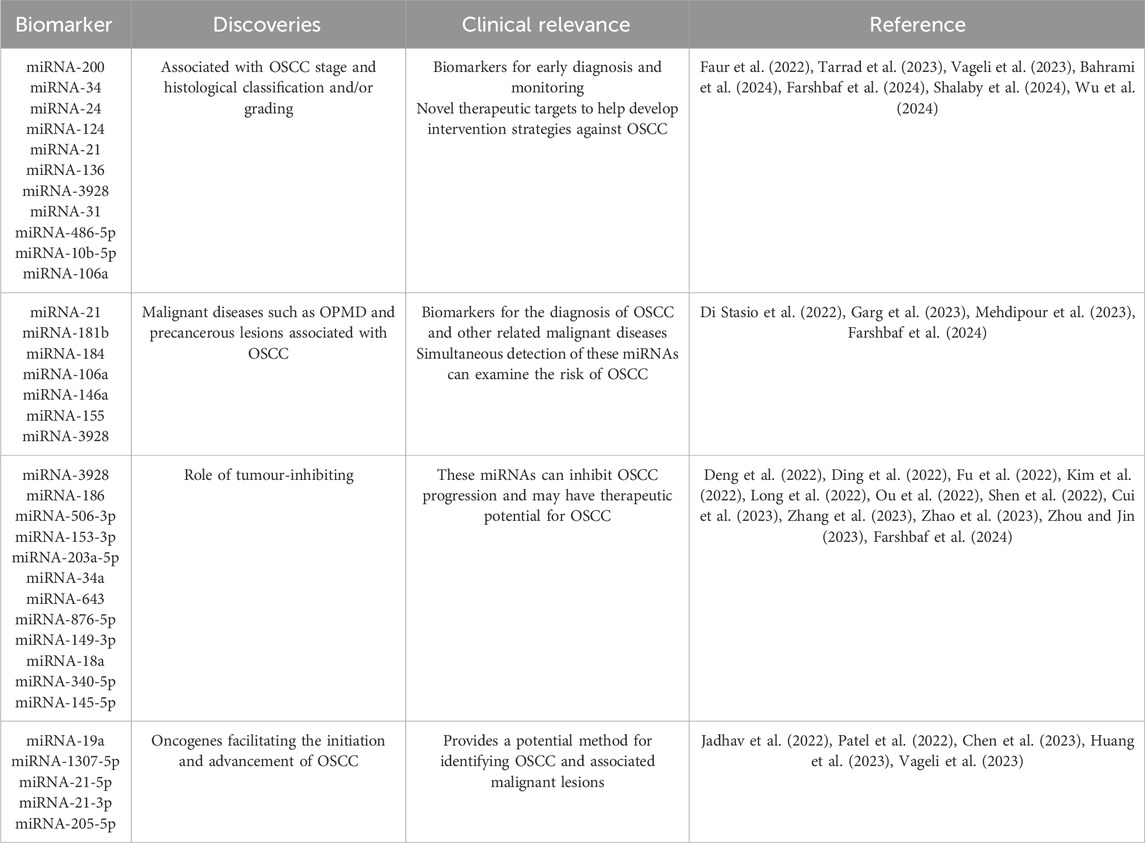
Table 1. Salivary miRNAs associated with OSCC diagnosis and prognosis.
2.1 miRNAs: tumour biomarkersIn 2008, Lawrie et al. compared the levels of tumour-associated miRNAs in the sera of diffuse large B-cell lymphoma (DLBCL) patients with those of healthy controls, and found that the levels of miRNAs in the patients were significantly higher than those in the controls, which suggested for the first time that circulating miRNAs are expected to be used as non-invasive markers for DLBCL (Lawrie et al., 2008). Subsequently, by studying miRNA stability, site differences, etc., scholars have shown that miRNAs of tumour origin detected in plasma or serum have great potential as circulating biomarkers for detecting common human cancer types (Mitchell et al., 2008). Then, miRNAs have been suggested as diagnostic and prognostic biomarkers for the treatment of several tumours, including breast and ovarian cancers, and OSCC (Zahran et al., 2015; Tung et al., 2020; Sempere et al., 2021; Coskunpinar et al., 2023). In recent years, the analysis of abnormal miRNAs profile levels in OSCC patients has been reviewed (Ghafouri-Fard et al., 2020; Manzano-Moreno et al., 2021; Li et al., 2024; Romani et al., 2024; Sanesi et al., 2024). Moreover, in this year’s Nobel Prize in Physiology or Medicine, the discovery of miRNAs was honored, which may represent another new high point in the upcoming research related to miRNAs. In summary, by analyzing miRNAs information in cancer patients, new biomarkers can be developed for OSCC clinical diagnosis. In addition, they are significant regulators of gene transcription and may be potential biomarkers for forecasting clinical outcomes of OSCC.
miRNAs can either increase or inhibit the expression of target genes by directly binding to the mRNAs of those genes. They also influence the stability of mRNA (Hudder and Novak, 2008). Aberrant miRNA regulation can significantly contribute to cancer development (Shi et al., 2008). The expression of some miRNAs is associated with the dysregulation of oncosuppressors or oncogenes and contributes to tumour development or suppression. miRNAs with oncogenic functions are upregulated and silence tumour-inhibiting genes that can facilitate cancer cell progression. Conversely, miRNAs with tumour-inhibiting functions are downregulated, thereby reducing the regulation of oncogenes and maintaining malignancy (Lee and Dutta, 2006). It is necessary to further elucidate the carcinogenic and tumour-inhibiting functions of different miRNAs and their specific mechanisms using new technologies such as bioinformatics or single-cell sequencing analysis, and establish the stable correlation between miRNAs and OSCC progression, which will help to identify specific biomarkers for the diagnosis and prognosis of OSCC.
2.2 Diagnostic value of salivary miRNAs for OSCCThe diagnostic value of salivary miRNAs in identifying diagnostic markers has been extensively studied in recent years, which will possess the important clinical significance and bring greater translational benefits with the deepening of research.
2.2.1 Salivary miRNAs from OSCC patientsRecent studies have shown that salivary miRNAs can be used for early diagnosis and clinical staging of OSCC. Researchers and technicians are committed to improving the salivary miRNA analysis platform in sample collection and pretreatment, RNA extraction technology, detection methods, data analysis algorithms, and quality control systems (Yoshizawa and Wong, 2013; Fortuna et al., 2020; McKenna and Dubey, 2022). Vageli et al. reported statistically significant differences in salivary miRNA-21, miRNA-136 and miRNA-3928 expression between early tumours and healthy controls (p < 0.05) (Vageli et al., 2023) (Table 1). Bahrami et al. found that in saliva samples from OSCC patients, the expression levels of two biomarkers, miRNA-200 and miRNA-34, were reduced relative to healthy individuals, whereas the expression level of miRNA-24 was increased (Bahrami et al., 2024) (Table 1). Shalaby et al. showed that miRNA-124 expression was significantly decreased in the saliva of OSCC patients (p < 0.001) (Shalaby et al., 2024) (Table 1). Tarrad et al. claim that miRNA-106a expression helps to differentiate between OSCC grade II and grade III (Tarrad et al., 2023) (Table 1). Wu et al. developed a dual 3D nanorobot capable of rapidly, sensitively and specifically analyzing miRNA-31, a diagnostic biomarker for OSCC, in saliva samples (Wu et al., 2024) (Table 1). Scheurer et al. compared saliva from healthy and OSCC samples using different isolation techniques and identified 11 differentially expressed miRNAs (p < 0.05): miRNA-1183, 128-1-5p, 3646, 3648, 383-5p, 4300, 4638-5p, 486-5p, 5189-5p, 6076 and 6784-5p (Scheurer et al., 2024). Meanwhile, in the study by Momen-Heravi et al., it was found that of the more than 700 miRNAs that were analyzed, 13 were identified as being significantly dysregulated in saliva samples from patients with OSCC when compared to healthy controls. 11 of these were found to be significantly underexpressed (miRNA-136, -147, −1250, −148a, −632, −646, −668, −877), Conversely, two miRNAs (miRNA-24, -27b) demonstrated significant overexpression (p < 0.05) (Momen-Heravi et al., 2014). The diagnostic value of these miRNAs in the saliva of OSCC patients needs to be further verified, providing potential research directions for future studies.
2.2.2 Salivary miRNAs from OSCC precancerous lesionsFurthermore, miRNA detection in saliva also exhibits the important diagnostic value for oral potentially malignant disease (OPMD) and precancerous lesions that may develop into OSCC. Garg et al. reported that miRNA-21 and miRNA-184 are biomarkers of OSCC and OPMD, where miRNA-21 was significantly increased in OSCC and OPMD, while miRNA-184 was significantly decreased (Garg et al., 2023) (Table 1). Di Stasio et al. noted that miRNA-181b showed upregulation (p = 0.006) in the saliva of patients in the OPMD high-grade dysplasia group, which was positively correlated with the degree of dysplasia and negatively correlated with OSCC (Di Stasio et al., 2022) (Table 1).
2.2.3 Salivary miRNAs for OSCC differential diagnosisIn addition, some salivary miRNAs can be used for the differential diagnosis between OSCC and other oral diseases. Tarrad et al. suggested that low salivary miRNA-106a levels may indicate malignancy and may also be used to diagnostically differentiate OSCC from oral lichen planus (OLP) (Tarrad et al., 2023). Mehdipour et al. reported increased expression of miRNA-146a in saliva of OLP patients (p = 0.004) (Mehdipour et al., 2023) (Table 1). In addition, miRNA-155 was significantly upregulated only in saliva specimens from OLP patients (p = 0.009). Farshbaf et al. found that the expression level of miRNA-3928 in saliva of OLP patients (p = 0.01) was different from that of OSCC patients (p < 0.0001) compared to healthy individuals, and the expression level of miRNA-3928 in saliva of OSCC patients was markedly downregulated (Farshbaf et al., 2024) (Table 1). The above studies have shown that salivary miRNA-106a, 146a, 155, and 3928 may serve as potential biomarkers for the differential diagnosis between OSCC and OLP.
2.2.4 Salivary exosomal miRNAs from OSCC patientsExosomes are small, single-membrane, secreted organelles of ∼30 to∼200 nm in diameter that have the same topology as the cell and are enriched in selected proteins, lipids, nucleic acids, and glycoconjugates (Pegtel and Gould, 2019). Exosomes, which are one of the smallest extracellular vesicles released from cells, have been shown to carry different nucleic acids, including miRNAs (Krylova and Feng, 2023). The significant advantage of salivary exosomal miRNAs lies in convenient sampling and better stability. Salivary exosomal miRNAs can be extracted from a very small amount of saliva and have been successfully used to detect OSCC. Faur et al. noted that salivary exosomal miRNA-486-5p was raised, while miRNA-10b-5p was diminished in OSCC relative to healthy controls. miRNA-486-5p expression levels are elevated in stage II OSCC (Faur et al., 2022) (Table 1). Patel et al. found that the expression level of miRNA-1307-5p was significantly elevated in tissue and salivary exosome samples from individuals diagnosed with OSCC when compared to samples from individuals without cancer. From a clinical perspective, the upregulation of miRNA-1307-5p has been linked to a number of unfavourable outcomes, including patient survival, disease progression, aggressiveness, and chemotherapy resistance (Patel et al., 2022).
2.2.5 Combined analysis of salivary miRNAsIt has also been shown that the combined analysis of multiple miRNAs in saliva is necessary to distinguish OSCC patients from healthy individuals. Scholtz et al. showed that a panel consisting of miRNA-345, miRNA-31-5p and miRNA-424-3p could be used to diagnose OSCC. In addition, IL-6 and miRNA-31 are synergistic in cancer stem cell activity, with higher specificity of salivary IL-6 for OSCC, and improved specificity by the combination of IL-6 mRNA and miRNA-31 as biomarkers (Scholtz et al., 2022). Meanwhile, Di Stasio et al. showed that salivary miRNA-27b and miRNA-181b can be used as potential biomarkers of oral dysplasia, and the combination of the two has implications for the diagnosis of the disease (Di Stasio et al., 2022) (Table 1). A more accurate diagnosis of OSCC would be achieved by using multiple miRNAs as a panel, with simultaneous corresponding changes, that is to say, panel changes.
2.3 miRNAs: tumour-inhibiting effectIn the past 3 years, a few studies have been conducted on the tumour suppressive effects of salivary miRNAs. Farshbaf et al. noted that miRNA-3928, which acts as a tumour-inhibiting in OSCC pathobiology, was significantly reduced in patient saliva (Farshbaf et al., 2024) (Table 1). Given the prospective tumour-inhibiting function of miRNA-3928 in OSCC, this miRNA may play an important role in early diagnosis, screening and targeted therapy of OSCC (Figure 1).
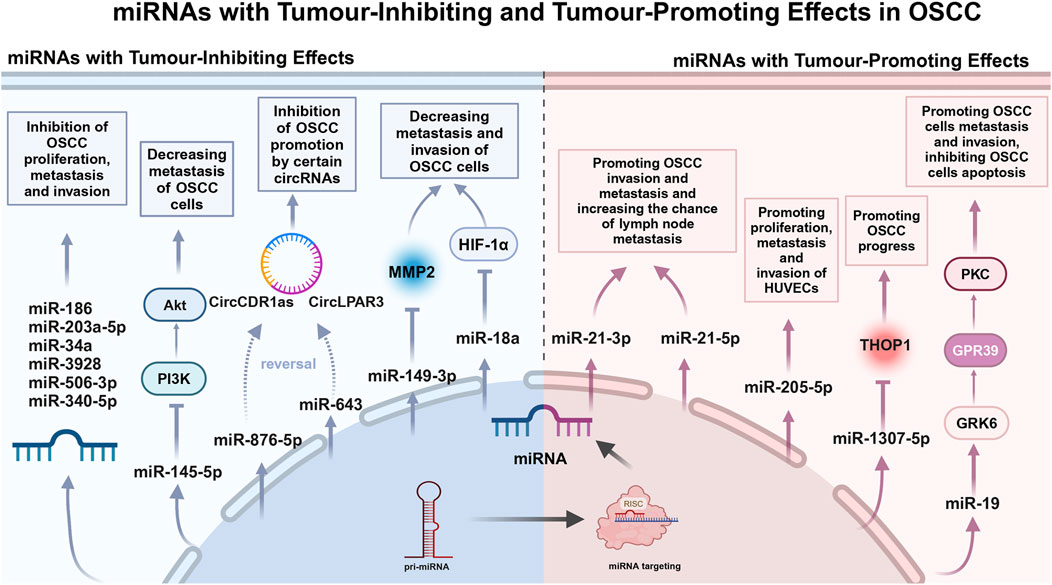
Figure 1. miRNAs with Tumour-Inhibiting and Tumour-Promoting Effects in OSCC.
The extensive studies on tumour-inhibiting miRNAs in serum or tissue have been conducted by multiple research groups, which may provide the potential targets for future studies on tumour-inhibiting effects of salivary miRNAs. Zhao et al. declared that miRNA-186 reduces OSCC cell proliferation, migration and invasion and promotes apoptosis (Zhao et al., 2023) (Table 1). Ding et al. suggested that miRNA-506-3p retards OSCC cell progression at elevated concentrations (Ding et al., 2022) (Table 1). Long et al. identified miRNA-153-3p as a tumour-inhibiting in OSCC (Long et al., 2022) (Table 1). Zhang et al. reported that miRNA-203a-5p was elevated to inhibit OSCC cell invasion and migration (Zhang et al., 2023) (Table 1). Deng et al. identified miRNA-34a as an inhibitor of OSCC cell proliferation, migration and invasion (Deng et al., 2022) (Table 1). Numerous tumour-inhibiting miRNAs work by counteracting the expression of circular RNAs (CircRNAs) that facilitate cell proliferation. For example, miRNA-643 reverses the promotion of OSCC progression by CircLPAR3 (Fu et al., 2022) (Table 1). miRNA-876-5p mitigates the impact of increased circCDR1as on autophagy, cell cycle progression, proliferation, motility, and death in OSCC cells through its overexpression (Cui et al., 2023) (Table 1). Several miRNAs can exert tumour-inhibiting effects by regulating tumour-associated factors. Shen et al. reported inhibition of matrix metalloproteinase-2 (MMP-2) by miRNA-149-3p, which was associated with a reduction in the invasive process in OSCC cells (Shen et al., 2022) (Table 1). Kim et al. constructed a miRNA-18a mimic and observed that it inhibited HIF-1α expression. In addition, they found that it reduced the metastatic and invasive potential of OSCC cells (Kim et al., 2022) (Table 1). Ou et al. found that downregulation of miRNA-340-5p enhances invasion and proliferation of oral cancer cells (Ou et al., 2022) (Table 1). Zhou et al. showed that miRNA-145-5p inhibited the PI3K/AKT pathway and hindered cell migration (Zhou and Jin, 2023) (Table 1).
2.4 miRNAs: tumour-promoting effectThe mechanism of salivary miRNAs promoting OSCC progression has gradually become a research focus. Chen et al. reported that overexpression of miRNA-19a is regulated through the miRNA-19a/GRK6/GPR39/PKC signaling pathway and thereby inhibiting apoptosis and promoting cell migration and invasion. It is a key regulator of OSCC progression and provides a possible avenue for developing novel therapies targeting downstream signaling pathways (Chen et al., 2023) (Table 1). Patel et al. found that miRNA-1307-5p expression was elevated in tissues and salivary exosomes of patients with oral cancer, which may promote oral cancer progression through mechanisms such as inhibition of Thimet Oligopeptidase 1 (THOP1). Increased expression of this miRNA is associated with decreased patient survival, disease progression, invasiveness, and chemotherapy resistance, and may serve as a reliable prognostic marker for predicting adverse outcomes in oral cancer (Patel et al., 2022) (Table 1). Jadhav et al. found that miRNA-21 is an important oncogenic miRNA associated with tumour invasion and metastasis. Sensitivity and specificity analyses showed that the specificity of salivary miRNA-21-5p was 90.30%, while that of salivary miRNA-21-3p was 83.90% (Jadhav et al., 2022) (Table 1). miRNA-21 was significantly overexpressed in OSCC smokers compared to non-smokers (p = 0.006), suggesting an association between smoking and the oncogenic effects of OSCC (Vageli et al., 2023) (Table 1). Huang et al. suggested a significant increase of exosomal miRNA-205-5p in saliva of OSCC patients. It was shown to promote proliferation, metastasis and invasion of HUVECs (Huang et al., 2023) (Table 1). Therefore, the study on the pro-carcinogenic mechanism of miRNAs may provide the experimental basis and theoretical basis for the clinical diagnosis and prognosis of OSCC.
3 Salivary cytokines associated with OSCC diagnosis and prognosisSalivary cytokines are important regulators involved in the inflammatory response of the body and the regulation of the tumour microenvironment. In OSCC patients, variations in salivary cytokine concentrations are tightly associated with tumour occurrence and their biological behavior. Studies have shown that dysregulation of specific cytokine levels is associated with the emergence of different cancer types and is considered an important biomarker for the diagnosis of tumours (Chechlinska et al., 2008; Kartikasari et al., 2021; Uchendu et al., 2023). Increasing research indicates that analyzing salivary cytokine levels can enhance the early identification of OSCC and assist doctors in formulating personalized treatment strategies (Ferrari et al., 2021). These findings offer novel insights into salivary cytokines for the diagnostic and prognostic evaluation of OSCC and may aid in the identification of new biomarkers.
3.1 TNF-αTNF-α is recognized as a key factor in many inflammatory, viral, metabolic, and neoplastic diseases (van Loo and Bertrand, 2023). Meanwhile, the role of TNF-α can be seen in many human malignancies, including breast (Cruceriu et al., 2020), gastric (Zhao et al., 2010), pancreatic (Egberts et al., 2008), ovaries (Gupta et al., 2016), endometrial (Morgado et al., 2016), prostate (Tse et al., 2012), bladder (Sethi et al., 2012; Ting et al., 2021), colorectal (Li et al., 2016), oral (Tang et al., 2017) and liver cancer (Roderburg et al., 2012). TNF-α has also been associated with the growth, proliferation, and immune evasion of tumour cells (Ben-Baruch, 2006).
Increased levels of TNF-α were observed in saliva of patients with oral submucous fibrosis (OSMF) and OSCC. Abdul Aziz Shaikh et al. found that salivary TNF-α concentrations were significantly higher in patients diagnosed with OSMF stage 3, and the difference was statistically significant compared to patients in other stages (Abdul Aziz Shaikh et al., 2024) (Table 2). (Azimudin et al., 2024) (Table 2) suggested a potential role of TNF-α in OSCC, especially in diabetic patients, by suggesting that the mean level of TNF-α in saliva of diabetic (DM) patients and OSCC patients was significantly higher compared to that of the healthy population and OSCC patients without diabetes mellitus (Figure 2).
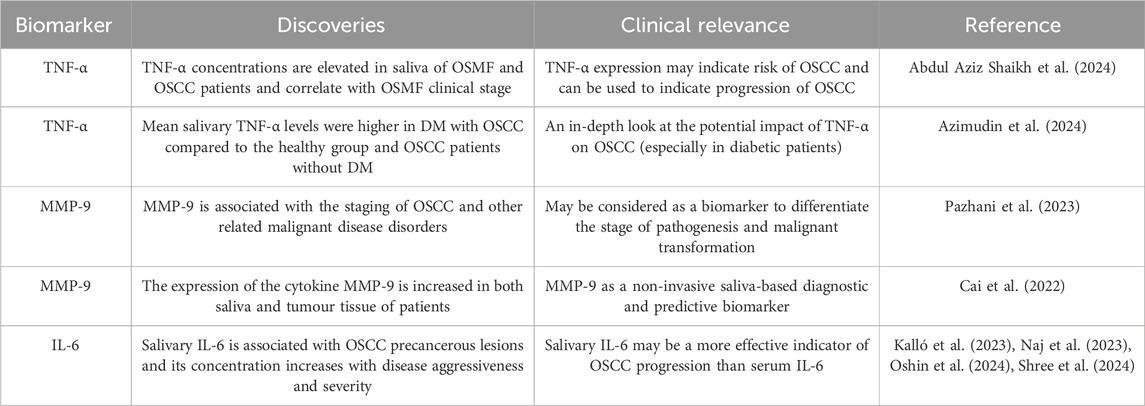
Table 2. Salivary cytokines associated with OSCC diagnosis and prognosis.
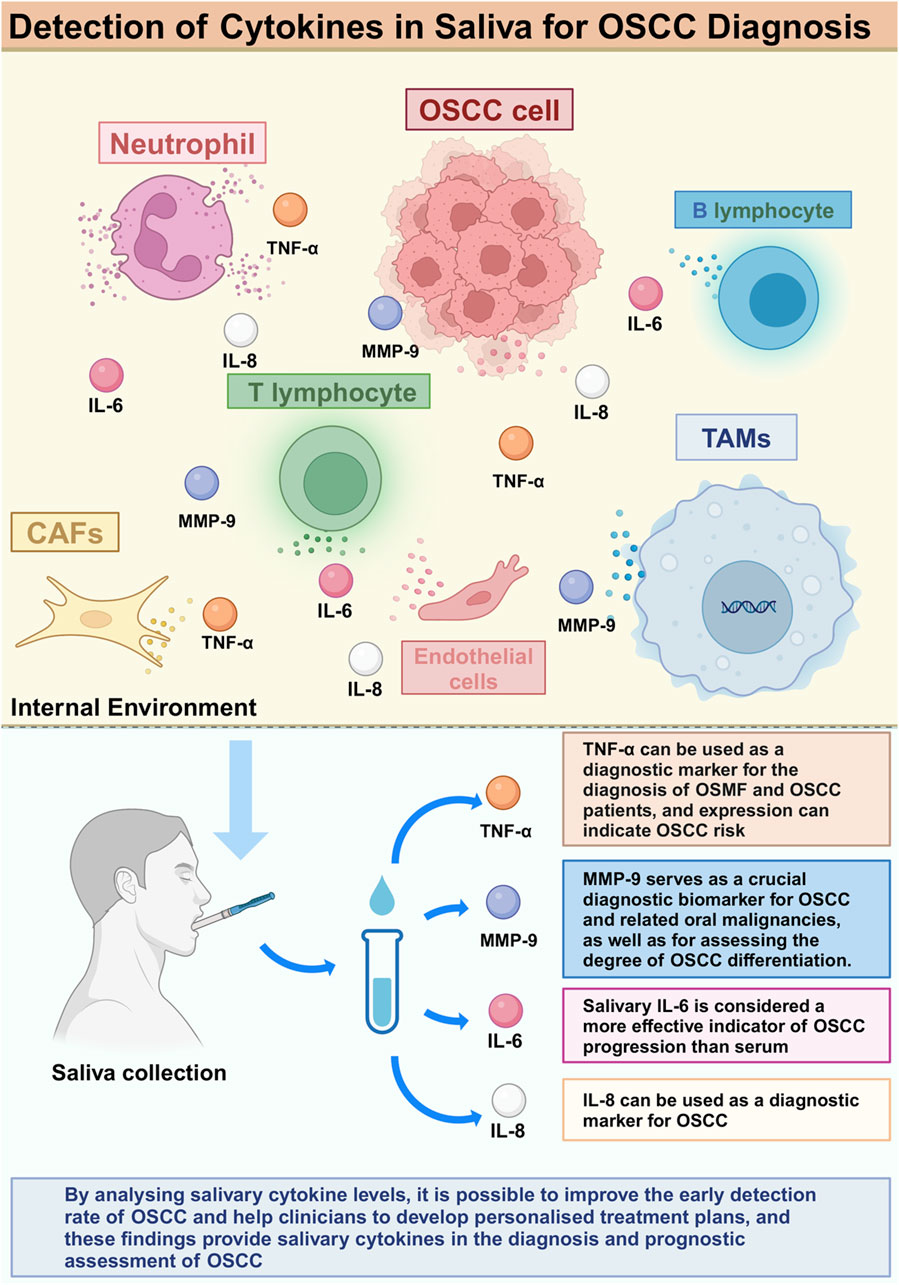
Figure 2. Detection of Cytokines in Saliva for OSCC diagnosis.
3.2 MMP-9Matrix metalloproteinase-9 (MMP-9), also known as 92 kDa type IV collagenase, has an important role in tumorigenesis (Gobin et al., 2019). In various studies, the expression of MMP-9 has been shown to be an important component of oral cancer tissue (Patel et al., 2007), serum (Lotfi et al., 2015) and saliva (Shpitzer et al., 2009) diagnostic markers in the sample.
The secretion level of MMP-9 in saliva samples was correlated with OSCC differentiation. Pazhani et al. reported that salivary MMP-9 concentrations were significantly higher in OSCC patients, and were higher in poorly differentiated OSCC patients compared to highly differentiated and moderately differentiated OSCC patients. Increased amounts of this and other macromolecules have been identified in precancerous lesions linked to the advancement of OSCC, while salivary concentrations of MMP-9 are reported to be higher in patients with oral leukoplakia compared to those with OSCC. Elevated MMP-9 levels in the saliva of subjects with oral epithelial dysplasia are detectable even in mild and moderate instances, with an increase related to disease advancement. Salivary MMP-9 concentrations are notably higher in severe oral epithelial dysplasia, attributed to the increased likelihood of malignant transformation in these patients (Pazhani et al., 2023) (Table 2). Therefore, salivary MMP-9 can be used as a reliable indicator of malignant transformation.
Cai et al. found that the expression level of MMP-9 was not only increased in saliva, but also in tumour tissues of patients (Cai et al., 2022) (Table 2). Bioinformatics analysis showed that MMP-9 was associated with survival and tumour function in patients with head and neck cancer, suggesting that MMP-9 may be a saliva-based noninvasive diagnostic biomarker and a prognostic therapeutic target for OSCC.
3.3 IL-6Interleukin 6 (IL-6) is important in several organs and systems, has a major impact on the immune response, is produced by cells such as epithelial cells and mast cells, and belongs to a major class of pro-inflammatory cytokines. When inflammation occurs, the concentration of IL-6 is usually elevated (Tanaka et al., 2014; Mauer et al., 2015; Hop et al., 2019; Kang et al., 2019). Increased salivary IL-6 levels have been documented in individuals with OSCC (Kalló et al., 2023; Naj et al., 2023; Oshin et al., 2024) (Table 2).
The level of IL-6 in saliva of OSCC patients has important diagnostic and prognostic significance. Shree et al. found a significant reduction in IL-6 expression after chemoradiotherapy (p < 0.0001), with significant changes in expression at different treatment intervals (Shree et al., 2024) (Table 2). This further confirms that IL-6 can be used as a biomarker for OSCC. It is present in precancerous lesions associated with OSCC and its pathogenesis (Naj et al., 2023; Oshin et al., 2024) (Table 2).
Moreover, the elevated levels of IL-6 are associated with the aggressiveness and severity of OSCC, leading to reduced survival and increased recurrence rates. It is noteworthy that salivary IL-6 is considered a more reliable marker of OSCC progression than serum IL-6 (Oshin et al., 2024) (Table 2). IL-6, mainly derived from macrophages, mediates the interaction among cells and is a very important OSCC-promoting cytokine in the tumour microenvironment, which makes salivary IL-6 an increasingly powerful biomarker for the diagnosis of OSCC.
3.4 Other cytokinesInterleukin-1β (IL-1β) plays a crucial role in immune response and inflammation, stimulates signaling pathways, promotes recruitment of inflammatory cells and is associated with a variety of diseases. Lee et al. (2018) concluded that the expression of this cytokine is elevated in both saliva and plasma of patients, particularly in the initial stages of OSCC, and its potential as a biomarker warrants greater consideration. Interleukin-8 (IL-8) is a significant chemokine with crucial immunological activities (Kim, 2020). Numerous publications have reported increased IL-8 levels in the saliva of patients diagnosed with OSCC (Csősz et al., 2017; Dikova et al., 2021). Other authors have also reviewed the value of IL-1β and IL-8 in saliva as biomarkers (Benito-Ramal et al., 2023; Kalló et al., 2023; Bastías et al., 2024; Khijmatgar et al., 2024). However, a large number of clinical trials are still required to further confirm their sensitivity and specificity as biomarkers in order to provide stronger evidence for early screening and personalized treatment of OSCC.
4 DiscussionRecent research indicates that assessing salivary miRNAs and cytokines is an effective approach for identifying and forecasting the outcomes of OSCC. These biomarkers facilitate the early detection of high-risk patients and furnish critical prognostic insights on the disease.
This review encapsulates the advancements of saliva-specific variables as biomarkers in the last 3 years, emphasizing the prospective significance of miRNAs and cytokines in OSCC. The significance of salivary miRNAs in diagnostic and prognostic applications has been a focal point of interest over the past 3 years, whereas there has been a relative scarcity of publications concerning salivary cytokines. Notably, the review mainly emphasized MMP-9, TNF-α, and IL-6, with IL-6 being specifically highlighted as a biomarker for OSCC. Studies related to IL-1, IL-8, and IL-10 were mostly concentrated in the last 3 years, while researches on diagnosis and prognosis were mainly performed before 2021, indicating that the role of cytokines as biomarkers has become more mature. But there is still a need to further optimize and standardize the relevant assays in order to improve the reliability of their clinical application. The amalgamation of several biomarkers is anticipated to improve the accuracy of early diagnosis and the efficacy of prognostic evaluation. In summary, salivary miRNAs and cytokines offer novel insights for the noninvasive diagnosis and personalized therapy of OSCC, warranting more investigation in the future.
Author contributionsYQ: Writing–original draft, Visualization. XD: Funding acquisition, Writing–review and editing. BL: Funding acquisition, Supervision, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by Natural Science Foundation of Jilin Province (YDZJ202401228ZYTS) and Innovative Training Program for College Students from Jilin University (X202410183456).
AcknowledgmentsFigure 1 created in BioRender. Yuxiao, Q. (2024) https://BioRender.com/z18s522. Figure 2 created in BioRender. Yuxiao, Q. (2024) https://BioRender.com/v27z650.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAbdul Aziz Shaikh, S., Denny E, C., Kumarchandra, R., Natarajan, S., Sunny, J., Shenoy, N., et al. (2024). Evaluation of salivary tumor necrosis factor α as a diagnostic biomarker in oral submucosal fibrosis and squamous cell carcinoma of the oral cavity and oropharynx: a cross sectional observational study. Front. Oral Health 5, 1375162. doi:10.3389/froh.2024.1375162
PubMed Abstract | CrossRef Full Text | Google Scholar
Ahmadi, N., Gao, K., Chia, N., Kwon, M. S., Palme, C. E., Gupta, R., et al. (2019). Association of PD-L1 expression in oral squamous cell carcinoma with smoking, sex, and p53 expression. Oral Surg. Oral Med. Oral Pathology Oral Radiology 128 (6), 631–638. doi:10.1016/j.oooo.2019.07.008
PubMed Abstract | CrossRef Full Text | Google Scholar
Azimudin, R., Palati, S., and R, P. (2024). Evaluating tumor necrosis factor (TNF)-Alpha pro-inflammatory cytokines in healthy and oral squamous cell carcinoma patients with and without diabetes. Cureus 16 (1), e52890. doi:10.7759/cureus.52890
PubMed Abstract | CrossRef Full Text | Google Scholar
Bahrami, N., Pirrafiee, M., Azadi, F., Azimnejad, R., Fotook Kiaei, S. Z., Abbasi, A. J., et al. (2024). Biomarkers for oral squamous cell carcinoma (miR-24, miR-200, and miR-34): screening and detection MicroRNA. Asian Pac J. Cancer Prev. 25 (7), 2265–2269. doi:10.31557/apjcp.2024.25.7.2265
PubMed Abstract | CrossRef Full Text | Google Scholar
Bastías, D., Maturana, A., Marín, C., Martínez, R., and Niklander, S. E. (2024). Salivary biomarkers for oral cancer detection: an exploratory systematic review. Int. J. Mol. Sci. 25 (5), 2634. doi:10.3390/ijms25052634
PubMed Abstract | CrossRef Full Text | Google Scholar
Ben-Baruch, A. (2006). Inflammation-associated immune suppression in cancer: the roles played by cytokines, chemokines and additional mediators. Semin. Cancer Biol. 16 (1), 38–52. doi:10.1016/j.semcancer.2005.07.006
PubMed Abstract | CrossRef Full Text | Google Scholar
Benito-Ramal, E., Egido-Moreno, S., González-Navarro, B., Jané-Salas, E., Roselló-Llabrés, X., and López-López, J. (2023). Role of selected salivary inflammatory cytokines in the diagnosis and prognosis of oral squamous cell carcinoma. A Systematic Review and Meta-analysis. Med. Oral, Patol. Oral Y Cirugia Bucal 28 (5), e474–e486. doi:10.4317/medoral.25889
PubMed Abstract | CrossRef Full Text | Google Scholar
Betel, D., Wilson, M., Gabow, A., Marks, D. S., and Sander, C. (2008). The microRNA.org resource: targets and expression. Nucleic Acids Res. 36 (Database issue), D149–D153. doi:10.1093/nar/gkm995
PubMed Abstract | CrossRef Full Text | Google Scholar
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 74 (3), 229–263. doi:10.3322/caac.21834
PubMed Abstract | CrossRef Full Text | Google Scholar
Cai, M., Zheng, Z., Bai, Z., Ouyang, K., Wu, Q., Xu, S., et al. (2022). Overexpression of angiogenic factors and matrix metalloproteinases in the saliva of oral squamous cell carcinoma patients: potential non-invasive diagnostic and therapeutic biomarkers. BMC cancer 22 (1), 530. doi:10.1186/s12885-022-09630-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Castellano, L. R. C., Cruz, S. B. S. C., Hier, M., Bonan, P. R. F., Alaoui-Jamali, M. A., and da Silva, S. D. (2022). Implications and emerging therapeutic avenues of inflammatory response in HPV+ head and neck squamous cell carcinoma. Cancers 14 (21), 5406. doi:10.3390/cancers14215406
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, J., Wang, L., Ma, D., Zhang, H., Fan, J., Gao, H., et al. (2023). miR-19a may function as a biomarker of oral squamous cell carcinoma (OSCC) by regulating the signaling pathway of miR-19a/GRK6/GPCRs/PKC in a Chinese population. J. Oral Pathol. Med. 52 (10), 971–979. doi:10.1111/jop.13478
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18 (10), 997–1006. doi:10.1038/cr.2008.282
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, Y.-T., Chen, H.-W., Wu, C.-F., Chu, L. J., Chiang, W.-F., Wu, C.-C., et al. (2017). Development of a multiplexed liquid chromatography multiple-reaction-monitoring mass spectrometry (LC-MRM/MS) method for evaluation of salivary proteins as oral cancer biomarkers. Mol. & Cell. proteomics MCP 16 (5), 799–811. doi:10.1074/mcp.M116.064758
PubMed Abstract | CrossRef Full Text | Google Scholar
Christianto, S., Li, K. Y., Huang, T. H., and Su, Y.-X. (2022). The prognostic value of human papilloma virus infection in oral cavity squamous cell carcinoma: a meta-analysis. Laryngoscope 132 (9), 1760–1770. doi:10.1002/lary.29996
PubMed Abstract | CrossRef Full Text | Google Scholar
Coskunpinar, E., Tiryakioglu, D. Z., Abaci, N., Tukenmez, M., and Pence, S. (2023). Investigation of the miR-637 and miR-523-5p as candidate biomarkers in breast cancer. Bratisl. Lek. Listy 124 (11), 814–820. doi:10.4149/BLL_2023_125
PubMed Abstract | CrossRef Full Text | Google Scholar
Cristaldi, M., Mauceri, R., Di Fede, O., Giuliana, G., Campisi, G., and Panzarella, V. (2019). Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: current status and perspectives. Front. Physiology 10, 1476. doi:10.3389/fphys.2019.01476
PubMed Abstract | CrossRef Full Text | Google Scholar
Cruceriu, D., Baldasici, O., Balacescu, O., and Berindan-Neagoe, I. (2020). The dual role of tumor necrosis factor-alpha (TNF-α) in breast cancer: molecular insights and therapeutic approaches. Cell. Oncol. Dordr. Neth. 43 (1), 1–18. doi:10.1007/s13402-019-00489-1
PubMed Abstract | CrossRef Full Text | Google Scholar
Csősz, É., Lábiscsák, P., Kalló, G., Márkus, B., Emri, M., Szabó, A., et al. (2017). Proteomics investigation of OSCC-specific salivary biomarkers in a Hungarian population highlights the importance of identification of population-tailored biomarkers. PloS One 12 (5), e0177282. doi:10.1371/journal.pone.0177282
PubMed Abstract | CrossRef Full Text | Google Scholar
Deng, W., Meng, Y., Wang, B., Wang, C.-X., Hou, C.-X., Zhu, Q.-H., et al. (2022). In vitro experimental study on the formation of microRNA-34a loaded exosomes and their inhibitory effect in oral squamous cell carcinoma. Cell CycleGeorget. Tex. 21 (16), 1775–1783. doi:10.1080/15384101.2022.2070832
PubMed Abstract | CrossRef Full Text | Google Scholar
Dikova, V., Jantus-Lewintre, E., and Bagan, J. (2021). Potential non-invasive biomarkers for early diagnosis of oral squamous cell carcinoma. J. Clin. Med. 10 (8), 1658. doi:10.3390/jcm10081658
PubMed Abstract | CrossRef Full Text | Google Scholar
Ding, Y., Duan, H., Lin, J., and Zhang, X. (2022). YY1 accelerates oral squamous cell carcinoma progression through long non-coding RNA Kcnq1ot1/microRNA-506-3p/SYPL1 axis. J. Ovarian Res. 15 (1), 77. doi:10.1186/s13048-022-01000-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Di Stasio, D., Romano, A., Boschetti, C. E., Montella, M., Mosca, L., and Lucchese, A. (2022). Salivary miRNAs expression in potentially malignant disorders of the oral mucosa and oral squamous cell carcinoma: a pilot study on miR-21, miR-27b, and miR-181b. Cancers 15 (1), 291. doi:10.3390/cancers15010291
PubMed Abstract | CrossRef Full Text | Google Scholar
Egberts, J. H., Cloosters, V., Noack, A., Schniewind, B., Thon, L., Klose, S., et al. (2008). Anti-tumor necrosis factor therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 68 (5), 1443–1450. doi:10.1158/0008-5472.Can-07-5704
PubMed Abstract | CrossRef Full Text | Google Scholar
Farshbaf, A., Mohajertehran, F., Aghaee-Bakhtiari, S. H., Ayatollahi, H., Douzandeh, K., Pakfetrat, A., et al. (2024). Downregulation of salivary miR-3928 as a potential biomarker in patients with oral squamous cell carcinoma and oral lichen planus. Clin. Exp. Dent. Res. 10 (2), e877. doi:10.1002/cre2.877
PubMed Abstract | CrossRef Full Text | Google Scholar
Fattahi, M., Rahdan, F., Shaterabadi, D., Zamani Sani, M., Alizadeh, M., Khatami, S. H., et al. (2024). MicroRNA biosensors for the detection of liver cancer. Clin. Chimica Acta; Int. J. Clin. Chem. 554, 117796. doi:10.1016/j.cca.2024.117796
PubMed Abstract | CrossRef Full Text | Google Scholar
Faur, C. I., Roman, R. C., Jurj, A., Raduly, L., Almășan, O., Rotaru, H., et al. (2022). Salivary exosomal MicroRNA-486-5p and MicroRNA-10b-5p in oral and oropharyngeal squamous cell carcinoma. Med. Kaunas. Lith. 58 (10), 1478. doi:10.3390/medicina58101478
PubMed Abstract | CrossRef Full Text | Google Scholar
Ferrari, E., Pezzi, M. E., Cassi, D., Pertinhez, T. A., Spisni, A., and Meleti, M. (2021). Salivary cytokines as biomarkers for oral squamous cell carcinoma: a systematic review. Int. J. Mol. Sci. 22 (13), 6795. doi:10.3390/ijms22136795
PubMed Abstract | CrossRef Full Text | Google Scholar
Fortuna, G., Aria, M., Piscitelli, A., Mignogna, M. D., and Klasser, G. D. (2020). Global research trends in complex oral sensitivity disorder: a systematic bibliometric analysis of the structures of knowledge. J. Oral Pathology & Med. 49 (6), 565–579. doi:10.1111/jop.13077
PubMed Abstract | CrossRef Full Text | Google Scholar
Fu, Y., Qiu, C., Yang, Y., Lu, J., and Qi, Y. (2022). CircLPAR3 acts as an oncogene in oral squamous cell carcinoma through regulating the miR-643/HMGB2 network. Biochem. Genet. 60 (3), 882–898. doi:10.1007/s10528-021-10134-y
PubMed Abstract | CrossRef Full Text | Google Scholar
Garg, A., Urs, A. B., Koner, B. C., Augustine, J., and Guru, S. A. (2023). Evaluation of diagnostic significance of salivary miRNA-184 and miRNA-21 in oral squamous cell carcinoma and oral potentially malignant disorders. Head Neck Pathology 17 (4), 961–968. doi:10.1007/s12105-023-01600-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Ghafouri-Fard, S., Gholipour, M., Taheri, M., and Shirvani Farsani, Z. (2020). MicroRNA profile in the squamous cell carcinoma: prognostic and diagnostic roles. Heliyon 6 (11), e05436. doi:10.1016/j.heliyon.2020.e05436
PubMed Abstract | CrossRef Full Text | Google Scholar
Gobin, E., Bagwell, K., Wagner, J., Mysona, D., Sandirasegarane, S., Smith, N., et al. (2019). A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 19 (1), 581. doi:10.1186/s12885-019-5768-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Guha, N., Warnakulasuriya, S., Vlaanderen, J., and Straif, K. (2014). Betel quid chewing and the risk of oral and oropharyngeal cancers: a meta-analysis with implications for cancer control. Int. J. Cancer 135 (6), 1433–1443. doi:10.1002/ijc.28643
留言 (0)