Breast cancer (BC) is the most common type of cancer among women worldwide and also the leading cause of cancer-related mortality—accounting for an estimated 2,268,333 new cases and 660,620 deaths in 2022 (1). In Brazil, 73,000 new cases of female BC were estimated in 2023, representing 30% of all neoplasms, and 17,000 deaths occurred in 2020 (2). Hereditary breast cancer (HBC) accounts for approximately 10% of all cases, of which approximately 50% are due to germline variants in BRCA1/2 (3, 4). Next-generation sequencing (NGS) panel tests have identified mutations in other cancer-associated genes in BRCA1/2-negative patients with suspected HBC, sometimes more than doubling the mutation detection rate (5–9). This technology has dramatically expanded the scope of HBC—other high and moderate penetrance (HMP) genes are becoming of increased relevance—and speed of genetic testing with reduced cost (10).
Worldwide, the estimated prevalence of germline pathogenic/likely pathogenic variants (P/LP) in HMP genes varies between 5% and 13% among women with BC in Caucasian-based studies (3–6, 8, 9). The prevalence and spectrum of BC P/LP variants are affected by age at diagnosis, race/ethnicity, ancestry, geographical region, and BC molecular subtype (9, 11, 12). A cross-sectional study that evaluated the prevalence of P/LP and variants of unknown significance (VUS) among individuals undergoing NGS panel testing for Hereditary Breast and Ovarian Cancer (HBOC) from Mexico, Central America, the Caribbean, South America, and self-reported Hispanic individuals from US showed an LP/P rate ranging from 9.1% to 18.3%. The South America rate of P/LP and VUS were 13.8% and 40.6%, respectively (13).
In Brazil, there are some published studies evaluating the prevalence of other HMP genes beyond BRCA1/2 with multigene panel testing (14–21). The prevalence of germline findings varies widely, depending on the selected demographic, clinical, and pathological factors as well as the number of genes included in the panel, some of which incorporate low-penetrance genes and non-HBC genes (14, 17, 20). One large study showed the prevalence of 17.5% in HMP genes in Brazilian BC patients tested in a single laboratory (21). However, the prevalence of HBC genes according to BC subtypes remains largely unexplored in Brazil. Many international cohorts revealed significant molecular heterogeneity for predisposition genes within BC subtypes (5, 22).
The aim of this study is to evaluate the prevalence of P/LP variants and VUS in BRCA1/2 as well as in other HMP BC genes, in the overall population diagnosed with BC referred to a central laboratory. In addition, we aimed to perform stratified analyses according to age and BC subtype.
MethodsStudy design and populationThis is a retrospective cross-sectional study that evaluates the germline profile of patients diagnosed with invasive BC (ICD-10 code C40) from 12 different sites in South, Southeast, Midwest, Northeast, and North regions of Brazil and were tested in a single reference laboratory [Oncoclinicas (OC) Medicina de Precisão (OCPM), São Paulo, Brazil] from 2019 and 2023. OCPM is a reference laboratory to the Oncoclinicas & CO group, which is the largest private healthcare provider of oncology care in Latin America. This study was approved with waiver of re-consent by the local Research Ethics Committee (CAAE: 70500223.8.0000.0227) in Rio de Janeiro.
Patients underwent testing with either a targeted HBOC panel of 35 genes or a broader germline panel covering 105 genes, at the discretion of the physician. Both assays cover HMP BC genes: BRCA1, BRCA2, PALB2, TP53, CDH1, NF1, PTEN, STK11, CHEK2, ATM, BARD1, RAD51C, and RAD51D. Irrespective of the panel, NGS followed the same protocols. Genomic DNA was obtained from a buccal swab or peripheral blood sample. NGS libraries and panel enrichment was prepared using SureSelect custom panel XTHS2 (Agilent). DNA sequencing was performed by Illumina platforms (NextSeq 550). The bioinformatic pipeline consisted of FastQ files (generated by Illumina’s pipeline) aligned to the reference genome GRCh37/UCSC hg19, low-quality and duplicate readings removal, and variants (SNPs/indels) calling with GATK HaplotypeCaller on the coding sequences and flanking regions (± 20 bp) of the target region. Copy number variations (CNVs) were identified at the exon level using both ExomeDepth and CNVkit. If a CNV was identified, a multiplex ligation-dependent probe amplification (MLPA) assay was performed as orthogonal confirmation. The variants were manually classified by internal molecular biologists and geneticists according to the guidelines of the American College of Medical Genetics and Genomics (ACMG)/Human Genome Variation Society (HGVS) considering the current literature (23).
Clinical databasePatient data, such as gender, age at testing, geographic region, and diagnosis of BC, were obtained from mandatory test requisition forms filled by ordering physicians and structured in the laboratory information management system (LIMS). The BC molecular subtype was also extracted from local LIMS for the subset of patients with information on hormone receptor (HR, both estrogen and progesterone) and HER2 status by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). BC samples were classified into three subtypes, HR+/HER2−, HER2+, or triple-negative breast cancer (TNBC), as per standard practice. All data were anonymized before analysis.
Statistical analysisDescriptive statistics were used to summarize the data. Categorical data were presented as frequency and percentages, and continuous data were expressed as medians and ranges. For comparisons of categorical and continuous variables, we used the chi-square test and Mann–Whitney U test, respectively. Statistical significance was assumed at p < 0.05. As the study was descriptive, estimation of sample size or statistical power was not applicable. All data were processed in Microsoft Office Excel along with R Programming Environment, version 4.0.5.
ResultsClinical and molecular characteristicsThis cohort involved 2,208 patients with BC from 2019 to 2023. Most patients were women (99%) from Southeastern Brazil (79.7%), followed by patients from the Midwestern (5.6%), Southern (5.0%), and Northeastern/Northern (4.8%) parts. The median age at genetic testing was 47 years; most patients (59.6%) were ≤50 years.
BC molecular subtype was available in 641 cases: 264 patients (41.2%) had HR+/HER2− BC, 116 (18.1%) were HER2+, and 261 (40.7%) had TNBC (Table 1).
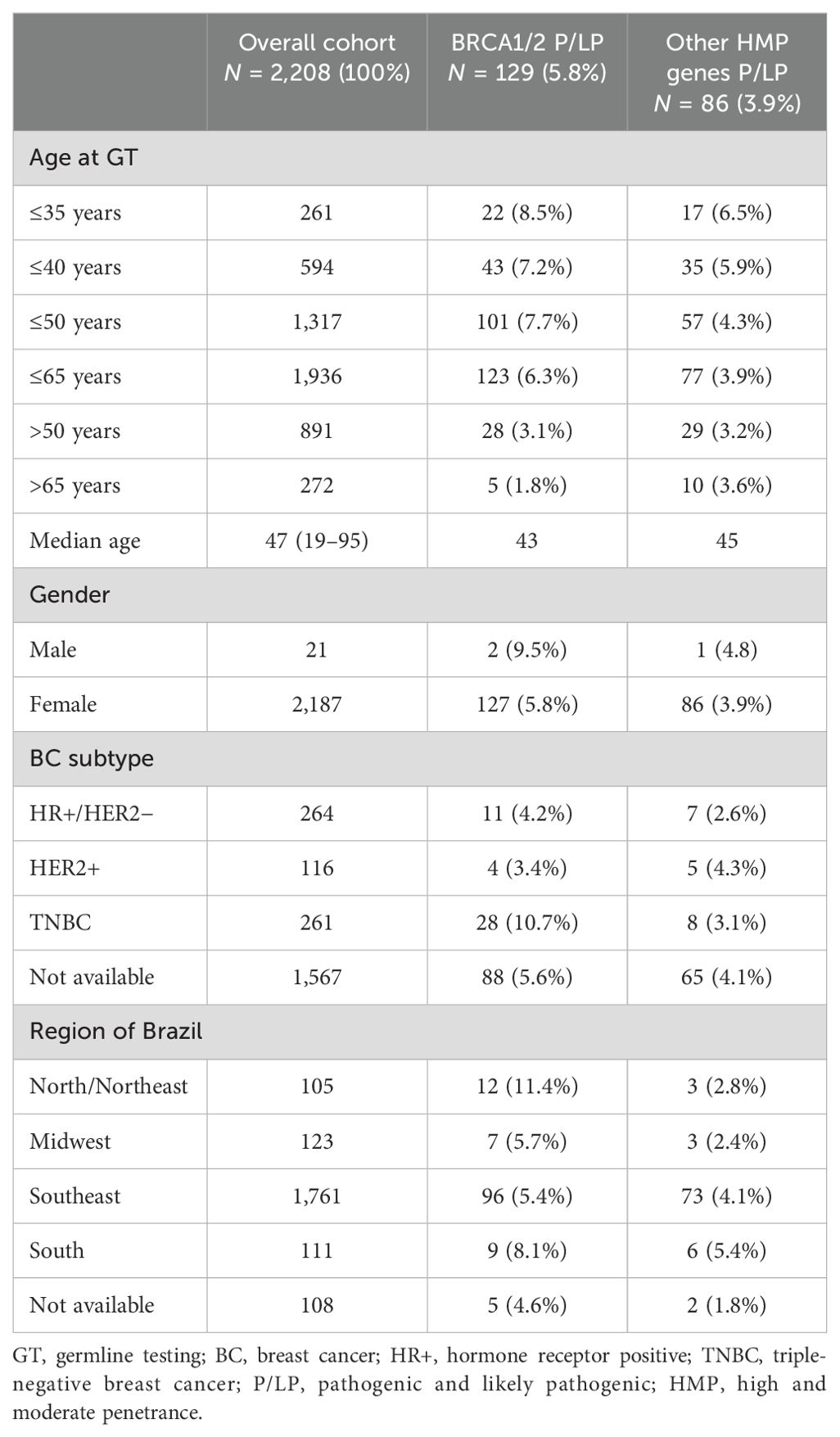
Table 1. Baseline characteristics.
Prevalence and spectrum of pathogenic and likely pathogenic variantsOverall, 215 (9.7%) had a P/LP in HMP genes, including 129 (5.8%) patients who had a BRCA1/2 P/LP variant and 86 (3.9%) who had a P/LP variant in other HMP BC genes (Table 1). The prevalence of BRCA1/2 P/LP variants was significantly higher in patients ≤50 years than in those >50 years (7.7% vs 3.1%; p = 0.009) as well as in those with TNBC (10.7%) when compared to HR+/HER2− (4.2%) and HER2+ (3.4%) (p < 0.001). Table 2 summarizes these results.
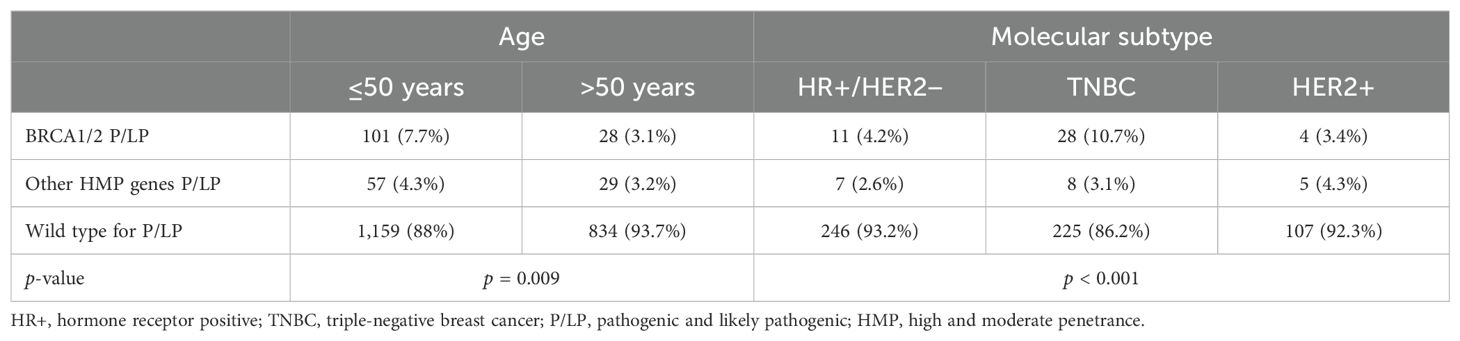
Table 2. Prevalence of germline mutations in selected subgroups of interest.
Of note, patients with TNBC who were 40 years or younger had 14.4% of deleterious variants in BRCA1/2 (Figure 1). In the population ≤65 years, per the new ASCO guideline cutoff for genetic testing recommendation in BC, the prevalence of BRCA1/2 P/LP variants was 5.6%, compared to 1.8% in those >65 years. Two patients (1%) had two P/LP variants involving a combination of BRCA1 and BRCA2 genes and were diagnosed with MINAS (Multi-locus Inherited Neoplasia Allele Syndrome) (Figure 2).
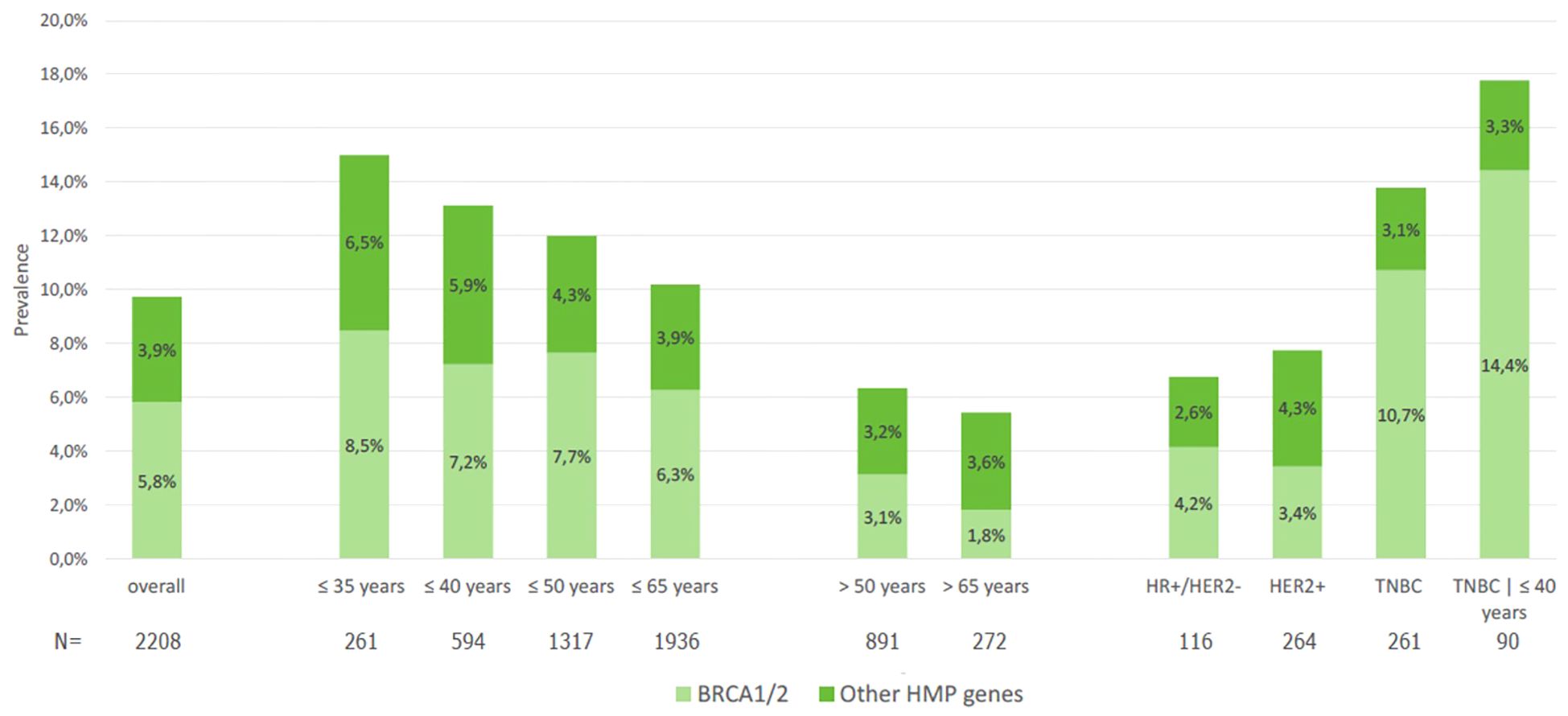
Figure 1. Prevalence of pathogenic or likely pathogenic variants in BRCA1/2 and other HMP genes in the overall population and according to age and breast cancer subtype.
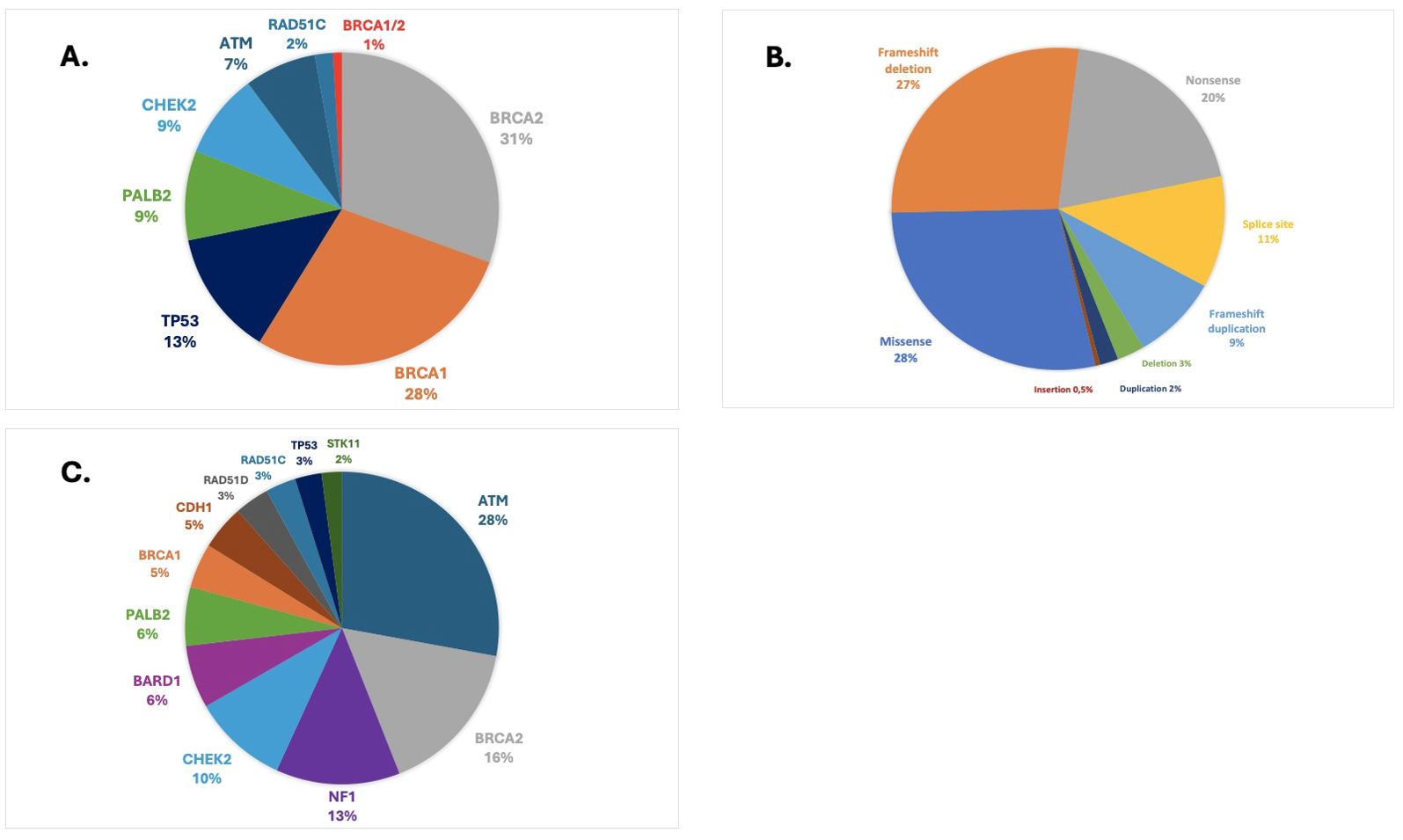
Figure 2. (A) Prevalence and spectrum of pathogenic or likely pathogenic variants in HMP genes. (B) Class of pathogenic or likely pathogenic variants. (C) Prevalence and spectrum of VUS in HMP genes.
On the other hand, the prevalence of deleterious variants in other HMP genes (excluding BRCA1/2) was 3.9% and did not significantly vary according to age categories (Table 1), although we observed numerically higher rates of P/LP variants in patients ≤35 years (6.5%) than in those >50 years (3.2%) or >65 years (3.6%) (Figure 1). In terms of BC molecular subtypes, the prevalence of P/LP variants in other HMP genes (excluding BRCA1/2) was 2.6% in HR+/HER2−, 4.3% in HER2+, and 3.1% in TNBC. Finally, among 21 male patients with BC, 2 (9.5%) had P/LP variant in BRCA2 and 1 in CHEK2 (4.8%), accounting for 15.3% of actionable findings.
The most frequent deleterious variants were found in BRCA2 (31%), BRCA1 (29%), TP53 (13%), PALB2 (9%), and CHEK2 (9%) (Figure 2A). The founder pathogenic variant in TP53 R337H accounted for 79% (22/28) of all TP53 variants, representing 1% of the overall BC population included in this study. We did not find P/LP variants in CDH1, NF1, PTEN, STK11, BARD1, or RAD51D. In the HR+/HER2− population, the most frequently mutated genes harboring P/LP variants were BRCA2 (33%), BRCA1 (27%), and PALB2 (22%). In HER2+ cases, these genes were BRCA2 (33%) and TP53 (22%). In patients with TNBC tumors, deleterious variants were most commonly found in BRCA1 (58%), BRCA2 (19%), or PALB2 (17%). The largest fraction of deleterious variants were missense variants (28%), followed by frameshift deletion (27%), nonsense (20%), splice site (11%), and frameshift duplications (9%). CNVs such as large deletions and duplications accounted for 5% of all variants (Figure 2B).
Prevalence and spectrum of variants of unknown significanceOverall, we found 495 VUS in HMP genes among 433 patients (19.6%). At least one VUS in BRCA1/2 was detected in 4.4% of the cases, while 15.9% of the patients carried at least one VUS in other HMP genes. The most frequent VUS were found in ATM (28%), BRCA2 (16%), NF1 (13%), and CHEK2 (10%) genes (Figure 2C). In the population who carried P/LP BRCA1/2 variants, 17% had at least one VUS in HMP genes.
DiscussionTo the best of our knowledge, this is the largest study to investigate a cohort of Brazilian patients with BC from the private healthcare system who received multi-gene NGS panels in the single reference laboratory and may not have been strictly selected for germline genetic testing (GGT) based on high-risk criteria for hereditary cancer [National Comprehensive Cancer Network (NCCN)]. We found that the prevalence of P/LP variants in 13 HMP genes was close to 10%, including 6% of deleterious variants in BRCA1/2 genes, which were significantly more prevalent in younger patients and in those with TNBC.
In Brazil, most published studies evaluating the prevalence of germline findings in BC patients were enriched for high-risk criteria for hereditary cancer—including low-penetrance genes and non-BC genes—and the prevalence varies between 15% and 20% (16–18, 20, 21). In the largest previous study, Guindalini et al. evaluated 1,663 Brazilian BC patients, who underwent germline multi-gene panels covering from 20 to 38 genes, which showed a 17.5% rate of deleterious variants in HMP genes, including 10.1% in BRCA1/2, 13.4% in high-penetrance (HP), and 4.1% in moderate-penetrance (MP) genes. Most mutated genes were BRCA1 (27.4%), BRCA2 (20.3%), and TP53 (10.5%) in the HP group and ATM (8.8%) and CKEK2 (6.2%) in the MP group. Of note, the R337H variant accounted for 70% of all TP53 pathogenic variants, representing 1% of the overall population (21). The worldwide prevalence of germline TP53 deleterious variants is estimated to be approximately one in every 3,500 to 10,000 individuals (24). However, in Brazil, a TP53-R337H founder PV in the South and Southern regions has a prevalence of approximately 0.3% of the healthy individuals (25). Among Brazilian women with BC, the prevalence of TP53 R337H varied from 0.9% to 12%, depending on the geographical region and age at diagnosis of BC (16, 21, 26–28).
Our results demonstrated lower prevalence of actionable germline findings, which could be explained by patient selection for NGS panel testing. Guindaline et al. selected for high-risk population as it included many patients referred to GGT in a laboratory (Mendelics Análise Genômica S.A., São Paulo, SP, Brazil) where testing costs could be reimbursed based on the restrictive coverage criteria (i.e., very high-risk criteria for GGT) defined by the Brazilian National Supplementary Health Agency (ANS—Agencia Nacional de Saude). In our study, all patients were tested in a single reference laboratory (OCPM, São Paulo, Brazil) using germline multigene panels where currently testing costs cannot be covered by health insurance and the GT recommendation was based on physician recommendation and out-of-pocket payment. Therefore, our study may have shown the prevalence of germline variants in a scenario closer to universal GGT as proposed by the American Society of Breast Surgeons (ASBrS) (29). According to the American Society of Clinical Oncology (ASCO) and Society of Surgical Oncology (SSO) guideline, BRCA1/2 testing should be offered to all women younger than age 65 at the time of BC diagnosis, as well as for all female BC patients who are eligible for PARP inhibitor therapy, have TNBC, have a second contralateral or ipsilateral primary BC, or have a personal or family history suggestive of hereditary cancer. BRCA1/2 testing is also recommended for all male BC patients. Finally, the guideline suggests that testing for HP genes beyond BRCA1/2 should be offered to selected patients (30). However, the most recent ASCO guideline on GGT panels in patients with cancer proposes that when GGT is indicated for a patient with cancer, multigene panel testing should be offered if more than one gene is relevant, including BRCA1, BRCA2, PALB2, CDH1, PTEN, STK11, and TP53 genes (31). In Brazil, we are far from implementing these guidelines, since GGT is not available in the public healthcare system, and in the private system, its coverage is only available for patients who fulfill restrictive criteria established by the ANS. Access to genetic testing is a challenge. Regulatory and policy actions, together with physician and patient education, are urgently needed to address these issues (32). This is a field of cancer care that should be substantially improved, since recent data revealed that only 26% of female patients with BC undergo GGT within the first 2 years after their diagnosis (33).
Our study demonstrated that the deleterious variants in HPM genes were more common in younger patients and in those with TNBC (14%) followed by HER2+ (9%) and HR+/HER2− (7%). Paixão et al. showed that the prevalence of P/LP variants in HMP was 22.8% in the TNBC subtype, 15.4% in HER2+, and 9.8% in HR+/HER2−. This study included high-risk patients with younger median age at diagnosis (44 years) and who met the NCCN criteria for GGT (18). The association between BC molecular subtype and germline findings is important not only from a testing perspective but also for interpretation of risk-reducing approaches. A clinical report suggests in some cases that germline pathogenic variants do not appear to play a major role in the tumorigenesis of BC (34). The CARRIERS study showed that the contralateral BC risk in PALB2 carriers was only statistically higher in HR− patients, highlighting the importance of tumor phenotype in genetic counseling (35).
The adequate evaluation of germline status is critical for BC patients as patients who test positive for HP genes may be considered for risk-reducing strategies including increased surveillance, chemoprevention, and surgical interventions, alongside preventive measures in family members (36). Therefore, adequate evaluation of germline status should be incorporated into clinical practice as a predictive biomarker with important implications for optimal treatment of BC in the early (37) and advanced (38, 39) settings. Of note, the GGT results may help personalize risk-reducing strategies such as bilateral mastectomy in young patients with BC who test negative, since a recent study suggests a low 10-year cumulative incidence (2.2%) of second primary BC in this population (40).
The present analysis has some limitations. First, this study involved a laboratory cohort with paucity of clinical data including high-risk features such as family history of cancer, reproductive and gynecological history, and modified risk factors. Second, we had complete information in BC molecular subtypes only for ~30% of the cohort. Third, there was underrepresentation of some Brazilian regions in the scenario of genetic diversity in miscegenated populations. Finally, our cohort may not have been entirely unselected on the basis of guidelines’ criteria given that physicians may have ordered the testing based on common clinical criteria, such as age at diagnosis, BC subtype, and family history.
In conclusion, this study is the largest cohort from the perspective of the Brazilian private health system involving the germline profile of P/LP variants in HMP BC genes in a population who might not have been selected based on high-risk criteria. This study provided a broader understanding of germline BC genes and has the potential to open support regulatory actions, healthcare provider and patient education, and policy recommendations toward broader testing access. Therefore, GGT incorporation into routine practice should be strongly considered by healthcare providers in BC.
Data availability statementThe dataset and materials used to conduct this research is available from the authors upon request to interested researchers.
Ethics statementThe studies involving humans were approved by the local Research Ethics Committee (CAAE: 70500223.8.0000.0227) in Rio de Janeiro, Rio de Janeiro. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because the patients have already consented at the time of germline genetic testing.
Author contributionsLO: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing, Supervision. AR: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. CF: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. FR: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. FK: Data curation, Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. AKS: Investigation, Writing – original draft, Writing – review & editing. TS: Investigation, Writing – original draft, Writing – review & editing. JF: Investigation, Writing – original draft, Writing – review & editing. MB: Investigation, Writing – original draft, Writing – review & editing. IN: Investigation, Writing – original draft, Writing – review & editing. ACS: Investigation, Writing – original draft, Writing – review & editing. IG: Investigation, Writing – original draft, Writing – review & editing. RM: Investigation, Writing – original draft, Writing – review & editing. RA: Investigation, Writing – original draft, Writing – review & editing. LG: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. DP: Investigation, Writing – original draft, Writing – review & editing. AC: Investigation, Writing – original draft, Writing – review & editing. MZ: Investigation, Writing – original draft, Writing – review & editing. BF: Investigation, Writing – original draft, Writing – review & editing. BG: Investigation, Writing – original draft, Writing – review & editing. RD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was sponsored by Pfizer. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
AcknowledgmentsPreliminary results of this study have been presented at the 2024 American Society of Clinical Oncology (ASCO) Annual Meeting - JCO 42, 10593-10593 (2024). https://doi.org/10.1200/JCO.2024.42.16_suppl.1059.
In memoriamAll authors would like to dedicate this publication in memoriam of Bernardo Garicochea who passed away during this article submission, a remarkable mentor and leader of the Oncoclinicas cancer genetics team and a true visionary in genetics and genomics. His kindness, leadership, and support have left an indelible mark on our group, inspiring countless individuals to pursue new ideas and insights.
Conflict of interestThe authors LO, AS, TS, JG, ML, IN, AS, IG, RM, VA, BF and BG were employed by the company Oncoclinicas&CO. The authors AR, CF, FR, FK, LG, MZ and RD were employed by the company Oncoclinicas Medicina de Precisão (OCPM). The authors DP and AC were employed by the company Pfizer Brasil.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References2. Câncer IN de. Estimativa 2023: incidência de câncer no Brasil. Rio de Janeiro, RJ: Instituto Nacional De Câncer (2023).
3. Kurian AW, Hughes E, Handorf EA, Gutin A, Allen B, Hartman AR, et al. Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol. (2017) 1):1–12. doi: 10.1200/PO.16.00066
PubMed Abstract | Crossref Full Text | Google Scholar
4. Suszynska M, Klonowska K, Jasinska AJ, Kozlowski P. Large-scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes — Providing evidence of cancer predisposition genes. Gynecol Oncol. (2019) 153:452–62. doi: 10.1016/j.ygyno.2019.01.027
PubMed Abstract | Crossref Full Text | Google Scholar
5. Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. (2017) 3:1190. doi: 10.1001/jamaoncol.2017.0424
PubMed Abstract | Crossref Full Text | Google Scholar
6. Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. (2017) 123:1721–30. doi: 10.1002/cncr.v123.10
PubMed Abstract | Crossref Full Text | Google Scholar
7. Hauke J, Horvath J, Groß E, Gehrig A, Honisch E, Hackmann K, et al. Gene panel testing of 5589 BRCA 1/2 -negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. (2018) 7:1349–58. doi: 10.1002/cam4.2018.7.issue-4
PubMed Abstract | Crossref Full Text | Google Scholar
8. Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, et al. Frequency of mutations in individuals with breast cancer referred for BRCA 1 and BRCA 2 testing using next-generation sequencing with a 25-gene panel. Cancer. (2015) 121:25–33. doi: 10.1002/cncr.v121.1
PubMed Abstract | Crossref Full Text | Google Scholar
9. Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. (2021) 384:440–51. doi: 10.1056/NEJMoa2005936
PubMed Abstract | Crossref Full Text | Google Scholar
11. Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. (2018) 39:593–620. doi: 10.1002/humu.2018.39.issue-5
Crossref Full Text | Google Scholar
12. Bhaskaran SP, Huang T, Rajendran BK, Guo M, Luo J, Qin Z, et al. Ethnic-specific BRCA1/2 variation within Asia population: evidence from over 78 000 cancer and 40 000 non-cancer cases of Indian, Chinese, Korean and Japanese populations. J Med Genet. (2021) 58:752–9. doi: 10.1136/jmedgenet-2020-107299
PubMed Abstract | Crossref Full Text | Google Scholar
13. Ossa Gomez CA, Achatz MI, Hurtado M, Sanabria-Salas MC, Sullcahuaman Y, Chávarri-Guerra Y, et al. Germline pathogenic variant prevalence among latin American and US hispanic individuals undergoing testing for hereditary breast and ovarian cancer: A cross-sectional study. JCO Glob Oncol. (2022) 8:e2200104. doi: 10.1200/GO.22.00104
PubMed Abstract | Crossref Full Text | Google Scholar
14. De Souza Timoteo AR, Gonçalves AÉMM, Sales LAP, Albuquerque BM, De Souza JES, De Moura PCP, et al. A portrait of germline mutation in Brazilian at-risk for hereditary breast cancer. Breast Cancer Res Treat. (2018) 172:637–46. doi: 10.1007/s10549-018-4938-0
PubMed Abstract | Crossref Full Text | Google Scholar
15. Da Costa E Silva Carvalho S, Cury NM, Brotto DB, De Araujo LF, Rosa RCA, Texeira LA, et al. Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: analysis of a 21 gene panel in the Brazilian population. BMC Med Genomics. (2020) 13:21. doi: 10.1186/s12920-019-0652-y
PubMed Abstract | Crossref Full Text | Google Scholar
16. Sandoval RL, Leite ACR, Barbalho DM, Assad DX, Barroso R, Polidorio N, et al. Germline molecular data in hereditary breast cancer in Brazil: Lessons from a large single-center analysis. PloS One. (2021) 16:e0247363. doi: 10.1371/journal.pone.0247363
PubMed Abstract | Crossref Full Text | Google Scholar
17. Barbalho D, Sandoval R, Santos E, Pisani J, Quirino C, Garicochea B, et al. Novel insights from the germline landscape of breast cancer in Brazil. Front Oncol. (2022) 11:743231. doi: 10.3389/fonc.2021.743231
PubMed Abstract | Crossref Full Text | Google Scholar
18. Paixão D, Torrezan GT, Santiago KM, Formiga MN, Ahuno ST, Dias-Neto E, et al. Characterization of genetic predisposition to molecular subtypes of breast cancer in Brazilian patients. Front Oncol. (2022) 12:976959. doi: 10.3389/fonc.2022.976959
PubMed Abstract | Crossref Full Text | Google Scholar
19. Pereira JZ, Carneiro JG, Vieira MS, Valente BM, De Oliveira PZ, Mello CL, et al. Frequency of germline genetic variants in women with a personal or family history of breast cancer from Brazil. Mol Biol Rep. (2022) 49:9509–20. doi: 10.1007/s11033-022-07840-0
PubMed Abstract | Crossref Full Text | Google Scholar
20. Felix GES, Guindalini RSC, Zheng Y, Walsh T, Sveen E, Lopes TMM, et al. Mutational spectrum of breast cancer susceptibility genes among women ascertained in a cancer risk clinic in Northeast Brazil. Breast Cancer Res Treat. (2022) 193:485–94. doi: 10.1007/s10549-022-06560-0
PubMed Abstract | Crossref Full Text | Google Scholar
21. Guindalini RSC, Viana DV, Kitajima JPFW, Rocha VM, López RVM, Zheng Y, et al. Detection of germline variants in Brazilian breast cancer patients using multigene panel testing. Sci Rep. (2022) 12:4190. doi: 10.1038/s41598-022-07383-1
PubMed Abstract | Crossref Full Text | Google Scholar
22. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. (2016) 13:674–90. doi: 10.1038/nrclinonc.2016.66
PubMed Abstract | Crossref Full Text | Google Scholar
23. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med Off J Am Coll Med Genet. (2015) 17:405–24. doi: 10.1038/gim.2015.30
PubMed Abstract | Crossref Full Text | Google Scholar
24. De Andrade KC, Strande NT, Kim J, Haley JS, Hatton JN, Frone MN, et al. Genome-first approach of the prevalence and cancer phenotypes of pathogenic or likely pathogenic germline TP53 variants. Hum Genet Genomics Adv. (2024) 5:100242. doi: 10.1016/j.xhgg.2023.100242
PubMed Abstract | Crossref Full Text | Google Scholar
26. Felix GE, Abe-Sandes C, MaChado-Lopes TM, Bomfim TF, Guindalini RSC, Santos VCS, et al. Germline mutations in BRCA1, BRCA2, CHEK2 and TP53 in patients at high-risk for HBOC: characterizing a Northeast Brazilian Population. Hum Genome Var. (2014) 1:14012. doi: 10.1038/hgv.2014.12
PubMed Abstract | Crossref Full Text | Google Scholar
27. Cury NM, Ferraz VE, Silva WA. TP53 p.R337H prevalence in a series of Brazilian hereditary breast cancer families. Hered Cancer Clin Pract. (2014) 12:8. doi: 10.1186/1897-4287-12-8
PubMed Abstract | Crossref Full Text | Google Scholar
28. Giacomazzi J, Graudenz MS, Osorio CABT, Koehler-Santos P, Palmero EI, Zagonel-Oliveira M, et al. Prevalence of the TP53 p.R337H mutation in breast cancer patients in Brazil. PloS One. (2014) 9:e99893. doi: 10.1371/journal.pone.0099893
PubMed Abstract | Crossref Full Text | Google Scholar
29. Manahan ER, Kuerer HM, Sebastian M, Hughes KS, Boughey JC, Euhus DM, et al. Consensus guidelines on genetic` testing for hereditary breast cancer from the American society of breast surgeons. Ann Surg Oncol. (2019) 26:3025–31. doi: 10.1245/s10434-019-07549-8
PubMed Abstract | Crossref Full Text | Google Scholar
30. Bedrosian I, Somerfield MR, Achatz MI, Boughey JC, Curigliano G, Friedman S, et al. Germline testing in patients with breast cancer: ASCO–society of surgical oncology guideline. J Clin Oncol. (2024) 42:584–604. doi: 10.1200/JCO.23.02225
PubMed Abstract | Crossref Full Text | Google Scholar
31. Tung N, Ricker C, Messersmith H, Balmaña J, Domchek S, Stoffel EM, et al. Selection of germline genetic testing panels in patients with cancer: ASCO guideline. J Clin Oncol. (2024) 42:2599–615. doi: 10.1200/OP.24.00278
PubMed Abstract | Crossref Full Text | Google Scholar
32. Achatz MI, Caleffi M, Guindalini R, Marques RM, Nogueira-Rodrigues A, Ashton-Prolla P. Recommendations for advancing the diagnosis and management of hereditary breast and ovarian cancer in Brazil. JCO Glob Oncol. (2020) 6:439–52. doi: 10.1200/JGO.19.00170
PubMed Abstract | Crossref Full Text | Google Scholar
34. Rezqallah A, Torres-Esquius S, Llop-Guevara A, Cruellas M, Martinez MT, Romey M, et al. Two germline pathogenic variants in cancer susceptibility genes and their null implication in breast cancer pathogenesis: the importance of tumoral homologous recombination deficiency testing. JCO Precis Oncol. (2024) 8):e2300446. doi: 10.1200/PO.23.00446
PubMed Abstract | Crossref Full Text | Google Scholar
35. Yadav S, Boddicker NJ, Na J, Polley EC, Hu C, Hart SN, et al. Contralateral breast cancer risk among carriers of germline pathogenic variants in ATM, BRCA1, BRCA2, CHEK2, and PALB2. J Clin Oncol. (2023) 41:1703–13. doi: 10.1200/JCO.22.01239
PubMed Abstract | Crossref Full Text | Google Scholar
36. Tung NM, Boughey JC, Pierce LJ, Robson ME, Bedrosian I, Dietz JR, et al. Management of hereditary breast cancer: American society of clinical oncology, American society for radiation oncology, and society of surgical oncology guideline. J Clin Oncol. (2020) 38:2080–106. doi: 10.1200/JCO.20.00299
PubMed Abstract | Crossref Full Text | Google Scholar
37. Geyer CE, Garber JE, Gelber RD, Yothers G, Taboada M, Ross L, et al. Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer. Ann Oncol. (2022) 33:1250–68. doi: 10.1016/j.annonc.2022.09.159
PubMed Abstract | Crossref Full Text | Google Scholar
38. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. (2017) 377:523–33. doi: 10.1056/NEJMoa1706450
留言 (0)