Lipoma is a prevalent subcutaneous benign tumor arising from adipose tissue, typically surrounded by a thin fibrous tissue. Variants such as Benign Lipoma (BL), and BLs affecting skin and subcutaneous tissues of the trunk (TS-BL), limbs (LS-BL), and head, face, and neck (HFNS-BL) are common (Kosztyuova and Shim, 2017). Lipomas may occur not only superficially but also within deep soft tissues, especially in the back, upper limbs, and thighs, where they predominantly exhibit an infiltrative growth pattern (Inamura et al., 2021; Ameloot et al., 2022). Rare variants may also appear in muscles, within joints, or bronchi (Seo et al., 2018; Wang et al., 2019; Nader et al., 2012), manifesting as either solitary or multiple lesions at any age. Studies suggest that genetic predisposition and external factors such as ionizing radiation, chlorophenol, and viruses may contribute to lipoma development (Palitot DE Melo et al., 2022). Currently, lipomas and their subtypes constitute over half of all soft tissue tumors, with a prevalence rate of 2.1% (Marteau et al., 2020), and approximately 5% of cases present as multiple lipomatosis (Charifa et al., 2024). Furthermore, traditional treatments, including surgical removal and vacuum aspiration, face challenges due to high recurrence rates and difficulties in complete removal, underscoring the importance of understanding their pathological characteristics and exploring potential treatment options (Pereira and Schonauer, 2001). Lipoma remains one of the most commonly encountered soft tissue tumors in clinical practice, yet its precise causes and underlying mechanisms, particularly its potential associations with the GM, are not well-understood. Therefore, a deeper investigation into the pathological mechanisms of lipomas, especially their relationship with the microbiome, is crucial for elucidating their etiology and developing new therapeutic approaches.
Compelling evidence suggests that the GM plays a role in the regulation of metabolic disorders, psychiatric conditions, and cancers, including gout (Tong et al., 2022), Alzheimer’s disease (Varesi et al., 2022), and lymphoma (Ml and Rh, 2014). Recent findings indicate that microbiome-induced tumors in the gastrointestinal tract and uterus are well-documented. As part of the tumor microenvironment (TME), the GM may influence the formation and progression of lipomas (Kovács et al., 2020). Dysbiosis of the GM can affect systemic immune functions, compromise mucosal barriers, and modulate immune responses that may promote or inhibit tumor development (Cs and Mj, 2011; Kawai and Akira, 2009). Additionally, moderate gut dysbiosis observed in lipoma patients can lead to reduced microbial diversity (Tkachuk et al., 2023).
Given the costs, time constraints, and ethical considerations associated with clinical trials, Mendelian Randomization (MR) has emerged as a robust method (Song et al., 2024). Commonly used to examine causal relationships between exposures and outcomes, MR employs single nucleotide polymorphisms (SNPs) derived from genome-wide association study (GWAS) as instrumental variables to establish causality. This method has been utilized to explore potential causal links between the GM and various diseases (Sekula et al., 2016). Based on existing research, this study hypothesized that an imbalance in the GM is causally related to the development of lipomas. By applying MR analysis, we aimed to uncover the potential role of the GM in lipoma formation, providing new insights into the pathological mechanisms underlying lipomas.
2 Materials and methods2.1 Study overviewThis research conducted a two-sample MR analysis to explore the causal relationships between the GM and four types of lipomas. Comprehensive GWAS data were obtained for both the GM and lipomas. Each genus level of the GM was considered an independent exposure factor, with BLs and three other location-specific lipomas as outcome factors. To ensure the reliability of our findings, this MR study adhered to three critical assumptions: 1) SNPs, used as instrumental variables (IVs), are closely associated with the exposure (GM); 2) SNPs are independent of confounding factors; 3) SNPs affect lipoma risk solely through the GM and not through other pathways (VanderWeele et al., 2014; Liu Y. et al., 2022). The workflow of this study is depicted in Figure 1.
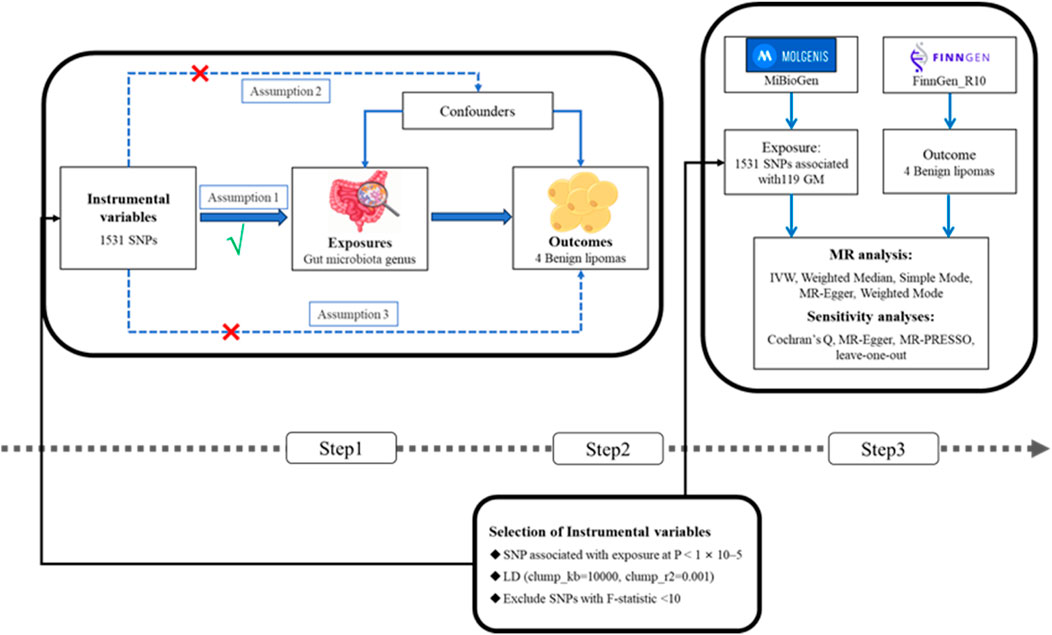
Figure 1. (Step1) Three assumptions of MR. I, Correlation assumption; II, independence assumption; III, exclusionary restriction assumption; (Step2) Selection of instrumental variables; (Step3) Flowchart of this MR study. MR, Mendelian randomization; SNP, single nucleotide polymorphism.
2.2 Data sourcesThe GWAS data for the GM were initially sourced from the MiBioGen consortium, involving a large-scale, ethnically diverse analysis of 18,340 individuals (Kurilshikov et al., 2021). We excluded 12 genera with unknown classifications, ultimately analyzing 119 genera (Xia et al., 2023). Additionally, GWAS data for BL, TS-BL, LS-BL, and HFNS-BL were obtained from the FinnGen R10 database (FinnGen provides genetic insights from), with all data derived from European populations. All summary data utilized in this study are publicly available and have received ethical approval from their respective institutions (Table 1).
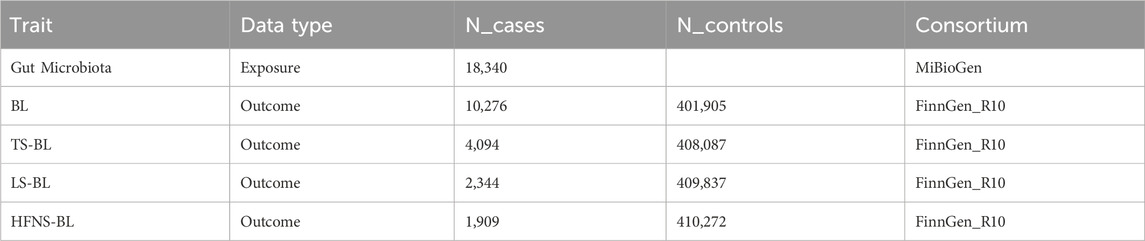
Table 1. Details of the GWASs included in the Mendelian Randomization.
2.3 Selection of instrumental variablesTo validate the accuracy of the causal relationship between the GM and lipomas, this study implemented rigorous quality control procedures. Initially, SNPs associated with specific genera were selected as IVs using a p-value threshold (p < 1 × 10−5). Linkage disequilibrium (LD) analysis was then conducted on a European genomic dataset (clump_kb = 10,000, clump_r2 = 0.001) (Chen et al., 2023), excluding palindromic SNPs to mitigate allelic biases. The strength of the IVs was assessed by calculating the F-statistic, where an F > 10 indicated robust instruments and IVs with F ≤ 10 were excluded. The F-statistic was calculated as follows: F = β2_exposure/SE2_exposure (Wang et al., 2024).
2.4 Statistical methods and sensitivity analysisFive MR analysis methods were utilized to thoroughly investigate the potential causal relationship between the GM and lipomas. These methods included inverse variance weighted (IVW), weighted median, simple mode, MR-Egger, and weighted mode, with IVW serving as the primary method (Lee et al., 2016). IVW, a commonly used approach, combines the effects of each genetic variant by weighting them inversely to their estimated uncertainty to maximize statistical efficiency. The weighted median method estimates causal effects while addressing pleiotropy, assigning weights to each IV, and estimating the causal effect as the weighted median. Simple mode estimates causal effects by selecting the most common effect direction among IVs, assuming a cluster around a true causal effect value. MR-Egger regression detects and adjusts for horizontal pleiotropy by allowing for pleiotropy among IVs and adjusting the causal effect estimate through the Egger regression approach, with the intercept testing for systematic pleiotropy. Lastly, the weighted mode method assigns weights to different IVs and integrates their effects, assuming the most significant IVs contribute the greatest weight and their mode represents the causal effect.
Cochran’s Q test assessed result heterogeneity, with a p < 0.05 indicating significant heterogeneity. Depending on heterogeneity, either a random-effects model or a fixed-effects model was applied. Horizontal pleiotropy was evaluated using MR-Egger and MR-PRESSO analyses, with an intercept p > 0.05 indicating no significant horizontal pleiotropy. A leave-one-out test was employed to identify individual SNPs with a significant impact on the MR analysis. Additionally, funnel plots and forest plots were utilized to enhance the robustness of the findings. All statistical analyses were conducted using Rstudio (version 4.3.2), utilizing the TwoSampleMR package.
3 Results3.1 Selection of instrumental variablesSNPs related to the GM were sourced from the MiBioGen database. After applying a p-value threshold (p < 1 × 10−5), a total of 14,587 SNPs associated with the GM were identified. These SNPs underwent further scrutiny through LD analysis. Utilizing the selection criteria for IVs, 1531 SNPs linked to 119 GM taxa were selected as IVs, all demonstrating F-statistics greater than 10 (Yao et al., 2024), suggesting a minimal likelihood of weak instrument bias. For more detailed information, (Supplementary Table S1). Figure 2 depicts the MR causal relationships between the 119 GM taxa and four types of lipomas, with separate results listed in Supplementary Tables S2–S5.

Figure 2. The circus plot showing the MR results of all gut microbiota. IVW, inverse-variance weighted; SM, Simple mode; WM, Weighted median; WMOED, Weighted mode; P, p-value; OR, odds ratio.
3.2 Causal relationship between GM and 4 types of lipoma3.2.1 Causal relationship between GM and BLIVW analysis identified protective associations between six gut microbial genera and the risk of BL. Specifically, the Eubacterium rectale group (OR = 0.848, 95% CI: 0.731–0.985), Desulfovibrio (OR = 0.842, 95% CI: 0.740–0.959), Ruminococcus1 (OR = 0.870, 95% CI: 0.760–0.994), Clostridium sensu stricto1 (OR = 0.789, 95% CI: 0.680–0.916), and Lachnospiraceae UCG001 (OR = 0.888, 95% CI: 0.795–0.993) exhibited protective effects against BL. Conversely, Lachnospiraceae UCG008 (OR = 1.141, 95% CI: 1.031–1.262) was linked to an increased risk of BL (Figure 3). Scatter plots from the MR analysis displayed the estimated effects of GM SNPs on BL (Supplementary Figure 1A).
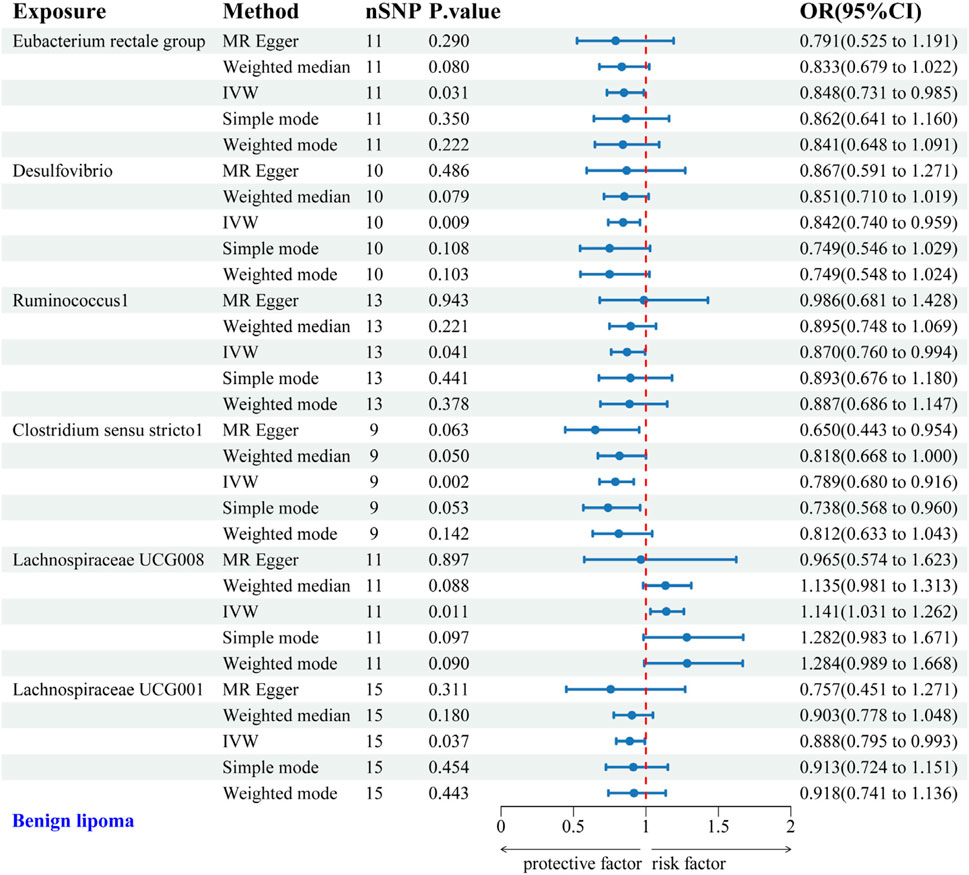
Figure 3. Forest plot of the associations between genetically predicted GM and BL risk using IVW methods. IVW, inverse-variance weighted. OR, odds ratio.
Results from Cochran’s Q, MR Egger, and MR-PRESSO analyses indicated no evidence of heterogeneity or horizontal pleiotropy among the six genera associated with BL (Table 2). Leave-one-out analysis, funnel plots, and forest plots confirmed the reliability of the MR findings (Supplementary Figures 1B, C).

Table 2. Sensitivity analysis of the MR analysis results of the GM and BL.
3.2.2 Causal relationship between GM and TS-BLIVW analysis revealed associations between five gut microbial genera and the risk of TS-BL. Desulfovibrio exhibited a protective effect against TS-BL (OR = 0.793, 95% CI: 0.647–0.972), whereas increased risks were associated with the Family XIII AD3011 group (OR = 1.382, 95% CI: 1.127–1.695), Eubacterium coprostanoligenes group (OR = 1.278, 95% CI: 1.008–1.621), Lachnospiraceae NK4A136 group (OR = 1.254, 95% CI: 1.041–1.512), and Parasutterella (OR = 1.209, 95% CI: 1.028–1.421) (Figure 4). Scatter plots from the MR analysis illustrate the estimated effects of GM SNPs on TS-BL (Supplementary Figure 2A).

Figure 4. Forest plot of the associations between genetically predicted GM and TS-BL risk using IVW methods. IVW, inverse-variance weighted. OR, odds ratio.
Results from Cochran’s Q, MR Egger, and MR-PRESSO analyses revealed no significant heterogeneity or horizontal pleiotropy among the five genera related to TS-BL (Table 3). Leave-one-out analysis, funnel plots, and forest plots indicated the robustness of the MR study results (Supplementary Figures 2B, C).
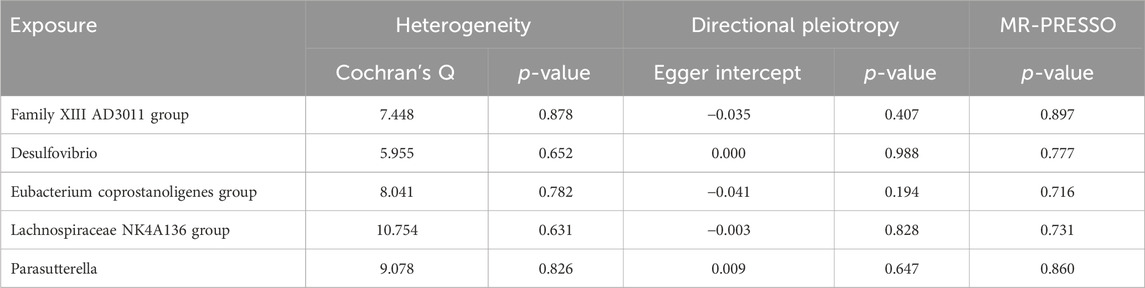
Table 3. Sensitivity analysis of the MR analysis results of the GM and TS-BL.
3.2.3 Causal relationship between GM and LS-BLIVW analysis identified six gut microbial genera associated with the risk of LS-BL. Protective effects against LS-BL were demonstrated by Clostridium innocuum group (OR = 0.841, 95%CI: 0.709–0.999), Eubacterium rectale group (OR = 0.728, 95%CI: 0.535–0.992), Anaerotruncus (OR = 0.724, 95%CI: 0.538–0.974), Ruminiclostridium6 (OR = 0.769, 95%CI: 0.601–0.984), and Lachnospiraceae UCG001 (OR = 0.801, 95%CI: 0.645–0.995). Conversely, an increased risk was associated with Lachnospiraceae UCG008 (OR = 1.282, 95%CI: 1.040–1.579) (Figure 5). Scatter plots from the MR analysis displayed the estimated effects of GM SNPs on LS-BL (Supplementary Figure 3A).
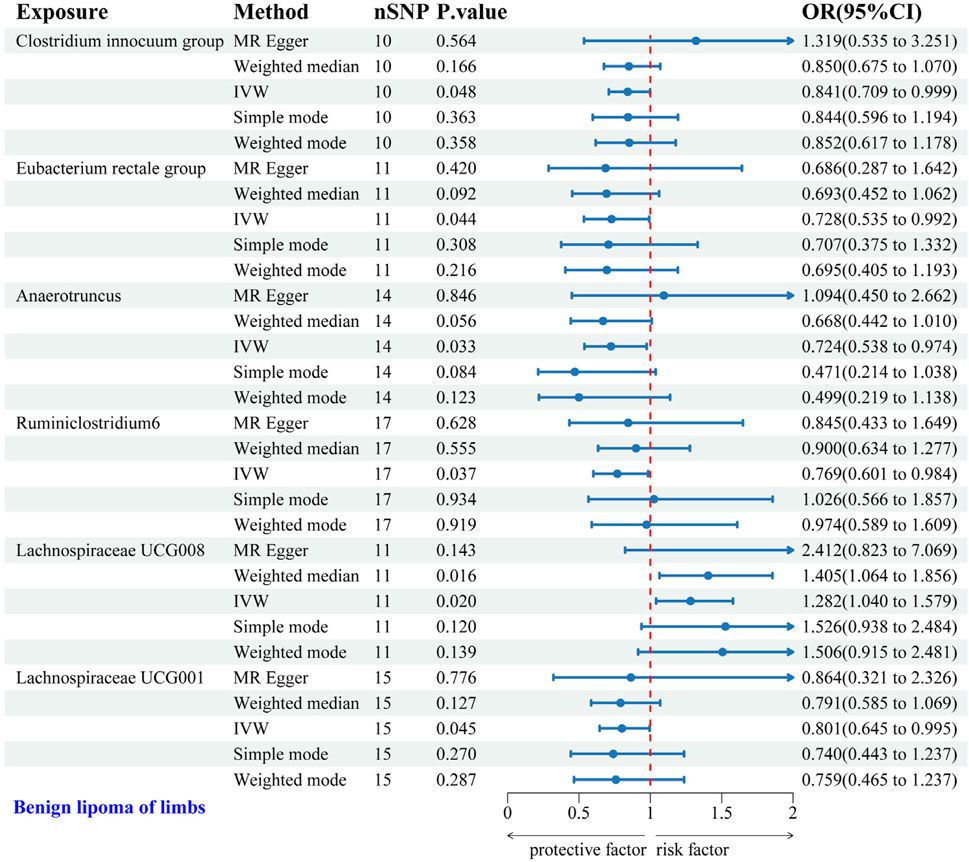
Figure 5. Forest plot of the associations between genetically predicted GM and LS-BL risk using IVW methods. IVW, inverse-variance weighted. OR, odds ratio.
Results from Cochran’s Q, MR Egger, and MR-PRESSO analyses revealed no significant heterogeneity or horizontal pleiotropy among the six genera related to LS-BL (Table 4). Leave-one-out analysis, funnel plots, and forest plots showed no significant outliers, underscoring the reliability of the MR study results (Supplementary Figures 3B, C).

Table 4. Sensitivity analysis of the MR analysis results of the GM and LS-BL.
3.2.4 Causal relationship between GM and HFNS-BLIVW analysis identified six gut microbial genera associated with the risk of HFNS-BL. Protective effects were shown by Eubacterium brachy group (OR = 0.810, 95% CI: 0.671–0.979), Odoribacter (OR = 0.559, 95% CI: 0.325–0.962), Butyricimonas (OR = 0.751, 95% CI: 0.579–0.975), Subdoligranulum (OR = 0.716, 95% CI: 0.515–0.994), and Clostridium sensu stricto1 (OR = 0.586, 95% CI: 0.416–0.826), each indicating a protective role against HFNS-BL. Conversely, Ruminococcaceae UCG005 was associated with an increased risk (OR = 1.411, 95% CI: 1.080–1.843) (Figure 6). Scatter plots from the MR analysis displayed the estimated effects of GM SNPs on HFNS-BL (Supplementary Figure 4A).
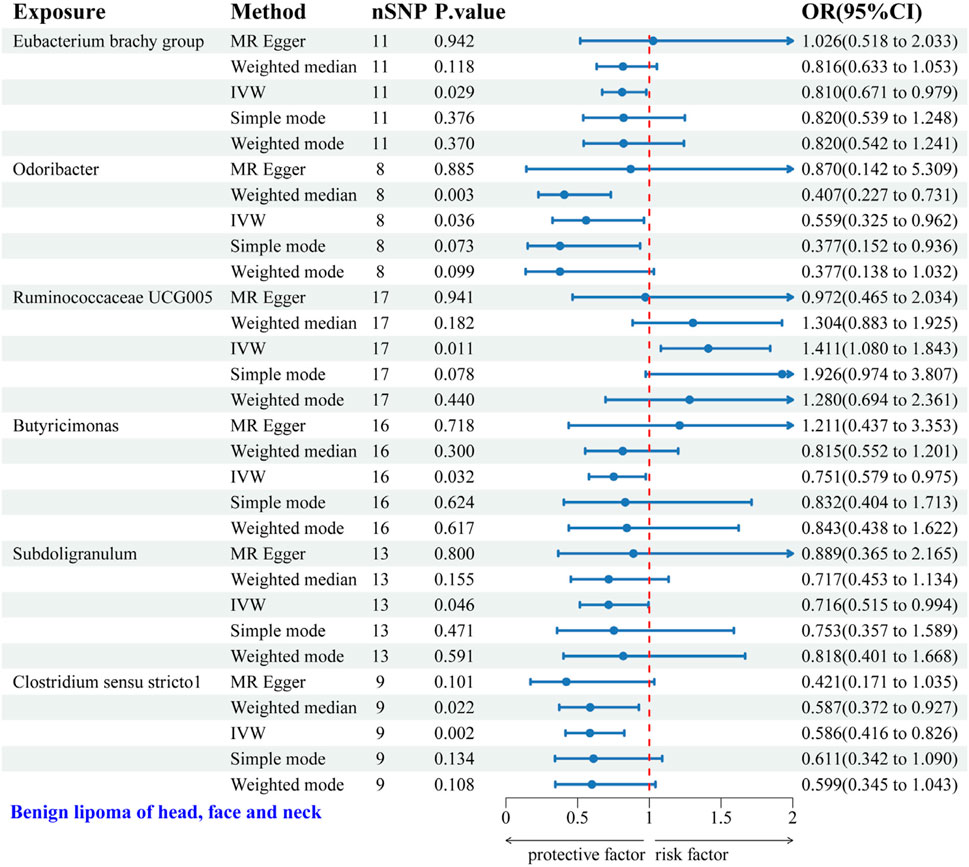
Figure 6. Forest plot of the associations between genetically predicted GM and HFNS-BL risk using IVW methods. IVW, inverse-variance weighted. OR, odds ratio.
Results from Cochran’s Q, MR Egger, and MR-PRESSO analyses indicated no significant heterogeneity or horizontal pleiotropy among the six genera related to HFNS-BL (Table 5). Leave-one-out analysis, funnel plots, and forest plots confirmed the absence of significant outliers, corroborating the reliability of the MR study results (Supplementary Figures 4B, C).
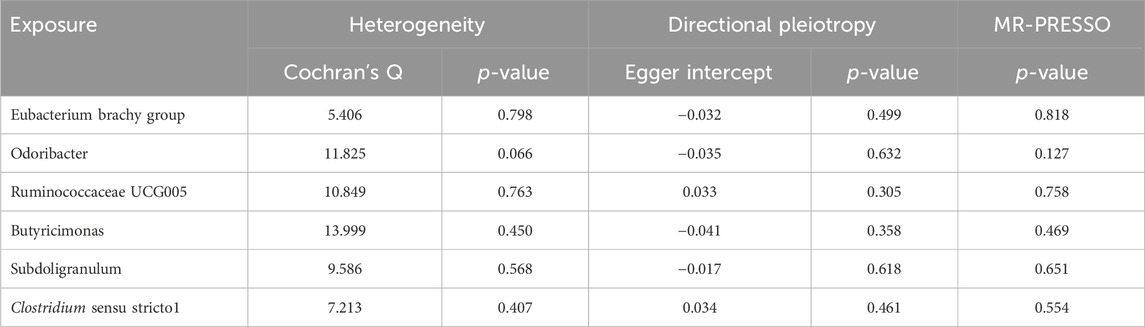
Table 5. Sensitivity analysis of the MR analysis results of the GM and HFNS-BL.
4 DiscussionTo our knowledge, supported by the GWAS database, this is the inaugural study investigating the potential causal relationship between GM and lipomas across various anatomical sites using MR, thus addressing a gap in this field from a novel perspective. This study employed MR to explore potential causal links between GM and lipomas. According to our clues, eighteen types of GM may influence the progression of lipomas, providing new directions for future treatments targeting specific GM in managing lipomas.
The GM, comprising trillions of bacteria within the gastrointestinal tract, plays critical roles, including enhancing intestinal permeability, mediating oxidative stress responses, and participating in tumor immunoregulation (Jiang and Zhang, 2024). Recent studies have established a close link between the composition and diversity of GM and various cancer-related diseases, such as renal tumors (Zhang et al., 2024), aneurysms (Qiu et al., 2024), and neuroblastoma (Zexin et al., 2024). Additionally, the relationship between the GM and the TME has attracted increasing attention. Research (Li et al., 2019) demonstrates that the microbiota affects cancer initiation, progression, and treatment response by regulating the host’s immune system. A review of previous research on GM and cancer-related diseases discusses how GM and their metabolites impact the host immune system and contribute to the development of the TME (Qiu et al., 2020). The TME, crucial for tumor growth, not only controls tumor proliferation but also supports tumor invasion, metastasis, and immune evasion (Khan et al., 2020; Zeng et al., 2016). Within the TME, the GM and its metabolites alter tumor tissues by influencing the intestinal epithelium, either promoting or hindering tumor progression. This highlights the emerging importance of the GM as a critical regulator in oncology (Erdman and Poutahidis, 2015). Moreover, existing studies show that extracellular vesicles (EV) are vital in cell-to-cell communication, especially within the TME. Extracellular vesicle long RNA (exLR) in EV exhibit stability and are rich in diagnostic information. They demonstrate high sensitivity and specificity in the early diagnosis of various cancer types, such as breast cancer (BC) and colorectal cancer (CRC) (Li et al., 2020; Li et al., 2021; Liu C. et al., 2022). exLRs can influence the behavior of tumor-infiltrating lymphocytes through immunoregulatory factors and impact tumor progression by modulating immune responses (Guo et al., 2022; Su et al., 2021).
Our MR analysis indicates that the Eubacterium rectale group, Desulfovibrio, Ruminococcus1, Clostridium sensu stricto1, and Lachnospiraceae UCG001 may provide protective effects against BL, while Lachnospiraceae UCG008 could be a risk factor. Within these microbial communities, the Eubacterium rectale group and Lachnospiraceae UCG001 act as protective factors against LS-BL, whereas Lachnospiraceae UCG008 heightens the risk. Desulfovibrio affords protection against TS-BL, and Clostridium sensu stricto1 guards against HFNS-BL.
The Eubacterium rectale group, an anaerobic Gram-positive bacterium, is prevalent in human fecal samples. While no studies directly associate this group with lipomas, research suggests its involvement in other tumor types. For instance, an animal study (Lu et al., 2022) found that the Eubacterium rectale group may prevent intestinal lymphoma by mitigating chronic inflammation and minimizing the B cell response to gut bacteria. Specifically, it prevents lymphoma by modulating the TNF-induced TLR4/MyD88/NF-κB axis, thereby reducing lymphoma incidence in Eμ-Myc mice. Moreover, treatment with 20 ng/mL Eubacterium rectale Lipopolysaccharide for 2.5 h significantly increased NF-κB expression in the nuclei of HCoEpiC and NCM460 cells (Wang et al., 2021), indicating its potential as a target for preventing both BL and LS-BL due to NF-κB’s role in tumor progression (Wullaert et al., 2011).
The TME plays a crucial role in lipoma formation. Clinical trials (Atzeni et al., 2021) have shown that Lachnospiraceae UCG001 contributes to the production of short-chain fatty acids (SCFAs), while research by Matsumoto et al. (N et al., 2021) demonstrates a strong link between Lachnospiraceae UCG008 and saturated fatty acids. These studies reveal how SCFAs and saturated fatty acids differentially influence the TME, affecting tumor development and progression. SCFAs, such as butyrate, propionate, and acetate, typically exhibit anti-inflammatory, anti-proliferative, epigenetic regulatory, and immunomodulatory effects within the TME, inhibiting tumor cell proliferation and inducing apoptosis (Dong et al., 2023; Carolin et al., 2024; Gomes et al., 2023; Tian et al., 2020). In contrast, saturated fatty acids are generally associated with negative effects in the TME, promoting inflammation, increasing tumor cell proliferation and survival, and suppressing immune functions, thus aiding tumor growth and survival (Westheim et al., 2023).
Overall, SCFAs are considered protective within the TME, while saturated acids may facilitate tumor development. In research examining skin and GM against a melanoma backdrop (Mekadim et al., 2022), Clostridium sensu stricto 1 was significantly linked to the TME, potentially influencing immune responses and cancer dynamics. Desulfovibrio, recognized as a protective factor in BLs, could also significantly affect the TME. Research shows that Desulfovibrio can enhance antitumor immunity within the TME (Li et al., 2024; Kim et al., 2023). Furthermore, studies have noted a correlation between Desulfovibrio concentration and Parkinson’s disease severity. Desulfovibrio produces hydrogen sulfide (H₂S), which affects cell signaling in neuronal cells at low concentrations and causes severe toxicity at higher concentrations (Murros et al., 2021).
In research exploring the causal relationship between GM and cancer (Yiwen et al., 2023), Ruminococcus1 was linked to head and neck cancer, indicating its role in tumor genesis. However, its association with lipomas warrants further experimental research. Conversely, in other conditions like non-alcoholic fatty liver disease (NAFLD), Ruminococcus1 may have exacerbated the progression of NAFLD (Zhang et al., 2023).
This study boasts several strengths. Firstly, it introduces a novel genus-level analysis of GM providing a theoretical foundation for further investigation into specific microbiota mechanisms influencing lipoma development. Secondly, leveraging genetic data from a large population sample enhances the reliability of the findings. Additionally, MR analysis helps reduce the influence of confounding factors.
However, some limitations remain. Due to stringent thresholds, some microbiota were excluded, potentially omitting critical data. The study was limited to a European population, and further research is needed to generalize these findings. Additionally, potential contamination from environmental factors was not controlled. Moreover, numerous variants with small effect sizes increase the risk of false positives. Lastly, comprehensive data on other GM are required to fully assess the causal relationship between GM and lipoma risk.
5 ConclusionIn summary, this study assesses the causal relationship between the GM and lipomas, identifying potentially pathogenic bacterial taxa. These findings hold significant implications for clinical practice, especially in the prevention and management of lipomas. Future studies are necessary to confirm these results through mechanistic studies and to explore potential therapeutic interventions targeting the GM.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statementThe studies involving humans were approved by the Finnish Genome Center Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributionsYL: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Writing–original draft, Writing–review and editing. JC: Conceptualization, Investigation, Methodology, Software, Writing–original draft, Writing–review and editing. HY: Conceptualization, Data curation, Investigation, Methodology, Writing–original draft, Writing–review and editing. XX: Data curation, Methodology, Writing–original draft, Writing–review and editing. XZ: Data curation, Methodology, Writing–original draft, Writing–review and editing. YW: Funding acquisition, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing. WW: Funding acquisition, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Nos 81960874, 82260935 and 82260937), Key Project of Jiangxi Provincial Natural Science Foundation (No. 20202ACB206010), Jiangxi University of Chinese Medicine Science and Technology Innovation Team Development Program (No. CXTD22009), Zhejiang Provincial Natural Science Foundation of China under Grant (No. LBY21H270002) and Provincial Postgraduate Innovation Special Funds of Jiangxi University of Chinese Medicine (YC2023-S771).
Conflict of interestThe authors affirm that there were no commercial or financial relationships involved that could be interpreted as a potential conflict of interest during the conduct of this research.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1430671/full#supplementary-material
ReferencesAmeloot, E., Cordier, F., Van Dorpe, J., and Creytens, D. (2022). Update of pediatric lipomatous lesions: a clinicopathological, immunohistochemical and molecular overview. J. Clin. Med. 11, 1938. doi:10.3390/jcm11071938
PubMed Abstract | CrossRef Full Text | Google Scholar
Atzeni, A., Galié, S., Muralidharan, J., Babio, N., Tinahones, F. J., Vioque, J., et al. (2021). Gut microbiota profile and changes in body weight in elderly subjects with overweight/obesity and metabolic syndrome. Microorganisms 9, 346. doi:10.3390/microorganisms9020346
PubMed Abstract | CrossRef Full Text | Google Scholar
Carolin, T., Patrick, T., Katja, H., Zuqin, Y., Susanne, K., Denis, T., et al. (2024). Short-chain fatty acids induced lung tumor cell death and increased peripheral blood CD4+ T cells in NSCLC and control patients ex vivo. Front. Immunol. 15, 15. doi:10.3389/fimmu.2024.1328263
CrossRef Full Text | Google Scholar
Chen, S., Zhou, G., Han, H., Jin, J., and Li, Z. (2023). Causal effects of specific gut microbiota on bone mineral density: a two-sample Mendelian randomization study. Front. Endocrinol. (Lausanne) 14, 1178831. doi:10.3389/fendo.2023.1178831
PubMed Abstract | CrossRef Full Text | Google Scholar
Dong, Y., Zhang, K., Wei, J., Ding, Y., Wang, X., Hou, H., et al. (2023). Gut microbiota-derived short-chain fatty acids regulate gastrointestinal tumor immunity: a novel therapeutic strategy? Front. Immunol. 14, 1158200. doi:10.3389/fimmu.2023.1158200
PubMed Abstract | CrossRef Full Text | Google Scholar
Gomes, S., Rodrigues, A. C., Pazienza, V., and Preto, A. (2023). Modulation of the tumor microenvironment by microbiota-derived short-chain fatty acids: impact in colorectal cancer therapy. Int. J. Mol. Sci. 24, 5069. doi:10.3390/ijms24065069
PubMed Abstract | CrossRef Full Text | Google Scholar
Guo, T.-A., Lai, H.-Y., Li, C., Li, Y., Li, Y.-C., Jin, Y.-T., et al. (2022). Plasma extracellular vesicle long RNAs have potential as biomarkers in early detection of colorectal cancer. Front. Oncol. 12, 829230. doi:10.3389/fonc.2022.829230
PubMed Abstract | CrossRef Full Text | Google Scholar
Inamura, E., Kitamura, S., Maeda, T., and Yanagi, T. (2021). A rare site of subcutaneous lipoma on the middle finger: case report and analysis of affected sites in 126 cases at a single institution. Dermatol Ther. 34, e14973. doi:10.1111/dth.14973
PubMed Abstract | CrossRef Full Text | Google Scholar
Jiang, H., and Zhang, Q. (2024). Gut microbiota influences the efficiency of immune checkpoint inhibitors by modulating the immune system (Review). Oncol. Lett. 27, 87. doi:10.3892/ol.2024.14221
PubMed Abstract | CrossRef Full Text | Google Scholar
Khan, M. A. W., Ologun, G., Arora, R., McQuade, J. L., and Wargo, J. A. (2020). Gut microbiome modulates response to cancer immunotherapy. Dig. Dis. Sci. 65, 885–896. doi:10.1007/s10620-020-06111-x
PubMed Abstract | CrossRef Full Text | Google Scholar
Kim, J., Kim, Y., La, J., Park, W. H., Kim, H.-J., Park, S. H., et al. (2023). Supplementation with a high-glucose drink stimulates anti-tumor immune responses to glioblastoma via gut microbiota modulation. Cell. Rep. 42, 113220. doi:10.1016/j.celrep.2023.113220
PubMed Abstract | CrossRef Full Text | Google Scholar
Kovács, T., Mikó, E., Ujlaki, G., Sári, Z., and Bai, P. (2020). The microbiome as a component of the tumor microenvironment. Adv. Exp. Med. Biol. 1225, 137–153. doi:10.1007/978-3-030-35727-6_10
PubMed Abstract | CrossRef Full Text | Google Scholar
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi:10.1038/s41588-020-00763-1
PubMed Abstract | CrossRef Full Text | Google Scholar
Kurki, M. I., Karjalainen, J., Palta, P., Sipilä, T. P., Kristiansson, K., Donner, K. M., et al. (2024). FinnGen provides genetic insights from FinnGen provides genetic insights from a well-phenotyped isolated population - PubMed. Available at: https://pubmed.ncbi.nlm.nih.gov/36653562/[Accessed May 9, 2024]
Lee, C. H., Cook, S., Lee, J. S., and Han, B. (2016). Comparison of two meta-analysis methods: inverse-variance-weighted average and weighted sum of Z-scores. Genomics Inf. 14, 173–180. doi:10.5808/GI.2016.14.4.173
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, G., Liu, H., Yu, Y., Wang, Q., Yang, C., Yan, Y., et al. (2024). Desulfovibrio desulfuricans and its derived metabolites confer resistance to FOLFOX through METTL3. EBioMedicine 102, 105041. doi:10.1016/j.ebiom.2024.105041
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y., He, X., Li, Q., Lai, H., Zhang, H., Hu, Z., et al. (2020). EV-origin: enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 18, 2851–2859. doi:10.1016/j.csbj.2020.10.002
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y., Li, Y., Yu, S., Qian, L., Chen, K., Lai, H., et al. (2021). Circulating EVs long RNA-based subtyping and deconvolution enable prediction of immunogenic signatures and clinical outcome for PDAC. Mol. Ther. Nucleic Acids 26, 488–501. doi:10.1016/j.omtn.2021.08.017
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y., Zhao, J., Yu, S., Wang, Z., He, X., Su, Y., et al. (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 65, 798–808. doi:10.1373/clinchem.2018.301291
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, C., Chen, J., Liao, J., Li, Y., Yu, H., Zhao, X., et al. (2022b). Plasma extracellular vesicle long RNA in diagnosis and prediction in small cell lung cancer. Cancers (Basel) 14, 5493. doi:10.3390/cancers14225493
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, Y., Xu, H., Zhao, Z., Dong, Y., Wang, X., and Niu, J. (2022a). No evidence for a causal link between Helicobacter pylori infection and nonalcoholic fatty liver disease: a bidirectional Mendelian randomization study. Front. Microbiol. 13, 1018322. doi:10.3389/fmicb.2022.1018322
PubMed Abstract | CrossRef Full Text | Google Scholar
Lu, H., Xu, X., Fu, D., Gu, Y., Fan, R., Yi, H., et al. (2022). Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell. Host Microbe 30, 1139–1150.e7. doi:10.1016/j.chom.2022.07.003
PubMed Abstract | CrossRef Full Text | Google Scholar
Marteau, É., Le Nail, L.-R., Rosset, P., de Pinieux, G., Laulan, J., Roulet, S., et al. (2020). Epidemiological, clinical and histological features of adipose tumors in the hand and wrist: findings from a continuous series of 37 cases. Orthop. Traumatol. Surg. Res. 106, 329–334. doi:10.1016/j.otsr.2019.12.011
PubMed Abstract | CrossRef Full Text | Google Scholar
Mekadim, C., Skalnikova, H. K., Cizkova, J., Cizkova, V., Palanova, A., Horak, V., et al. (2022). Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol. 22, 63. doi:10.1186/s12866-022-02458-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Ml, Y., and Rh, S. (2014). Intestinal microbiome and lymphoma development. Cancer J. (Sudbury, Mass), 20. doi:10.1097/PPO.0000000000000047
CrossRef Full Text | Google Scholar
Murros, K. E., Huynh, V. A., Takala, T. M., and Saris, P. E. J. (2021). Desulfovibrio bacteria are associated with Parkinson’s disease. Front. Cell. Infect. Microbiol. 11, 652617. doi:10.3389/fcimb.2021.652617
PubMed Abstract | CrossRef Full Text | Google Scholar
Nader, S., Nikakhlagh, S., Rahim, F., and Fatehizade, P. (2012). Endolaryngeal lipoma: case report and literature review. Ear Nose Throat J. 91, E18–E21. doi:10.1177/014556131209100218
PubMed Abstract | CrossRef Full Text | Google Scholar
N, M., J, P., R, T., H, K., K, H., K, M., et al. (2021). Relationship between nutrient intake and human gut microbiota in monozygotic twins. Med. Kaunas. Lith., 57. doi:10.3390/medicina57030275
留言 (0)