Juvenile neuronal ceroid lipofuscinosis (JNCL), also known as Batten disease, is a neurodegenerative disorder that affects young individuals (Lerner et al., 1995). Afflicted children display progressive vision loss without any precursor symptoms. Within a span of 2–4 years, patients experience personality changes, behavioral problems, and cognitive decline, followed by the loss of motor functions, ultimately leading to premature death, often before the age of 25 (Nita et al., 2016). The NCL diseases are characterized by the accumulation of ceroid lipofuscin within lysosomes. One suggested consequence of this accumulation is the disruption of vital cellular processes, resulting in cell death and subsequent neurodegeneration (Cooper, 2003). However, the precise mechanisms underlying Batten disease at the cellular level, particularly in stem cells, remain largely unknown.
Researches have identified mutated genes relevant to Batten disease named ceroid-lipofuscinosis, neuronal (CLN) 1-14 (Nita et al., 2016). Among them, CLN3 is responsible for JNCL and is the most common type of NCLs. The gene CLN3 has been found to be highly conserved across yeast, fruit fly, mouse and human. The coding product, CLN3 protein, is highly hydrophobic, with six transmembrane helices and is predicted to be constitutively expressed (Mirza et al., 2019). Studies have confirmed its subcellular localization within the membrane of lysosomes or endosomes (Kyttala et al., 2004). Numerous investigations have suggested that CLN3 is involved in various cellular functions, including endocytosis (Fossale et al., 2004; Metcalf et al., 2008), lysosomal homeostasis (Chattopadhyay et al., 2000; Gachet et al., 2005), mitochondrial function (Fossale et al., 2004; Luiro et al., 2006) and autophagy (Cao et al., 2006). Our previous research has demonstrated the requirement of CLN3 in the response to oxidative stress (Tuxworth et al., 2011). However, the functions of CLN3 in adult stem cells still remain unexplored.
Adult stem cells play a crucial role in maintaining tissue and organ homeostasis by continuously replacing functional cells (Post and Clevers, 2019). When tissues experience injury or loss of functional cells, there is a requirement for adult stem cells to rapidly proliferate and differentiate in order to replenish the damaged areas. The adult Drosophila digestive tract has been established as a useful system for the study of adult stem cell proliferation due to the advantages of simple cellular component and feasible genetic manipulation (Nászai et al., 2015). Similar to the mammalian intestine, the Drosophila midgut epithelium undergoes constant replenishment through intestinal stem cells (ISCs) (Ohlstein and Spradling, 2005; Micchelli and Perrimon, 2005). Upon activation, a quiescent ISC can undergo symmetric division to expand the population of stem cells, or asymmetric division to generate a new ISC and a committed progenitor cell called an enteroblast (EB), which subsequently differentiates into a mature polyploid enterocyte (EC) (Ohlstein and Spradling, 2007). On rare occasions, an ISC can undergo asymmetric division and produce a distinct type of progenitor cell known as an enteroendocrine mother cell (EMC). The EMC then goes through symmetric division, resulting in the formation of a pair of enteroendocrine (EE) cells that contribute to the maintenance of homeostasis (Guo and Ohlstein, 2015). To date, no research on the role of CLN3 in ISCs has been published.
In this study, we aimed to investigate the impact of Cln3 loss in Drosophila intestinal stem cells. Under conditions of homeostasis, the loss of Cln3 does not affect ISCs. However, when the midgut is exposed to stress, Cln3 loss results in the failure of ISC activation. This phenotype is cell-autonomous and caused by the lack of activating of the JAK/STAT pathway in ISCs. These results shed light on a novel role of Cln3 in adult stem cells and provide a potential mechanism underlying Batten disease.
2 Results2.1 Cln3 does not affect midgut homeostasis under normal condition in DrosophilaTo explore the function of Cln3 in Drosophila midgut, we previously generated a Cln3 mutant line named Cln3ΔMB1 by imprecise excision of a transposable element (Tuxworth et al., 2011). This allele exhibites a deletion of 1,534 bp and can be viewed as a null (Tuxworth et al., 2011). This homozygote mutant survived to adulthood and showed no phenotype in the midgut epithelium (a model of cell lineage was shown in Figure 1A) from the following aspects: (Lerner et al., 1995): escargot+ (esg+, a promotor activating in stem and progenitor cells) cells (Figures 1B–D); (Nita et al., 2016) Delta+ (Dl+, the ligand of Notch signaling) intestinal stem cells (ISCs) (Figures 1B, E); (Cooper, 2003) Prospero+ (Pros) enteroendocrine cells (EEs) (Figures 1C, F); (Mirza et al., 2019) polyploid enterocytes (ECs) (Figures 1B, C, G). Also, phosphorylated histone H3 (pH3, marks mitotic cells) staining revealed no difference in ISC proliferation between Cln3ΔMB1 mutant and wild type flies (Figure 1H). These data suggested that Cln3 is not a necessary gene under homeostasis condition in the Drosophila midgut.
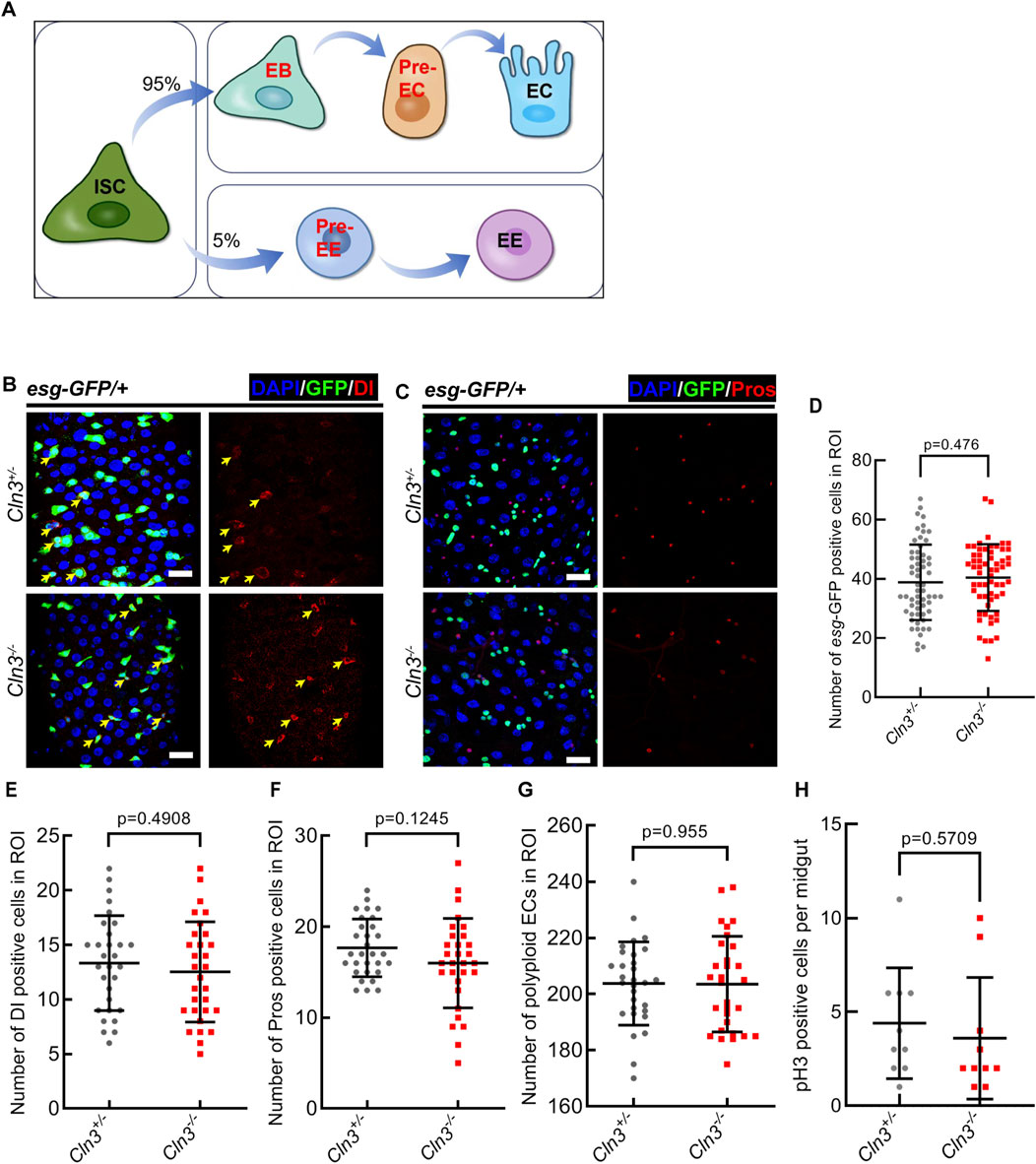
Figure 1. Cln3 is not necessary in Drosophila midgut homeostatsis maintainence. (A) A model of cell lineage in Drosophila midgut epithelium. (B) Representative immunofluorescence of midguts with esg-GFP (green) and Dl (red) staining from control (Cln3+/−) flies and Cln3 mutant (Cln3−/−) flies. Yellow arrows indicate Dl+ ISCs. (C) Representative immunofluorescence of midguts with esg-GFP (green) and Pros (red) staining from control (Cln3+/−) flies and Cln3 mutant (Cln3−/−) flies. (D–G) Quantification of the number of esg+ cells (D), Dl+ cells (E), Pros+ cells (F) and polyploid ECs (G) per region of interest (ROI) of midguts in experiments (B, C). ROI size 84,100 μm2. (H) Quantification of the number of pH3+ cells per midgut in experiments (B, C). DAPI-stained nuclei (blue). Scale bar, 25 μm. Bars are mean ± SD. Statistics were measured by two-tailed, unpaired Student’s t-test. n = 60 and 60 ROIs from 30 midguts each in (D), n = 30 and 30 ROIs from 15 midguts each in (E–G), n = 10 and 10 midguts in (H), respectively. Each experiment was repeated for 3 times.
2.2 Cln3 is required for the activation of ISCs upon tissue damage in DrosophilaSince we previously demonstrated that Cln3ΔMB1 mutant flies showed no phenotype in normal condition but were hypersensitive to oxidative stress (Tuxworth et al., 2011), we asked whether Cln3 also functions only under stress condition in the midgut. To test this, we fed Drosophila with bleomycin (Figure 2A) and Erwinia carototovovora carototovovora 15 (Ecc15) (Supplementary Figure S1), which induced DNA break of ECs and infection of the midgut, respectively, and subsequent activation of ISCs to replenish the damage of epithelium (Guo et al., 2013). We first tested the RNA expression level of Cln3 in the midgut and found it upregulated after bleomycin and Ecc15 treatment (Figure 2B; Supplementary Figure S1A). This suggested that Cln3 may play a role in the regeneration process. Then, we checked the changes of ISC behavior. As a result, while the percentage of esg+ and Dl+ cells increased in wild type flies after bleomycin and Ecc15 treatment, Cln3ΔMB1 mutant failed to exhibit this increase (Figures 2C–E; Supplementary Figures S1B–D). Moreover, pH3 staining revealed fewer proliferative stem cells in the Cln3ΔMB1 mutant (Figure 2F; Supplementary Figure S1E). These results indicated that Cln3 plays a role in the proliferation of ISCs during regeneration. Since both of bleomycin and Ecc15 treatment revealed similar results, we used only bleomycin for further investigation.
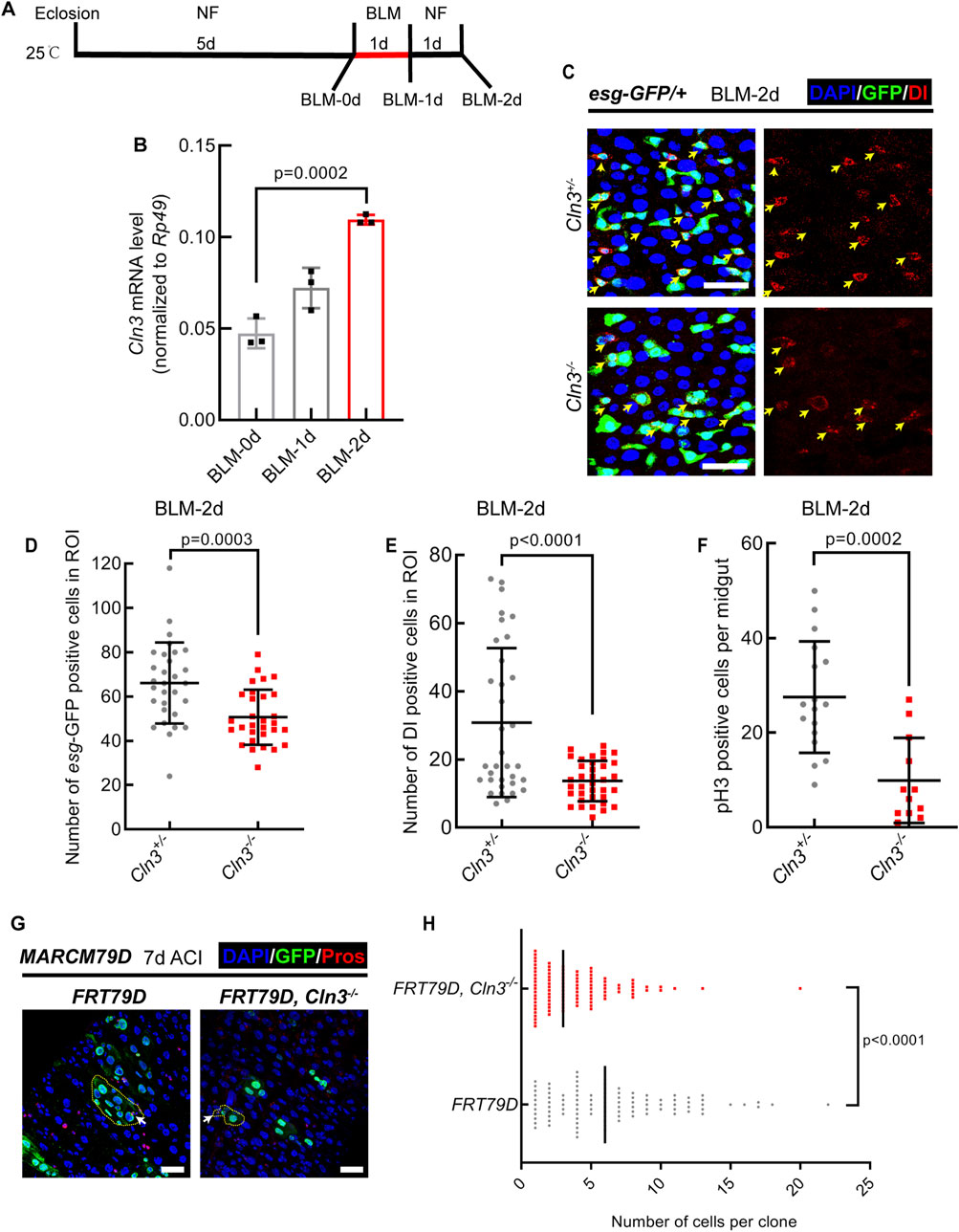
Figure 2. Cln3 activates ISC proliferation during regeneration. (A) Diagram of the experimental procedure of fly treatment. (B) The relative mRNA expression level of Cln3 from whole midguts of wild type flies (w1118) at indicated time points. (C) Representative immunofluorescence of midguts with esg-GFP (green) and Dl (red) staining from control (Cln3+/−) flies and Cln3 mutant (Cln3−/−) flies at the time point of BLM-2d. Yellow arrows indicate Dl+ ISCs. (D, E) Quantification of the number of esg+ cells (D) and Dl+ cells (E) per ROI of midguts in experiment (C). ROI size 84,100 μm2. (F) Quantification of the number of pH3+ cells per midgut in experiment (C). (G) Immunofluorescence analysis of clones induced by MARCM79D of control (FRT79D) and Cln3 mutant (FRT79D, Cln3−/−). Pros (red) staining was used to indicate EEs. Yellow dotted lines indicated the border of clones. White arrows indicated Pros+ EEs inside the clones. (H) Quantification of the number of cells per clone in experiment (G). DAPI-stained nuclei (blue). Scale bar, 25 μm. Bars are mean ± SD. Statistics were measured by one-way ANOVA in (B) and two-tailed, unpaired Student’s t-test in (D, E) and (H). n = 30 and 30 ROIs from 15 midguts each in (D), n = 34 and 36 ROIs from 17 to 18 midguts in (E), n = 17 and 12 midguts in (F), n = 100 and 100 clones from 20 midguts each in (H), respectively. Each experiment was repeated for 3 times.
To further support this, we used mosaic analysis with a repressible cell marker (MARCM) system (Lee and Luo, 1999), in which only active ISCs are able to generate a clone. Results revealed that compared to wild type clones, Cln3ΔMB1 mutant clones were smaller (Figures 2G, H), suggesting weaker mitotic ability of mutated ISCs. Noticed that both wild type clones and mutant clones contained Pros+ EEs (Figure 2G), which further supported the conclusion that Cln3 did not affect ISC differentiation.
2.3 ISCs are responsible for their activation during regeneration upon Cln3 lossNext, we sought to figure out which cell type is responsible for this failure of ISC activation. We first tested the efficiency of two Cln3 RNAi lines. Results revealed that both of these two RNAi lines driven by whole body expressed tub-gal4 exhibited over 60% of Cln3 mRNA depletion (Figure 3A), suggesting high RNAi efficiency. Then, we used different kinds of cell type specific-gal4 to drive Cln3 RNAi (stem and progenitor cell-specific esg-gal4; ISC-specific esg-gal4, NRE-gal80, tub-gal80ts (shortened as ISCts-gal4); EE-specific 386Y-gal4; EC-specific Mex-gal4 and muscle-specific How-gal4). These results combinely suggested that the failure of ISC activation during regeneration was cell-autonomous, because only esg-gal4 and ISCts-gal4 driven Cln3 RNAi led to the failure of the increase of esg+ cells, Dl+ cells and pH3+ cells upon bleomycin treatment (Figures 3B–F).
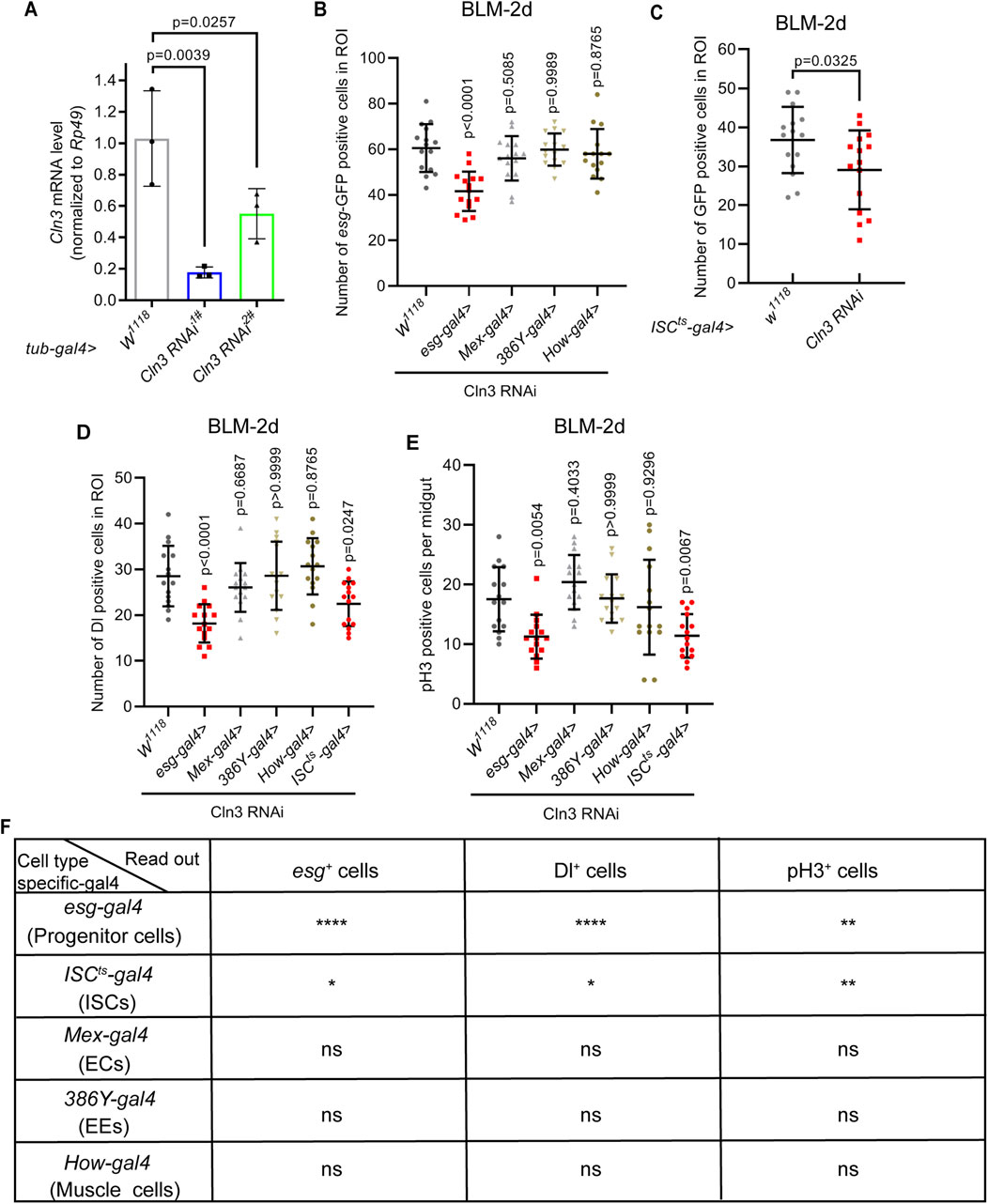
Figure 3. Gal4 screening reveals ISCs responsible for their activation upon Cln3 loss during regenaration. (A) The relative mRNA expression level of Cln3 from whole flies of control (tub-gal4, w1118) and Cln3 depletion (tub-gal4>Cln3 RNAi1#, tub-gal4>Cln3 RNAi2#). Each experiment was repeated for 3 times. (B–D) Quantification of the number of esg-GFP+ cells (B), ISCts-gal4>GFP+ cells (indicate esg+ cells without NRE expression) (C) and Dl+ cells (D) per ROI of midguts with indicated genotypes. ROI size 84,100 μm2. (E) Quantification of the number of pH3+ cells per midgut with indicated genotypes. (F) Summary of the results of gal4 screening. Only esg-gal4 and ISCts-gal4 driven Cln3 RNAi led to significant differences. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, not significant. Statistics were measured by one-way ANOVA in (A, B) and (D, E), and two-tailed, unpaired Student’s t-test in (C). n = 15 ROIs from 8 midguts each in all groups of (B–D), and n = 15 midguts in all groups of (E). Each experiment was repeated for 3 times.
2.4 Cln3 activates ISC proliferation during regeneration through JAK/STAT signaling pathwayThen, we asked the mechanism underlying how Cln3 regulates ISC proliferation during regeneration. JAK/STAT signaling is well reported to regulate ISC proliferation (Jiang et al., 2009; Zhai et al., 2017; Osman et al., 2012). During regeneration, ECs that undergo apoptosis secreate the ligand of JAK/STAT signaling, Unpaired 3, which binds to the receptor Domeless expressed through the plasma membrane of ISCs, to activate ISC proliferation (Osman et al., 2012). To test whether Cln3 depletion has an impact on JAK/STAT signaling in ISCs, 10 × STAT-GFP reporter line was used (Bach et al., 2007). During regeneration, while the control group showed more cells with 10 × STAT-GFP positive signal, stronger 10 × STAT-GFP signal per cell, and higher GFP protein level, these changes were not significant in the Cln3 depletion group (Figures 4A–D). Moreover, we tested the mRNA level of two STAT-target genes, Socs36E and Dpp, which revealed similar results (Supplementary Figure S2A). These data suggested that JAK-STAT signaling was not able to switch on with Cln3 depletion.
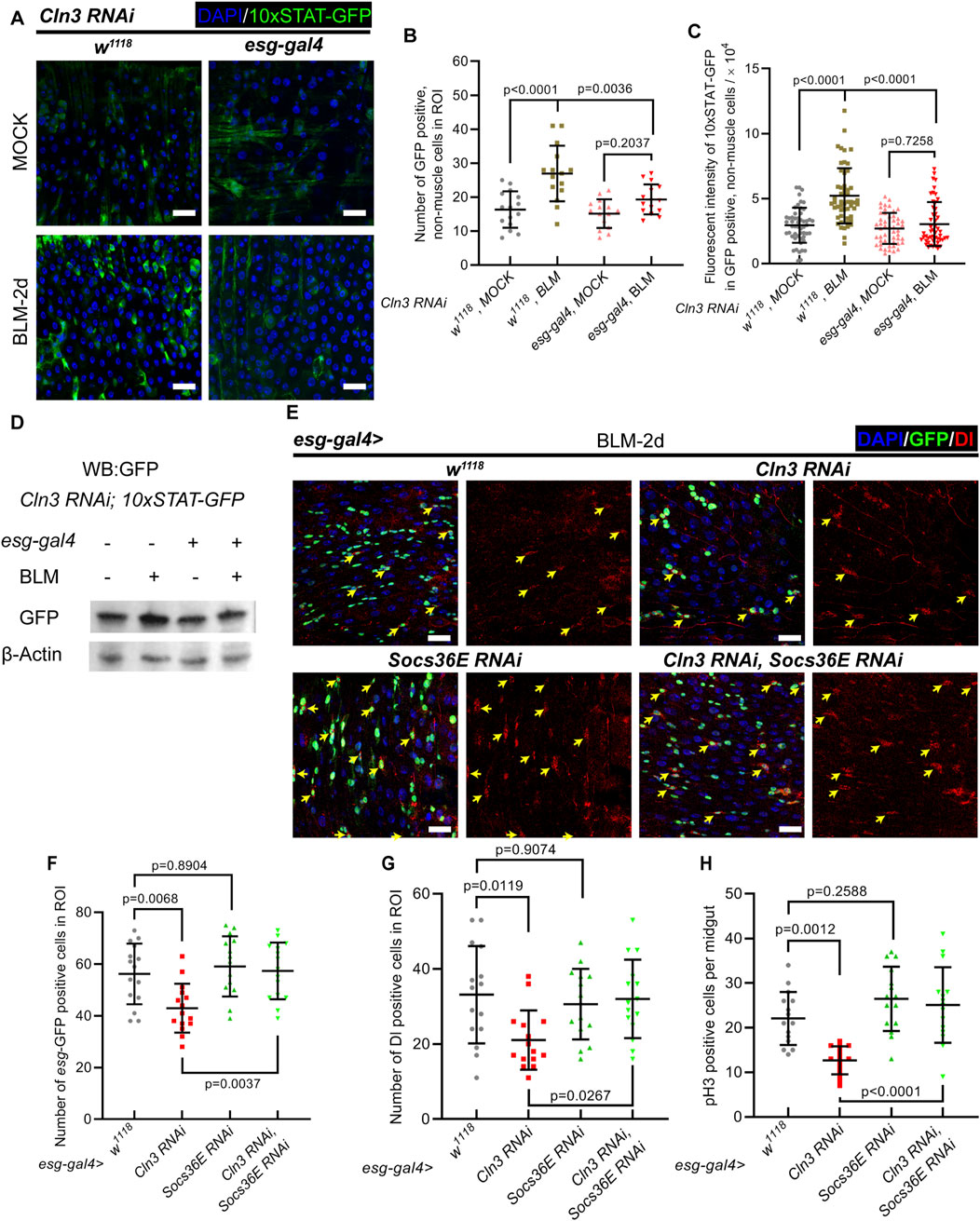
Figure 4. JAK/STAT signaling functions downstream of Cln3 to activate ISCs during regeneration. (A) Representative immunofluorescence images of midguts from control (w1118, Cln3 RNAi, 10 × STAT-GFP) and Cln3 depletion (esg-gal4>Cln3 RNAi, 10 × STAT-GFP) flies with MOCK (sucrose) or BLM treatment. 10 × STAT-GFP staining indicated the activity of JAK/STAT signaling pathway. (B, C) Quantification of the number of 10 × STAT-GFP+, non-muscle cells per ROI (B) and fluorescent intensity of 10 × STAT-GFP in 10 × STAT-GFP+, non-muscle cells (C). (D) Western blot analysis of 10 × STAT-GFP protein level from whole midgut in experiment (A). (E) Representative immunofluorescence of midguts with esg-GFP (green) and Dl (red) staining from control (esg-gal4, w1118), or flies expressing UAS-Cln3 RNAi or UAS-Socs36E RNAi single depletion, and UAS-Cln3 RNAi, UAS-Socs36E RNAi double depletion driven by esg-gal4. Yellow arrows indicate Dl+ ISCs. (F, G) Quantification of the number of esg+ cells (F) and Dl+ cells (G) per ROI of midguts in experiment (H). ROI size 84,100 μm2. (H) Quantification of the number of pH3+ cells per midgut in experiment (E). DAPI-stained nuclei (blue). Scale bar, 25 μm. Bars are mean ± SD. Statistics were measured by one-way ANOVA. n = 15 ROIs from 8 midguts each in all groups of (B) and (F, G); n = 53, 52, 50 and 50 cells from 8 midguts each in (C), respectively and n = 15 midguts in all groups of (H). Each experiment was repeated for 3 times.
To further verify the function of JAK/STAT signaling upon Cln3 depletion, we tested the genetic relationship between JAK/STAT signaling and Cln3. Apart from the function of reporting JAK/STAT signaling, Socs36E is also a vital negative regulator which mediates degradation of positive regulator STAT92E (Issigonis et al., 2009). We drove Socs36E RNAi to upregulate JAK/STAT signaling. During regeneration, while wild type flies exhibited increased esg+ cells, Dl+ cells and pH3+ cells, these numbers reached a lower level in Cln3 depletion group (Figures 4E–H). In the Cln3 and Socs36E double depletion group, the esg+ cell number, Dl+ cell number and pH3+ cell number showed the same level to Socs36E depletion group (Figures 4E–H). These data indicated that JAK/STAT signaling pathway functioned downstream of Cln3 in response to stress.
3 DiscussionJNCL is an uncurable inherited metabolic disorder caused by CLN3 mutation. It is meaningful to uncover more details about the disease. To date, the function of CLN3 is not clearly demonstrated (Mirza et al., 2019). Particularly, the mechanisms underlying how CLN3 loss develops the disease remains largely unknown. Using the Drosophila midgut system as a model, here we demonstrate that Drosophila Cln3 is required for the activation of ISCs during regeneration. Upon damage of midgut, ISCs of Cln3 mutant fail to undergo fast proliferation to replenish the loss of functional cells. Genetic analysis reveals that this failure is ISC-autonomous through JAK/STAT signaling pathway (Supplementary Figure S3).
We have previously reported that in Drosophila, Cln3 mutant exhibits hypersensitive to oxidative stress (Tuxworth et al., 2011). In this study, we uncover that Cln3 mutant fails to activate ISCs upon tissue damage. These two findings suggest that in Drosophila, Cln3 is not necessary for homeostasis maintainance, but becomes vital under stress condition. To validate this, further investigations should focus on the survival performance and stem cell behavior of Cln3 mutant under various stress conditions, though we have proved some (Tuxworth et al., 2011). In particular, whether adult stem cells are able to mediate regeneration promptly, is urgent to be demonstrated. If this applies to more types of stem cells and more kinds of stresses, it will safely comes to the conclusion that Cln3 is required for adult stem cell activation during regeneration. This will further shed light on the mechanisms underlying how CLN3 mutation develops JNCL in human. It is possible that CLN3 mutation limits adult stem cells to produce new progenies after cell death caused by accumulation of ceroid lipofuscin, which subsequently results in more severe organ degeneration.
We have previously demonstrated the behavior of ISCs during regeneration in the Drosophila midgut (Du et al., 2020). In wild type flies, the proliferation rate of ISCs first increases sharply, and gradually slows down to the level before exposed to stress. The time point BLM-2d in this study shows the highest ISC proliferation rate in the regeneration model (Du et al., 2020), which sidely supports the role of Cln3 in the Drosophila midgut. Failure of the activation of ISCs to proliferate results in the longer existence of injury, as well as a shortened lifespan. This may also contribute, at least in part, to the development of Batten disease.
Specifically, our previous work has revealed that RAB7-mediated endocytosis functions upstream of JAK/STAT signaling pathway to regulate ISC regeneration (Du et al., 2020). Combined with the hint that CLN3 was reported to be involved in endocytosis (Fossale et al., 2004; Metcalf et al., 2008), we tried to find the direct relationship between Cln3 and RAB7 by generating a Cln3-HA-knock in line. However, while DNA sequence revealed by PCR showed correct knock in, the HA reporter cannot be detected even through western blot analysis. This may suggest complicated epigenetic regulation of the Cln3 locus. Therefore, the direct mechanisms underlying how CLN3 regulates JAK/STAT signaling and subsequent phenotype of Batten disease should be investigated by other methods or in other organisms.
The role of JAK/STAT signaling in the development of Batten disease still remains largly unknown. Based on the reported functions of JAK/STAT signaling pathway, 4 possible mechanisms could be put forward: (Lerner et al., 1995): Inflammation and immune response. Batten disease is associated with chronic inflammation in the brain. The JAK/STAT pathway plays a crucial role in regulating immune responses and inflammatory processes. Dysregulation of this pathway may contribute to the immune dysfunction observed in Batten disease, leading to increased inflammation and subsequent neuronal damage (Nita et al., 2016). Neuroprotection and cell survival. Activation of the JAK/STAT pathway has been shown to promote cell survival and protect against apoptosis. In Batten disease, the progressive loss of neurons contributes to the symptoms. Dysfunctional JAK/STAT signaling could impair neuroprotection mechanisms, making neurons more susceptible to degeneration (Cooper, 2003). Neuronal development and maturation. The JAK/STAT pathway is involved in neurodevelopmental processes such as neuronal proliferation, differentiation, and migration. A disruption in the JAK/STAT pathway in Batten disease may result in impaired neuronal development and maturation, contributing to the progressive neurodegeneration observed in affected individuals (Mirza et al., 2019). Oxidative stress and antioxidant defense. Batten disease is associated with increased oxidative stress, which can lead to cellular damage. The JAK/STAT pathway has been implicated in regulating antioxidant defense mechanisms, which help counteract oxidative stress. Dysregulation of this pathway may compromise the antioxidant defense system, further aggravating oxidative damage in Batten disease.
Nevertheless, limitations about studying Batten disease using Drosophila model are obvious. The most important is that the Cln3 mutant Drosophila does not develop any JNCL-like phenotypes, nor accumulates intracellular autofluorescent materials (Tuxworth et al., 2011). Although CLN3 is structurally conservd from yeast to human, its functions, particularly in material transport through lysosome, may have changed during evolution. Therefore, more details about the functions of CLN3 relevant to lysosome should be investigated in other organisms.
4 Methods4.1 Drosophila stocksThe fly stocks used in this study are listed as follows: wild type line: w1118;; (BDSC: 3605); Cln3 mutant line: ;;Cln3ΔMB1 (Our lab); esg-GFP reporter line: ;esg-GFP/CyO; (From Allan Spradling); 10 × STAT-GFP reporter line: ;10 × STAT-GFP; (From Zheng Guo); tub-gal4 line: ;;tub-gal4/TM3.e.Sb (From Allan Spradling); esg-gal4 line: ;esg-gal4/CyO; (From Allan Spradling); ISCts-gal4 line: Su(H)-lacZ;esg-gal4.UAS-GFP/Kr-GFP.CyO;tub-gal80ts.GBE-gal80/MKRS (From Zheng Guo); Mex-gal4 line: ;Mex-gal4; (From Zheng Guo); 386Y-gal4 line: w[*];; P386Y (BDSC: 25410); How-gal4 line: w[*];; Phow[24B] (BDSC: 1767); Cln3 RNAi 1# line: TH201500324.S; Cln3 RNAi 2# line: THU1691; Socs36E RNAi line: THU1666; UAS-GFP reporter line: w[1118];; P8 (BDSC: 4776). FRT79D line: ;;FRT79D (From Zheng Guo); MARCM79D tool line: yw.hsflp.tub-gal4::GFP/FM7;;FRT79D, tub-gal80/TM6B (From Zheng Guo).
4.2 Drosophila husbandryAll flies were cultured at 25°C at a 12 h light/dark cycle. For the Gal4/Gal80ts system, the offspring were shifted to 29°C 5 days after eclosion for removing Gal80 inhibition and enabling Gal4 to drive UAS-linked transgene expression for another 5 days. For MARCM system, flies were raised at 25°C until 5 days after eclosion. Then the flies were transferred to a new vial and incubated in 37°C water bath for 1-h-heat shock. Flies were transferred back to 25°C for another 7 days for clone growth followed by dissection.
4.3 Bleomycin and Ecc15 treatmentDrosophila were transferred from the medium to empty bottles for 2 h. The filter paper was cut into 3.5 × 6.0 cm pieces and treated with 5% (wt/vol) sucrose with 25 μg/mL bleomycin (BLM). Then, the moist papers were added to empty bottles for 24 h. BLM-1d group flies were dissected at this point, and the rest flies were transferred into a new standard medium without BLM for another 24 h before dissection. For Ecc15 infection, an infection solution was prepared by mixing an equal volume of 100× concentrated pellet from an overnight culture of Ecc15 (OD600 = 200) with a solution of 5% sucrose. One piece of filter paper treated with this infection solution was placed into a bottle with standard food. After treatment with empty bottles for 2 h, Drosophila were transferred to the bottles with standard food and filter paper for 24 h followed by dissection. Filter paper treated with 5% sucrose were used as control.
4.4 Immunofluorescence microscopy for Drosophila tissuesAdult fly intestines were dissected at 4°C in PBS and fixed with a mixture of 100 µL 4% EM-grade paraformaldehyde fixation buffer and 100 µL n-Heptane for 20 min. The intestines were washed in 200 µL methanol for 2 times, 5 min each. They were subsequently rinsed in 200 µL PBS plus 0.1% Triton X-100 (Sigma, PBST) for 2 times, 5 min each. The tissues were then incubated with primary antibodies dissolved in 0.1% PBST for overnight at 4°C. Then they were washed with 0.1% PBST for 3 times, 5 min each before secondary antibodies and DAPI (1:1,000; Sigma) incubated for 2 h at room temperature. After secondary antibodies incubation, the intestines were washed with 0.1% PBST for 3 times, 5 min each. The sources and dilutions of antibodies used are listed as follows: Chicken anti-GFP (1:1000; ab13970, Abcam), Rabbit anti-GFP (1:1000; 50430-2-AP, Proteintech), Prospero antibody (1:200; MR1A, DSHB), Delta antibody, (1:50; C594,9B, DSHB), and Alexa Fluor secondary antibodies (1:2000; A11004, A11008, A11011, A11039, A32733 and A32933). Leica TCS-SP8 confocal microscope was used to acquire all immunofluorescence images. For each set of experiments, images were acquired as confocal stacks using the same settings.
4.5 RNA isolation and RT-qPCR for Drosophila50 adult midguts or 10 whole flies were collected into 4°C diethylpyrocarbonate (DEPC)-treated water-PBS solution. Samples were homogenized in RNA-easy Isolation Reagent (Vazyme, R701) for total RNA isolation and cDNA synthesis. RT-qPCR was performed on a CFX96 Touch Deep Well (Bio-Rad) using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711). The reference standard group was Rp49. The expression levels were counted by the 2−△△CT method. The primers used were listed as below:
Rp49-F: GCCCAAGGGTATCGACAACA.
Rp49-R: GCGCTTGTTCGATCCGTAAC.
CLN3-F: TCGTCGGGAAGAAACTGTCAC.
CLN3-R: GAGCTATACGGAAATTCACCCAA.
Socs36E-F: GCTGCCAGTCAGCAATATGT.
Socs36E-R: GACTGCGGCAGCAACTGT.
Dpp-F: TGGCGACTTTTCAAACGATTGT.
Dpp-R: CAGCGGAATATGAGCGGCAA.
4.6 Fluorescence intensity statisticsImmunofluorescence imaging results were analyzed based on z-stacks acquired with confocal microscopy. The fluorescence intensity of the region of interest (ROI) or cell was calculated using ImageJ software following the fomular:
Integrated Density=Integrated Density of ROI or cell—Integrated Density of background region/Area of background region×Area of ROI or cell.4.7 Western blotting50 midguts were collected for protein extraction. The primary antibodies used for western blotting in this study were: anti-GFP (rabbit, 1:5000, 50430-2-AP, Proteintech); and anti-β-actin (rabbit, 1:2000, PA5-85271, Invitrogen). The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit (1:5000, Jackson ImmunoResearch Labs, Cat# 211-032-171).
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributionsZY: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft, Writing–review and editing. JY: Investigation, Methodology, Writing–original draft. ZL: Investigation, Methodology, Writing–review and editing. HW: Writing–original draft, Writing–review and editing. GL: Supervision, Writing–review and editing. HC: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Basic Research Program of China (2020YFA0803602), the National Natural Science Foundation of China (92157109) (HC), the National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Z20201006) (HC), and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYYC20024) (HC). The funders had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation.
AcknowledgmentsWe thank BDSC and Tsinghua Fly Center for fly strains, and DSHB for antibodies.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1508714/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Cln3 activates ISC proliferation upon infection. (A) The relative mRNA expression level of Cln3 from whole midguts of wild type flies (w1118) with or without Ecc15 treatment. (B) Representative immunofluorescence of midguts with esg-GFP (green) and Dl (red) staining from control (Cln3+/-) flies and Cln3 mutant (Cln3-/-) flies with or without Ecc15 treatment. Yellow arrows indicate Dl+ ISCs. (C, D) Quantification of the number of esg+ cells (C) and Dl+ cells (D) per ROI of midguts in experiment (B). ROI size 84,100 μm2. (E) Quantification of the number of pH3+ cells per midgut in experiment (B). DAPI-stained nuclei (blue). Scale bar, 25 μm. Bars are mean ± SD. Statistics were measured by two-tailed, unpaired Student’s t-test. n = 30 and 30 ROIs from 15 midguts each in (C, D) and n = 15 and 15 midguts in (E), respectively. Each experiment was repeated for 3 times.
SUPPLEMENTARY FIGURE S2 | The expression of STAT-target genes reveals regulation by Cln3. (A) The relative mRNA expression level of Socs36E and Dpp from whole midguts of flies with indicated genotype with or without BLM treatment. The expression level was normalized to rp49. The relative expression level of the MOCK groups was further normalized to 1 and marked as the dotted line. Bars are mean ± SD. Statistics were measured by one-way ANOVA. Each experiment was repeated for 3 times.
SUPPLEMENTARY FIGURE S3 | Proposed model. Left: Cln3 loss does not affect homeostasis. Right: During regeneration, Cln3 loss represses the activation of ISCs through JAK/STAT signaling pathway, which may contribute to Battern disease.
ReferencesBach, E. A., Ekas, L. A., Ayala-Camargo, A., Flaherty, M. S., Lee, H., Perrimon, N., et al. (2007). GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7 (3), 323–331. doi:10.1016/j.modgep.2006.08.003
PubMed Abstract | CrossRef Full Text | Google Scholar
Cao, Y., Espinola, J. A., Fossale, E., Massey, A. C., Cuervo, A. M., MacDonald, M. E., et al. (2006). Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem. 281 (29), 20483–20493. doi:10.1074/jbc.M602180200
PubMed Abstract | CrossRef Full Text | Google Scholar
Chattopadhyay, S., Muzaffar, N. E., Sherman, F., and Pearce, D. A. (2000). The yeast model for batten disease: mutations in btn1, btn2, and hsp30 alter pH homeostasis. J. Bacteriol. 182 (22), 6418–6423. doi:10.1128/JB.182.22.6418-6423.2000
PubMed Abstract | CrossRef Full Text | Google Scholar
Cooper, J. D. (2003). Progress towards understanding the neurobiology of Batten disease or neuronal ceroid lipofuscinosis. Curr. Opin. Neurol. 16 (2), 121–128. doi:10.1097/01.wco.0000063762.15877.9b
PubMed Abstract | CrossRef Full Text | Google Scholar
Du, G., Xiong, L., Li, X., Zhuo, Z., Zhuang, X., Yu, Z., et al. (2020). Peroxisome elevation induces stem cell differentiation and intestinal epithelial repair. Dev. Cell. 53 (2), 169–184.e11. doi:10.1016/j.devcel.2020.03.002
PubMed Abstract | CrossRef Full Text | Google Scholar
Fossale, E., Wolf, P., Espinola, J. A., Lubicz-Nawrocka, T., Teed, A. M., Gao, H., et al. (2004). Membrane trafficking and mitochondrial abnormalities precede subunit c deposition in a cerebellar cell model of juvenile neuronal ceroid lipofuscinosis. BMC Neurosci. 5, 57. doi:10.1186/1471-2202-5-57
PubMed Abstract | CrossRef Full Text | Google Scholar
Gachet, Y., Codlin, S., Hyams, J. S., and Mole, S. E. (2005). btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell. Sci. 118 (Pt 23), 5525–5536. doi:10.1242/jcs.02656
PubMed Abstract | CrossRef Full Text | Google Scholar
Guo, Z., Driver, I., and Ohlstein, B. (2013). Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell. Biol. 201 (6), 945–961. doi:10.1083/jcb.201302049
PubMed Abstract | CrossRef Full Text | Google Scholar
Issigonis, M., Tulina, N., de Cuevas, M., Brawley, C., Sandler, L., and Matunis, E. (2009). JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 326 (5949), 153–156. doi:10.1126/science.1176817
PubMed Abstract | CrossRef Full Text | Google Scholar
Jiang, H., Patel, P. H., Kohlmaier, A., Grenley, M. O., McEwen, D. G., and Edgar, B. A. (2009). Cytokine/jak/stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 137 (7), 1343–1355. doi:10.1016/j.cell.2009.05.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Kyttala, A., Ihrke, G., Vesa, J., Schell, M. J., and Luzio, J. P. (2004). Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol. Biol. Cell. 15 (3), 1313–1323. doi:10.1091/mbc.e03-02-0120
PubMed Abstract | CrossRef Full Text | Google Scholar
Lee, T., and Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 (3), 451–461. doi:10.1016/s0896-6273(00)80701-1
PubMed Abstract | CrossRef Full Text | Google Scholar
Lerner, T. J., Boustany, R.-M. N., Anderson, J. W., D’Arigo, K. L., Schlumpf, K., Buckler, A. J., et al. (1995). Isolation of a novel gene underlying batten disease, CLN3. Cell. 82 (6), 949–957. doi:10.1016/0092-8674(95)90274-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Luiro, K., Kopra, O., Blom, T., Gentile, M., Mitchison, H. M., Hovatta, I., et al. (2006). Batten disease (JNCL) is linked to disturbances in mitochondrial, cytoskeletal, and synaptic compartments. J. Neurosci. Res. 84 (5), 1124–1138. doi:10.1002/jnr.21015
PubMed Abstract | CrossRef Full Text | Google Scholar
Metcalf, D. J., Calvi, A. A., Seaman, M. N. J., Mitchison, H. M., and Cutler, D. F. (2008). Loss of the batten disease gene CLN3 prevents exit from the TGN of the mannose 6-phosphate receptor. Traffic 9 (11), 1905–1914. doi:10.1111/j.1600-0854.2008.00807.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Mirza, M., Vainshtein, A., DiRonza, A., Chandrachud, U., Haslett, L. J., Palmieri, M., et al. (2019). The CLN3 gene and protein: what we know. Mol. Genet. Genomic Med. 7 (12), e859. doi:10.1002/mgg3.859
PubMed Abstract | CrossRef Full Text | Google Scholar
Nászai, M., Carroll, L. R., and Cordero, J. B. (2015). Intestinal stem cell proliferation and epithelial homeostasis in the adult Drosophila midgut. Insect Biochem. Mol. Biol. 67, 9–14. doi:10.1016/j.ibmb.2015.05.016
PubMed Abstract | CrossRef Full Text | Google Scholar
Ohlstein, B., and Spradling, A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science 315 (5814), 988–992. doi:10.1126/science.1136606
PubMed Abstract | CrossRef Full Text | Google Scholar
Osman, D., Buchon, N., Chakrabarti, S., Huang, Y. T., Su, W. C., Poidevin, M., et al. (2012). Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell. Sci. 125 (Pt 24), 5944–5949. doi:10.1242/jcs.113100
PubMed Abstract | CrossRef Full Text | Google Scholar
Post, Y., and Clevers, H. (2019). Defining adult stem cell function at its simplest: the ability to replace lost cells through mitosis. Cell. Stem Cell. 25 (2), 174–183. doi:10.1016/j.stem.2019.07.002
PubMed Abstract | CrossRef Full Text | Google Scholar
Tuxworth, R. I., Chen, H., Vivancos, V., Carvajal, N., Huang, X., and Tear, G. (2011). The Batten disease gene CLN3 is required for the response to oxidative stress. Hum. Mol. Genet. 20 (10), 2037–2047. doi:10.1093/hmg/ddr088
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhai, Z., Boquete, J. P., and Lemaitre, B. (2017). A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila. PLoS Genet. 13 (6), e1006854. doi:10.1371/journal.pgen.1006854
留言 (0)