Staphylococcus aureus, an important Gram-positive pathogen, can cause various infectious diseases including bacteremia and pneumonia (Bashabsheh et al., 2024). It represents a growing public health burden owing to the emergence and spread of antibiotic-resistant clones, particularly within the hospital environment (Kholaseh et al., 2023). Based on data from the China Antimicrobial Surveillance Network (CHINET), in 2022, S. aureus was the third most common clinical bacterial isolate, comprising 9.5% of all clinical bacterial isolates, and it had the highest detection rate in Gram-positive clinical bacteria (https://www.chinets.com/). Methicillin-resistant S. aureus (MRSA) is major causative agent for nosocomial infections and has become a difficult problem in treatment of infections (Lee et al., 2018).
S. aureus ST45 is a major global MRSA lineage with different regions, hosts, antimicrobial susceptibility, and clinical manifestations, which frequently causes severe invasive disease, such as bacteremia (Effelsberg et al., 2020). S. aureus ST45 was originally identified in Berlin in 1993 (Witte et al., 2001) and now widely distributed worldwide (Peng et al., 2024). Epidemiological surveillance has shown that ST45-MRSA was mainly associated with t116 in Hainan and Guangzhou of China, with 70% and 66.7%, respectively (Ge et al., 2019; Li et al., 2019).
The cause of MRSA resistance to beta-lactam antibiotics is mecA and its homologues (mecB, mecC, and mecD) carried by MRSA (Lakhundi and Zhang, 2018). The mec genes are widely disseminated among staphylococcal species due to the acquisition and insertion of the staphylococcal cassette chromosome mec (SCCmec) element into the chromosome of susceptible strains, which is responsible for conferring the broad-spectrum beta-lactam resistance (Lakhundi and Zhang, 2018; Uehara, 2022). The SCCmec is a mobile genetic element (MGE) composed of mec complex, ccr complex and J region (Wang et al., 2022b). SCCmec elements were classified into different types based on the combination of mec gene complex (five classes) (Liu et al., 2016) and ccr gene complex (nine classes) (Wu et al., 2015). To date, 15 SCCmec types (types I-XV) have been officially reported (Wang et al., 2022b). Notably, subtypes of type IV SCCmec vary more than other types, with the main subtypes as follows: IVa, IVb, IVc, IVd, IVg, IVh, IVi, IVj, IVk, IVl, IVm, IVn,and Ivo (Uehara, 2022).
The aim of the study was to report the whole-genome sequence of a MRSA ST45-t116 strain SA2107 harboring SCCmec type IVa, which was isolated from the sputum of a 5-year-old child with pneumonia in China.
2 Materials and methods2.1 Bacterial isolate and antimicrobial susceptibility testingThis sample was obtained from the sputum of a 5-year-old child with pneumonia at Zhuhai People’s Hospital, Guangdong Province, China. Strain identification was performed using the VITEK-2 Compact system (bioMérieux, France), which was also confirmed by sequencing of the entire 16S rRNA gene. Antimicrobial susceptibility testing was measured by the VITEK 2 COMPACT system, which used the following 15 antimicrobial agents: clindamycin, daptomycin, gentamicin, levofloxacin, moxifloxacin, rifampicin, teicoplanin, vancomycin, ceftaroline, erythromycin, linezolid, oxacillin, penicillin, sulfamethoxazole/trimethoprim, and tigecycline. The results of other antimicrobial agents were interpreted according to the Institute of Clinical and Laboratory Standards (CLSI M100–S33) (CLSI, 2023).
2.2 Genome sequencing, assembly, and annotationWhole-genome sequencing (WGS) of strain SA2107 was conducted by Genewiz Biotechnology Co. Ltd. (Suzhou, China). Genomic DNA was extracted from the strain SA2107 using a genomic DNA extraction kit (provided by Genewiz) according to the manufacturer’s instructions. WGS was performed using paired-end sequencing with Novaseq 6000 (Illumina, 2×150 bp paired-end reads) and long sequencing with PacBio Sequel IIe (Pacific Biosciences, 10-15 Kb insert whole-genome shotgun libraries). PacBio reads were assembled using Hifiasm (version 0.13-r308) (Cheng et al., 2021) and Canu (version 2.2) (Koren et al., 2017). Genome assembly polishing was performed with Pilon (version 1.22) (Walker et al., 2014) using Illumina reads. The assembled genome of S. aureus SA2107 was submitted to the NCBI GenBank database (Benson et al., 2018) and annotated using the NCBI Prokaryotic Annotation Pipeline (PGAP) (Tatusova et al., 2016).
2.3 Bioinformatics analysisAcquired antibiotic resistance genes (ARGs) of the genome of SA2107 were identified using ResFinder 4.1 software (Bortolaia et al., 2020), with a minimum identity of 90% and a minimum coverage of 60%. The PointFinder software (Zankari et al., 2017) was used to detect the chromosomal gene mutations mediating antimicrobial resistance. Virulence genes of the genome of SA2107 were identified using VirulenceFinder 2.0 (Joensen et al., 2014). Multilocus sequence typing (MLST) of SA2107 was performed using MLST 2.0 (Larsen et al., 2012). Plasmid replicon types were determined using PlasmidFinder 2.1 (Carattoli et al., 2014). SCCmec typing was performed using the web-based SCCmecFinder version 1.2 (Kaya et al., 2018). spa typing was performed using the wed-based SpaFinder version 1.0 (Bartels et al., 2014). MGEs of SA2107, including genomic islands and prophage, were identified by the VRprofile2 (Wang et al., 2022a). Sequence similarity searching was performed using MegaBLAST (Morgulis et al., 2008) scans against the GenBank non-redundant (nr) database. Easyfig software (Sullivan et al., 2011) and BLAST Ring Image Generator (BRIG) software (Alikhan et al., 2011) was used to compare and visualize the sequences.
3 Results3.1 Identification and antimicrobial susceptibility results of the strain SA2107The strain SA2107 was identified as S. aureus using the VITEK 2 COMPACT system, and BLASTN analysis against rRNA_typestrains/16S_ribosomal_RNA database indicated that the entire 16S rRNA gene of SA2107 showed highest similarity with that of S. aureus strain S33 R (GenBank accession NR_037007, 100% coverage and 99.87% identity). Based on the results of the antibiotic susceptibility test, strain SA2107 exhibited resistance to oxacillin, penicillin, clindamycin, and erythromycin as well as intermediate-level resistance to rifampicin (Supplementary Table S1).
3.2 Genomic characteristics of the strain SA2107Genome sequencing and analysis revealed that SA2107 contained one 2.83 Mb chromosome (CP104559) and two plasmids with sizes of 30,064 bp (pSA2107-1, CP104560) and 8,033 bp (pSA2107-2, CP104561), respectively. Results of PlasmidFinder indicated that plasmid pSA2107-1 contained two replicons (rep16 and repUS5), whereas the plasmid pSA2107-2 consists of repB family plasmid replication initiator (coordinate: 4572.5429). MLST analysis indicated that S. aureus strain SA2107 belonged to sequence type (ST) 45. spa typing indicated that SA2107 displayed spa-type t116.
ResFinder results indicated that SA2107 carried two kinds of beta-lactam resistance genes (mecA and blaZ), which only located on the chromosome of SA2107, the mecA gene is known to confer resistance to methicillin in MRSA isolates and the blaZ is the structural gene of the staphylococcal penicillinase. It’s worth noting that SA2107 was found to harbor six copies of bla complex (blaZ, blaR1, and blaI) on its chromosome, with coordinates as 283131.286210, 477278.480357, 893462.896541, 2018600.2021679, 2036382.2039461, and 2548907.2551986, respectively. PointFinder results indicated that the rifampin resistance of SA2107 may be due to H481N mutation in RpoB. However, no ARG or chromosomal point mutation linked to clindamycin and erythromycin resistance was found. By the way, PointFinder software also detected many unknown point mutations (Supplementary Table S2), some of which might be associated with resistance to clindamycin and erythromycin. VirulenceFinder results indicated that different virulence factors were also detected on the chromosome of SA2107 (Table 1), including toxins (hlgA, hlgB, hlgC, sec, seg, sei, sel, sem, sen, seo, and seu), exoenzyme (aur), and genes encoding immune-modulating protein, i.e., hostimm (sak and scn). However, neither ARG nor virulence gene was detected on the two plasmids of SA2107.
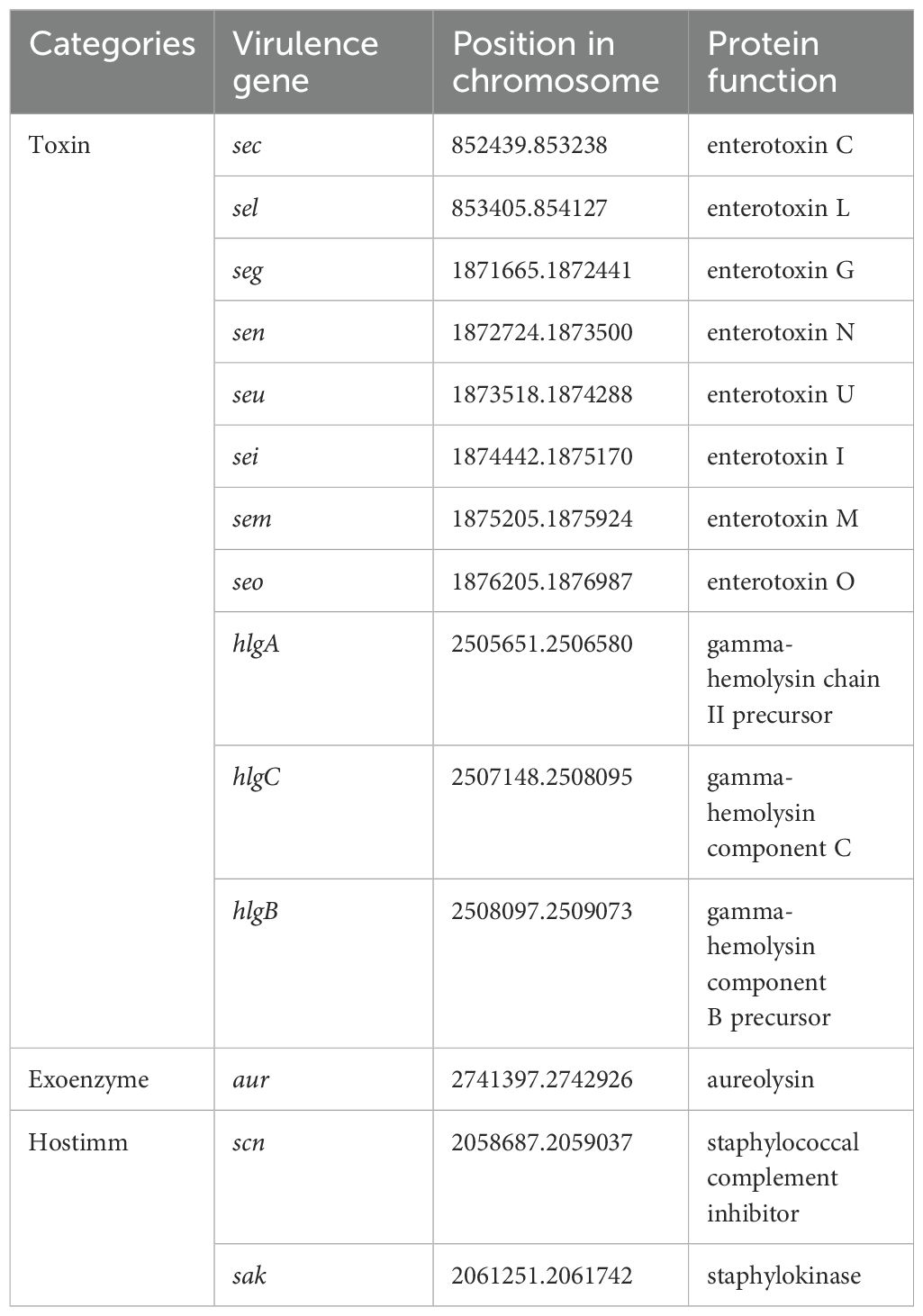
Table 1. Virulence genes harbored by the chromosome of SA2107.
Results of VRprofile2 indicated that three prophages, two integrative and conjugative elements (ICEs) and one SCCmec were identified on the chromosome of SA2107 (Table 2). Notably, the three prophages were found to carry the virulence genes (Table 2), indicating their role in the horizontal transfer of these virulence genes.
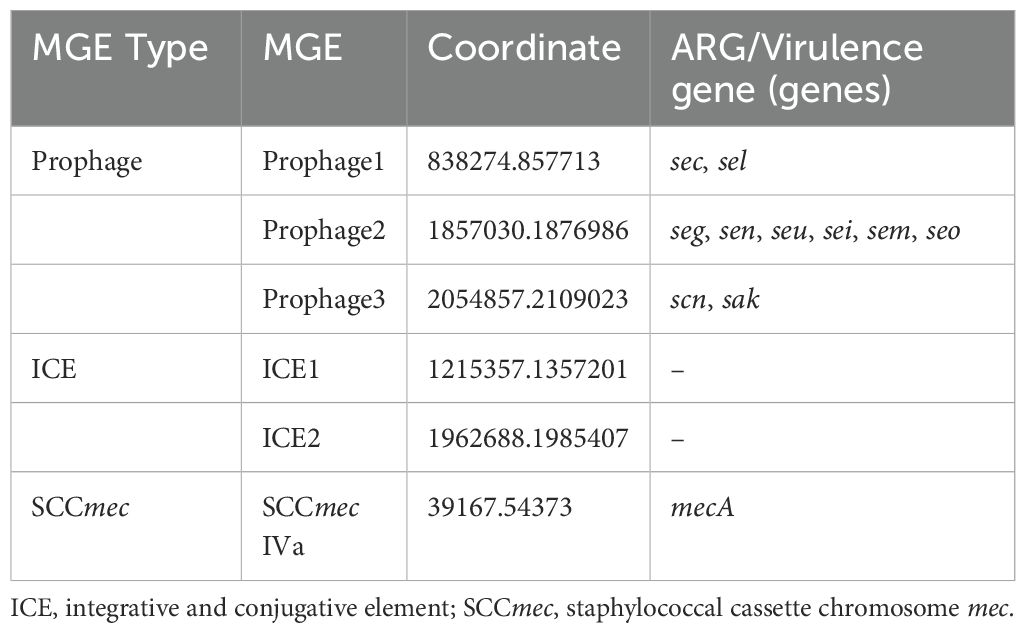
Table 2. MGEs carried by the chromosome of SA2107.
Based on the result of SCCmecFinder, the strain SA2107 carried the SCCmec type IVa (2B) on its chromosome. The SCCmec type IVa (2B) of SA2107 contained the class B mec complex, in which mecA and the truncated mecR1 gene encoding the signal transducer protein (ΔmecR1) were flanked by IS431 and IS1272, forming the structure IS431-mecA-ΔmecR1-IS1272 (Figure 1). In addition, the type 2 ccr gene complex (ccrA2 and ccrB2) was found in the SCCmec type IVa (2B) of SA2107 aligned with S. aureus JCSC1968, and other staphylococci species reference genomes (Figure 1). Notably, based on the results of the BLAST search hit from the nr database of GenBank, the whole SCCmec type IVa (2B) detected in strain SA2107 was not only present in the S. aureus, but also in the Staphylococcus caprae, Staphylococcus schleiferi, Staphylococcus epidermidis, Staphylococcus argenteus, and Staphylococcus warneri (Figure 1; Supplementary Table S3).
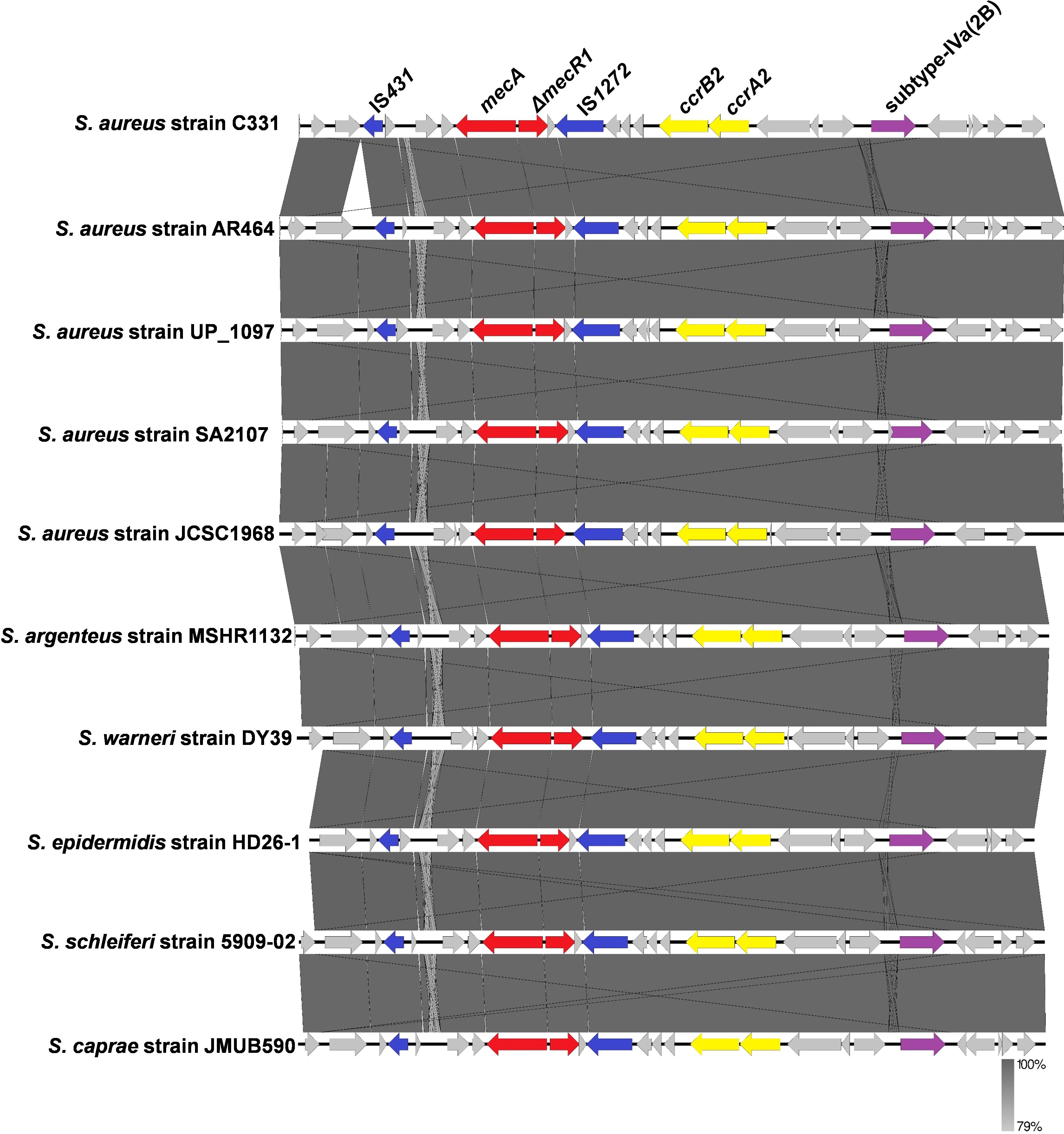
Figure 1. Comparative analysis of the SCCmec type IVa (2B) carried by the ST45-t116 MRSA strain SA2107 with other methicillin-resistant staphylococci generated by Easyfig.
3.3 An overview of ST45 SCCmec type IVa global strainsNCBI blast search of SCCmec type IVa (2B) of SA2107 against S. aureus (taxid: 1280), hit a total of 301 strains of S. aureus with complete genomes harboring SCCmec type IVa, which was widely present in more than 30 different STs of S. aureus (Figure 2). The most common ST amongst the SCCmec type IVa was ST8 (58.1%, n=175), followed by ST1 (7.6%, n=23), ST5 (7.3%, n=22), and ST59 (5.6%, n=17) (Figure 2). SA2107 in our study was the first report on the complete genome of the SCCmec type IVa (2B) in S. aureus ST45 in China (Figure 3). Another three strains of S. aureus ST45 with complete genomes containing the SCCmec type IVa (2B) were found in the United States (CP029084, AR464), Germany (CP047803, UP_1097), and Australia (CP127579, C331), respectively (Figure 3). The distribution of MGEs (prophages, ICEs, and SCCmec type IVa) carried by SA2107 was explored in other three S. aureus ST45 harboring SCCmec type IVa (Figure 4). Two prophages harboring virulence genes carried by SA2107, prophage2 (seg, sen, seu, sei, sem, seo) and prophage3 (scn, sak), were found to distributed in all other three ST45-SCCmec IVa strains. However, the prophage1 harboring sec and sel was only found in SA2107 and AR464. In addition, the ICE1 (coordinate: 1215357.1357201) carried by SA2107 was also found to distributed in all other three ST45-SCCmec IVa strains.
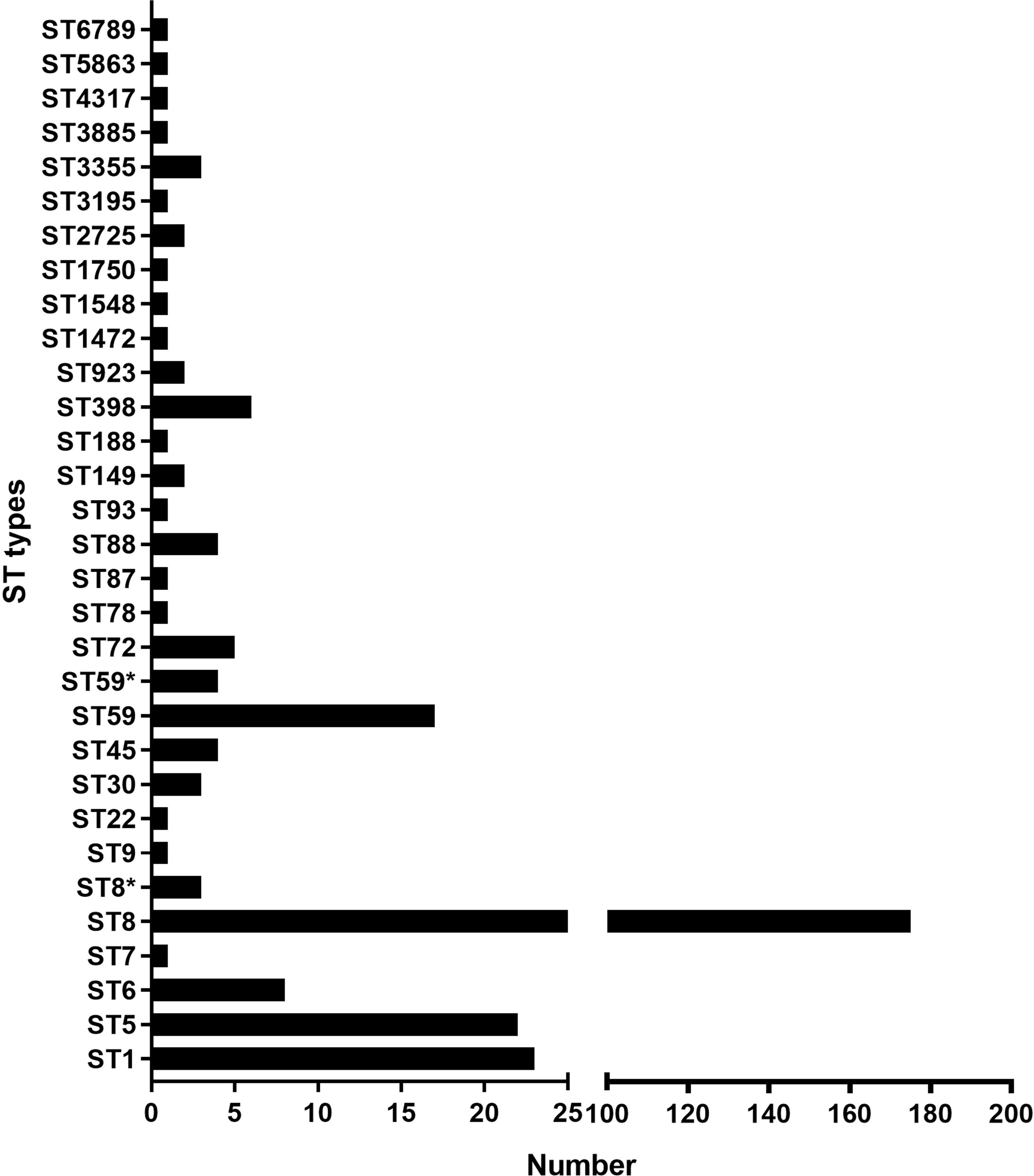
Figure 2. An overview of the ST types among the 301 of MRSA strains harboring the SCCmec type IVa. Histogram about number of the SCCmec type IVa distributed in different ST types of MRSA was drawn. * alleles with less than 100% identity found.
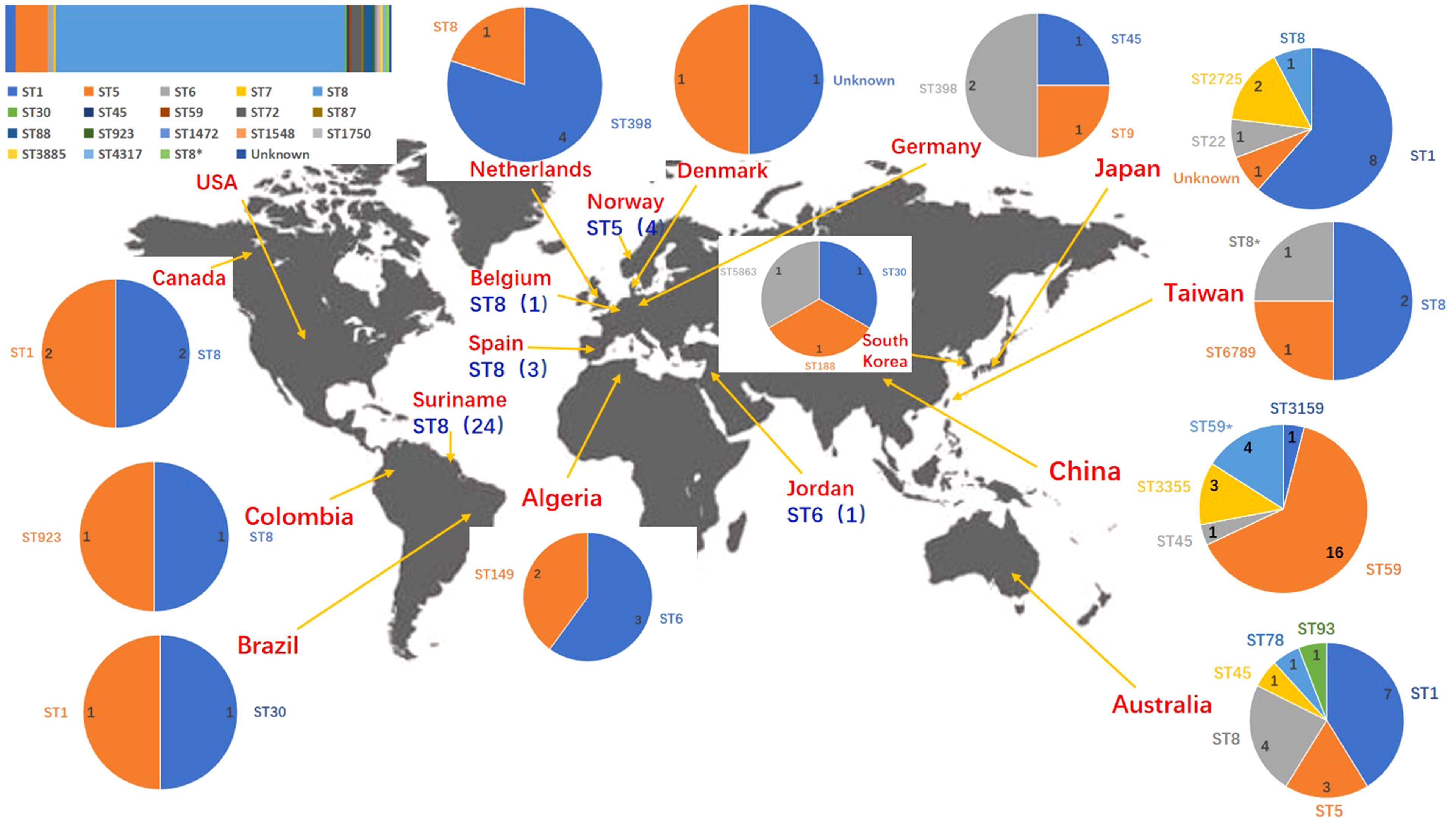
Figure 3. Geographic distribution of the MRSA strains with complete genomes harboring the SCCmec type IVa. ST types of different countries/regions were displayed by pie chart except the USA. ST types of the USA were displayed by columnar stacking diagram. * alleles with less than 100% identity found.
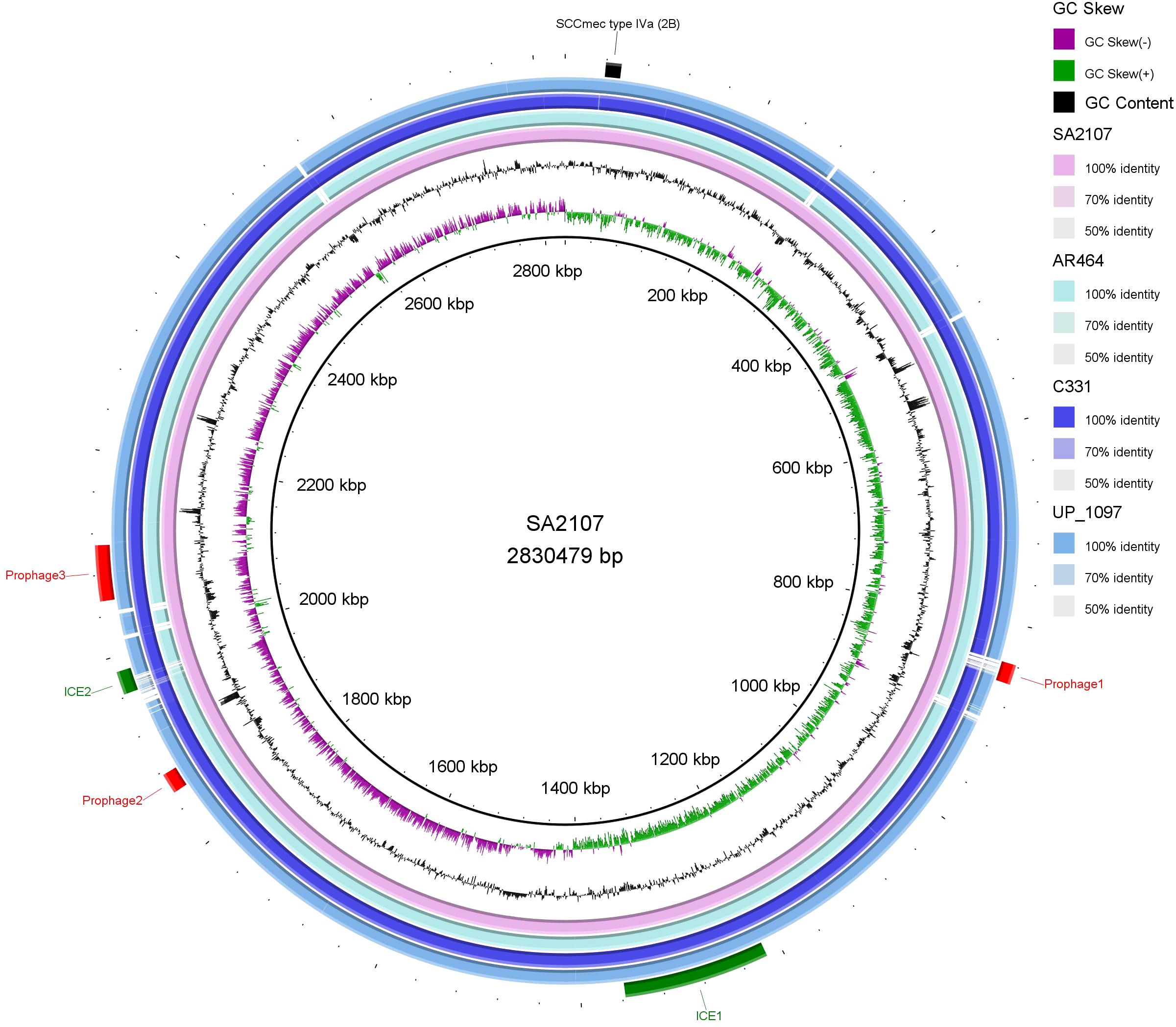
Figure 4. Comparative analysis of the MGEs, including the prophages, ICEs, and SCCmec type IVa (2B), carried by SA2107 with other three S. aureus ST45 harboring SCCmec type IVa (2B) generated by BRIG. Prophages, ICEs, and SCCmec are shown in red, green, and black arcs, respectively. GC Skew (+): the relative content of G is greater than C; GC Skew (-): the relative content of G is less than C.
4 DiscussionThe ST45-t116 MRSA strain SA2107 was found to contain the SCCmec type IVa (2B) on its chromosome. The SCCmec type IV has the combination of class B mec gene complex and a type 2 ccr gene complex (Hiramatsu et al., 2001), just as the “IS431-mecA-ΔmecR1-IS1272” and “ccrA2 and ccrB2” carried by the MRSA strain SA2107 in this study. The SCCmec type IVa was first reported in S. aureus CA05 (JCSC1968) isolated from the joint fluid of a patient with septic arthritis and osteomyelitis (Ma et al., 2002). Interestingly, only four strains of MRSA ST45-SCCmec IVa-t116 with complete genome worldwide harbor SCCmec type IVa, and SA2107 is the first report with complete genome in China.
For the SCCmec type IVa of MRSA SA2107, the mecA-ΔmecR1 was flanked by IS431 and IS1272. IS431, a staphylococcal insertion sequence (IS)-like element related to IS26 from Proteus vulgaris, was first described in 1987, which has been implicated in the transfer of antimicrobial resistance genes (e.g., mecA conferring methicillin resistance) (Barberis-Maino et al., 1987; Kobayashi et al., 2001). For the five classes of mec gene complexes have been described to date in MRSA (Liu et al., 2016), four were found to contain the IS431, including the class A mec gene complex (mecI-mecR1-mecA-IS431) (Ito et al., 2001; Liu et al., 2016), class B mec gene complex (IS431-mecA-ΔmecR1-IS1272) (Hiramatsu et al., 2001), the class C mec gene complex (IS431-mecA-ΔmecR1-IS431) (Katayama et al., 2001) and the class D mec gene complex (IS431-mecA-ΔmecR) (Liu et al., 2016). IS1272 was first described in Staphylococcus haemolyticus isolated in the United States in 1996 (Archer et al., 1996), has disseminated to other staphylococcal species and is prevalent in multi-resistant isolates (Wolska-Gębarzewska et al., 2023).
In this study, six copies of bla complex (blaZ, blaR1, and blaI) were found on the chromosome of SA2107. The beta-lactamase gene blaZ was the structural gene of the staphylococcal penicillinase, and the bla complex was necessary for penicillinase production (Massidda et al., 2006). The blaR1 gene encoded a signal-transducing membrane protein, and the blaI gene encoded a repressor protein (Hackbarth and Chambers, 1993). The expression of blaZ was regulated by the two adjacent genes, blaI and blaR1, the first being a blaZ transcription repressor, and the second an anti-repressor (Lowy, 2003).
Three categories of virulence genes were identified in the genome of SA2107, including toxin genes, exoenzyme genes, and hostimm genes. Until now, more than 24 staphylococcal enterotoxin (SE) genes have been identified from different outbreaks of staphylococcal food poisoning, clinical cases and strains isolated from animals (Lefebvre et al., 2022; Cieza et al., 2024). Eight SE genes (sec, seg, sei, sel, sem, sen, seo, and seu) were found in SA2107 and six genes (seg, sei, sem, sen, seo, and seu) were found on prophage2 of the SA2107. Schwendimann et al. reported that these six genes were also found on the S. aureus genomic island vSaβ (Schwendimann et al., 2021).
5 ConclusionThis study describes the genomic characteristics of a ST45-t116 MRSA strain SA2107 harboring SCCmec type IVa (2B), which is the first complete genome data (CP104559-CP104561) from China for ST45-SCCmec IVa (2B)-t116, and can be the reference genome for ST45-SCCmec IVa (2B)-t116 MRSA.
Data availability statementThe datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP104559–CP104561.
Ethics statementThe studies involving humans were approved by the Ethics Committee of Zhuhai People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributionsLH: Data curation, Formal analysis, Methodology, Software, Writing – original draft. RG: Formal analysis, Funding acquisition, Resources, Writing – original draft. JL: Data curation, Formal analysis, Methodology, Writing – original draft. XWL: Data curation, Formal analysis, Writing – original draft. ZL: Data curation, Writing – original draft. LZ: Data curation, Writing – original draft. WL: Conceptualization, Writing – original draft. RX: Data curation, Software, Writing – review & editing. CZ: Conceptualization, Resources, Supervision, Writing – review & editing. XF: Conceptualization, Resources, Supervision, Writing – review & editing. XBL: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported financially by the grants from the Guangdong Provincial Key Laboratory of Tumor Interventional Diagnosis and Treatment (Grant No. 2021B1212040004), the Science and Technology Projects of Social Development in Zhuhai (Grant No. 2220004000093), and the Xiangshan Talent Project of Zhuhai People’s Hospital (Grant No. 2020XSYC-02).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1413024/full#supplementary-material
ReferencesAlikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
PubMed Abstract | Crossref Full Text | Google Scholar
Archer, G. L., Thanassi, J. A., Niemeyer, D. M., Pucci, M. J. (1996). Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 40, 924–929. doi: 10.1128/aac.40.4.924
PubMed Abstract | Crossref Full Text | Google Scholar
Barberis-Maino, L., Berger-Bächi, B., Weber, H., Beck, W. D., Kayser, F. H. (1987). IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene 59, 107–113. doi: 10.1016/0378-1119(87)90271-x
PubMed Abstract | Crossref Full Text | Google Scholar
Bartels, M. D., Petersen, A., Worning, P., Nielsen, J. B., Larner-Svensson, H., Johansen, H. K., et al. (2014). Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 52, 4305–4308. doi: 10.1128/jcm.01979-14
PubMed Abstract | Crossref Full Text | Google Scholar
Bashabsheh, R. H. F., Al-Fawares, O., Natsheh, I., Bdeir, R., Al-Khreshieh, R. O., Bashabsheh, H. H. F. (2024). Staphylococcus aureus epidemiology, pathophysiology, clinical manifestations and application of nano-therapeutics as a promising approach to combat methicillin resistant Staphylococcus aureus. Pathog. Glob Health 118, 209–231. doi: 10.1080/20477724.2023.2285187
PubMed Abstract | Crossref Full Text | Google Scholar
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., et al. (2018). GenBank. Nucleic Acids Res. 46, D41–d47. doi: 10.1093/nar/gkx1094
PubMed Abstract | Crossref Full Text | Google Scholar
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
PubMed Abstract | Crossref Full Text | Google Scholar
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
PubMed Abstract | Crossref Full Text | Google Scholar
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H., Li, H. (2021). Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175. doi: 10.1038/s41592-020-01056-5
PubMed Abstract | Crossref Full Text | Google Scholar
Cieza, M. Y. R., Bonsaglia, E. C. R., Rall, V. L. M., Santos, M. V. D., Silva, N. C. C. (2024). Staphylococcal enterotoxins: description and importance in food. Pathogens 13, 676. doi: 10.3390/pathogens13080676
PubMed Abstract | Crossref Full Text | Google Scholar
Effelsberg, N., Stegger, M., Peitzmann, L., Altinok, O., Coombs, G. W., Pichon, B., et al. (2020). Global epidemiology and evolutionary history of staphylococcus aureus ST45. J. Clin. Microbiol. 59, e02198-20. doi: 10.1128/jcm.02198-20
PubMed Abstract | Crossref Full Text | Google Scholar
Ge, J., Zhong, X. S., Xiong, Y. Q., Qiu, M., Huo, S. T., Chen, X. J., et al. (2019). Methicillin-resistant Staphylococcus aureus among urban rodents, house shrews, and patients in Guangzhou, Southern China. BMC Vet. Res. 15, 260. doi: 10.1186/s12917-019-2012-8
PubMed Abstract | Crossref Full Text | Google Scholar
Hackbarth, C. J., Chambers, H. F. (1993). blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 37, 1144–1149. doi: 10.1128/aac.37.5.1144
PubMed Abstract | Crossref Full Text | Google Scholar
Hiramatsu, K., Cui, L., Kuroda, M., Ito, T. (2001). The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9, 486–493. doi: 10.1016/s0966-842x(01)02175-8
PubMed Abstract | Crossref Full Text | Google Scholar
Ito, T., Katayama, Y., Asada, K., Mori, N., Tsutsumimoto, K., Tiensasitorn, C., et al. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1323–1336. doi: 10.1128/aac.45.5.1323-1336.2001
PubMed Abstract | Crossref Full Text | Google Scholar
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/jcm.03617-13
PubMed Abstract | Crossref Full Text | Google Scholar
Katayama, Y., Ito, T., Hiramatsu, K. (2001). Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45, 1955–1963. doi: 10.1128/aac.45.7.1955-1963.2001
PubMed Abstract | Crossref Full Text | Google Scholar
Kaya, H., Hasman, H., Larsen, J., Stegger, M., Johannesen, T. B., Allesøe, R. L., et al. (2018). SCCmecFinder, a Web-Based Tool for Typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus Using Whole-Genome Sequence Data. mSphere 3, e00612-17. doi: 10.1128/mSphere.00612-17
PubMed Abstract | Crossref Full Text | Google Scholar
Kholaseh, S., Derakhshan, S., Abedini, M. (2023). A comparative study on antibiotic resistance and virulence properties of Staphylococcus aureus isolated from hospitalized patients and hospital environment. Am. J. Infect. Control 51, 859–865. doi: 10.1016/j.ajic.2022.12.006
PubMed Abstract | Crossref Full Text | Google Scholar
Kobayashi, N., Alam, M. M., Urasawa, S. (2001). Genomic rearrangement of the mec regulator region mediated by insertion of IS431 in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45, 335–338. doi: 10.1128/aac.45.1.335-338.2001
PubMed Abstract | Crossref Full Text | Google Scholar
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
PubMed Abstract | Crossref Full Text | Google Scholar
Lakhundi, S., Zhang, K. (2018). Methicillin-resistant staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, e00020-18. doi: 10.1128/cmr.00020-18
PubMed Abstract | Crossref Full Text | Google Scholar
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/jcm.06094-11
PubMed Abstract | Crossref Full Text | Google Scholar
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant staphylococcus aureus. Nat. Rev. Dis. Primers 4, 18033. doi: 10.1038/nrdp.2018.33
PubMed Abstract | Crossref Full Text | Google Scholar
Lefebvre, D., Blanco-Valle, K., Hennekinne, J. A., Simon, S., Fenaille, F., Becher, F., et al. (2022). Multiplex detection of 24 staphylococcal enterotoxins in culture supernatant using liquid chromatography coupled to high-resolution mass spectrometry. Toxins (Basel) 14, 249. doi: 10.3390/toxins14040249
PubMed Abstract | Crossref Full Text | Google Scholar
Li, X., Huang, T., Xu, K., Li, C., Li, Y. (2019). Molecular characteristics and virulence gene profiles of Staphylococcus aureus isolates in Hainan, China. BMC Infect. Dis. 19, 873. doi: 10.1186/s12879-019-4547-5
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, J., Chen, D., Peters, B. M., Li, L., Li, B., Xu, Z., et al. (2016). Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 101, 56–67. doi: 10.1016/j.micpath.2016.10.028
PubMed Abstract | Crossref Full Text | Google Scholar
Ma, X. X., Ito, T., Tiensasitorn, C., Jamklang, M., Chongtrakool, P., Boyle-Vavra, S., et al. (2002). Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46, 1147–1152. doi: 10.1128/aac.46.4.1147-1152.2002
PubMed Abstract | Crossref Full Text | Google Scholar
Massidda, O., Mingoia, M., Fadda, D., Whalen, M. B., Montanari, M. P., Varaldo, P. E. (2006). Analysis of the beta-lactamase plasmid of borderline methicillin-susceptible Staphylococcus aureus: focus on bla complex genes and cadmium resistance determinants cadD and cadX. Plasmid 55, 114–127. doi: 10.1016/j.plasmid.2005.08.001
PubMed Abstract | Crossref Full Text | Google Scholar
Morgulis, A., Coulouris, G., Raytselis, Y., Madden, T. L., Agarwala, R., Schäffer, A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. doi: 10.1093/bioinformatics/btn322
PubMed Abstract | Crossref Full Text | Google Scholar
Peng, K. T., Chen, P. C., Chen, J. L., Huang, T. Y., Peng, Y. H., Liu, J. F., et al. (2024). A comparative phenotypic and genomic analysis of methicillin-resistant Staphylococcus aureus ST45 isolates from cellulitis and from osteomyelitis in Taiwan. J. Infect. Dis. 230, e568-e578. doi: 10.1093/infdis/jiae096
PubMed Abstract | Crossref Full Text | Google Scholar
Schwendimann, L., Merda, D., Berger, T., Denayer, S., Feraudet-Tarisse, C., Kläui, A. J., et al. (2021). Staphylococcal enterotoxin gene cluster: prediction of enterotoxin (SEG and SEI) production and of the source of food poisoning on the basis of vSaβ Typing. Appl. Environ. Microbiol. 87, e0266220. doi: 10.1128/aem.02662-20
PubMed Abstract | Crossref Full Text | Google Scholar
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
PubMed Abstract | Crossref Full Text | Google Scholar
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al.
留言 (0)