Brucellosis is a longstanding global zoonosis that has been reported in over 170 countries and regions, with an estimated 500,000 new cases occurring annually worldwide. This disease represents a significant public health challenge on a global scale, garnering substantial attention within the public health sector (1). Presently, in China, the incidence of brucellosis is on the rise, attributed to rapid economic development and the increasing movement of animals and animal products. In 2023, the number of new cases in the country surpassed 70,400 (2, 3).
Despite advancements in existing diagnostic methods, including serological, pathogenetic, and molecular biology testing, there remain significant limitations in the practical application of each of these approaches (4, 5). Serological methods, such as the standard agglutination test (SAT), the rose bengal plate agglutination test (RBPT), and the enzyme-linked immunosorbent assay (ELISA), are widely employed in clinical diagnostics due to their operational simplicity and cost-effectiveness. However, these methods are not without their drawbacks. For instance, the specificity of serological diagnoses can be adversely affected by antigenic factors. A frequently utilized antigen, lipopolysaccharide (LPS), is susceptible to cross-reactivity with Yersinia enterocolitica O9, potentially leading to false-positive results. Furthermore, while pathogenicity tests are known for their high accuracy, they are complex to execute and present challenges for implementation in primary care settings (6–9).
Bioinformatics, defined as the integration of biology, computer science, and information technology, plays a crucial role in contemporary biomedical research, particularly within the domains of genomics and proteomics (10). This interdisciplinary field encompasses the development and application of computational tools and algorithms designed to analyze and interpret complex biological data, including DNA sequences, protein structures, and gene expression profiles. In the realm of vaccine design, bioinformatics has become essential, facilitating the identification of potential antigens, the prediction of immune responses, and the optimization of vaccine candidates (11). Through the analysis of genomic sequences, bioinformatics aids researchers in understanding the pathogenicity and antigenic characteristics of microorganisms, which is vital for the development of effective vaccines. Furthermore, the analysis of proteomic data using bioinformatics tools can identify key proteins that are differentially expressed in pathogenic strains, thereby assisting in the selection of specific epitopes for vaccine development. The application of bioinformatics in these contexts has resulted in significant advancements in the understanding of host-pathogen interactions and the rational design of vaccines with enhanced efficacy and safety (12).
This study addresses the current status and challenges associated with the diagnosis of brucellosis by employing proteomics and bioinformatics to develop multiepitope fusion proteins. The research involves a meticulous screening of key antigenic epitopes derived from wild-virulent strains of Brucella, which are subsequently fused into a single protein molecule. The objective is to construct a novel diagnostic antigen characterized by high specificity, aimed at facilitating the development of an indirect ELISA (iELISA) for the diagnosis of human brucellosis.
2 Materials and methods2.1 Serum samples and bacterial strainsA total of 100 positive and 96 negative serum samples were obtained from the Xuzhou Center for Disease Control and Prevention, all of which were confirmed as either positive or negative through the SAT. Furthermore, serum samples from 40 febrile patients infected with various pathogens (stored in the laboratory; detailed information is provided in Supplementary Material 1: Cross-Reactivity Assessment) were utilized to assess the cross-reactivity of the developed method. Additionally, the vaccine strain Brucella abortus A19 and the wild-type Brucella abortus DT21, both isolated and preserved by the China Animal Health and Epidemiology Center, were employed in this study.
2.2 TMT proteomics2.2.1 Bacterial cultureThe preserved bacterial strain was inoculated into 500 mL of tryptic soy broth (TSB) medium and incubated at 37°C with shaking for a duration of 24 to 48 hours. After the incubation period, 5 mL of 1% formaldehyde was introduced to inactivate the bacteria, which were subsequently stored at 4°C for future use.
2.2.2 TMT proteomics analysisTMT proteomics analysis was conducted in accordance with established protocols documented in the literature. This process encompassed several key steps, including protein extraction and quantification, protein digestion and TMT labeling, as well as LC-MS/MS analyses, followed by both qualitative and quantitative assessments of the proteins (13).
2.3 Preparation of fusion proteins2.3.1 Prediction of B-cell linear epitopesBased on the results obtained from TMT proteomics, proteins that exhibited high expression levels in the wild-type strain were selected for further analysis. The amino acid sequences of these proteins were retrieved from the NCBI protein database (http://www.ncbi.nlm.nih.gov/protein/). To enhance the accuracy of epitope prediction, four B-cell linear epitope prediction tools were employed: ABCpred (https://webs.iiitd.edu.in/raghava/abcpred/index.html, with a default threshold of 0.5), SVMTriP (http://sysbio.unl.edu/SVMTriP, with no threshold), BCPred (http://ailab-projects2.ist.psu.edu/bcpred/predict.html, with a default specificity threshold of 75%), and Bepipred Linear Epitope Prediction 2.0 (http://tools.iedb.org/bcell/, with a default threshold of 0.5) (14–17). The predicted B-cell epitopes from all tools were compared, and overlapping B-cell epitopes were selected as candidate epitopes. For each prediction tool, the prediction threshold was maintained at the default value, with the exception of SVMTriP, and epitopes with scores exceeding 0.5 were considered potential candidates.
2.3.2 Construction of the fusion protein amino acid sequenceThe predicted linear epitopes of B-cells were concatenated, incorporating a ‘GS’ linker between adjacent epitopes to construct the amino acid sequence of the fusion protein. This sequence was subsequently submitted to Beijing Protein Innovation Co., Ltd. for codon optimization, thereby rendering it suitable for prokaryotic expression. Gene synthesis was conducted, and a 6×His tag was incorporated to facilitate subsequent protein purification. The three-dimensional (3D) molecular model of the fusion protein was predicted using I-TASSER (https://zhanggroup.org/I-TASSER/), while the antigenicity of the fusion protein was assessed using VaxiJen (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), employing a default threshold of 0.4.
2.3.3 Prokaryotic expression of the fusion proteinThe gene encoding the synthesized fusion protein was subsequently cloned and inserted into the expression vector pET30a. This vector was then transformed into competent BL21 cells to facilitate IPTG-induced expression. The transformation process involved several steps. Initially, the competent BL21 cells were stored at -80°C and were slowly thawed on ice. They were then mixed with the pET30a vector and incubated on ice for 30 minutes. Following this, the samples underwent a heat shock at 42°C for 90 seconds, after which they were immediately cooled on ice for 2 minutes. Subsequently, 800 μL of LB medium was added to the mixture, which was incubated at 37°C for 45 minutes before being centrifuged at 5,000 rpm for 3 minutes. Most of the supernatant was discarded, leaving approximately 100–150 μL, after which the cells were resuspended. The resulting suspension was plated onto LB agar plates containing the appropriate antibiotic and incubated overnight at 37°C. The cultured bacterial mixture was then transferred to 250 mL of LB liquid medium supplemented with the corresponding antibiotic and incubated at 37°C with shaking at 200 rpm until the optical density at 600 nm (OD600) reached 0.6–0.8. Expression of the target protein was induced by the addition of 0.5 mM IPTG, followed by incubation at 37°C for 4 hours. The mixture was subsequently centrifuged at 8,000 rpm for 6 minutes, and the supernatant was discarded to collect the cells. The resulting pellet was resuspended in 20–30 mL of 10 mM Tris-HCl (pH 8.0) solution, and ultrasonic disruption was performed (500 W, 180 cycles, 5 seconds per cycle, with 5-second intervals). One hundred microliters of the disrupted bacterial suspension was then centrifuged at 12,000 rpm for 10 minutes. Following centrifugation, 50 μL of the supernatant was transferred to a separate Eppendorf tube, and the pellet was resuspended in 50 μL of 10 mM Tris-HCl (pH 8.0) solution. A 12% SDS-PAGE gel was employed to ascertain the presence of the target protein in either the supernatant or the pellet for subsequent purification.
2.3.4 Fusion protein purificationThe nickel affinity chromatography column (Ni Sepharose 6 Fast Flow, GE Healthcare) was initially washed with deionized water until the pH stabilized at 7.0. Subsequently, the column was equilibrated with approximately 100 mL of a 10 mM Tris-HCl buffer (pH 8.0). This was followed by further equilibration using approximately 50 mL of the same buffer supplemented with 0.5 M NaCl. The sample containing the target protein was then diluted and applied to the column. Following the loading of the sample, the column was washed with a 10 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl. The target protein was eluted using 10 mM Tris-HCl buffers (pH 8.0) containing imidazole concentrations of 15 mM, 60 mM, and 300 mM, along with 0.5 M NaCl. The eluted protein peaks were collected, and the purification efficiency was assessed through 12% SDS-PAGE electrophoresis. Additionally, the protein content was quantified using a BCA protein quantification kit.
2.4 Establishment of iELISA and serum detectionThe iELISA was conducted according to the following protocol. Initially, the purified protein was diluted in carbonate-bicarbonate buffer solution (CBS) to achieve a concentration of 10 µg/mL, and 100 µL of this solution was dispensed into each well of a 96-well microplate (Corning, USA). The microplate was then incubated overnight at 4°C. Following incubation, the wells were washed three times with PBST. Subsequently, 300 µL of blocking solution (5% skim milk in PBS) was added to each well, and the samples were incubated at 37°C for 2 hours. After another round of washing with PBST, human serum diluted in PBS (1:200) was introduced and incubated at 37°C for 1 hour. Following three additional washes with PBST, 100 µL of horseradish peroxidase (HRP)-conjugated rabbit anti-human IgG (diluted 1:10,000, Thermo Fisher, USA) was added to each well and incubated at 37°C for 1 hour. The plate was washed three times with PBST, after which the tetramethylbenzidine (TMB) substrate solution was added, and the plate was incubated in the dark for 10 minutes to allow for color development. The reaction was terminated by the addition of 2 M H2SO4, and the optical density at 450 nm (OD450) was measured using a microplate reader (Versa Max microplate reader, MD, USA). Laboratory-stored LPS (3 mg/mL, provided by the China Animal Health and Epidemiology Center) served as a control antigen, and serum samples were analyzed in triplicate following the same procedure. The sensitivity, specificity, area under the curve (AUC), and cutoff values were determined through receiver operating characteristic (ROC) curve analysis.
2.5 Cross-reactivity assessmentFollowing the aforementioned iELISA procedure, sera from febrile patients without brucellosis were analyzed by using the two antigens to evaluate the cross-reactivity of the constructed fusion protein. Cross-reactivity was assessed based on the cut-off value determined by the ROC curve.
2.6 Statistical methodsDot plot and ROC curve analyses were conducted using GraphPad Prism version 6.05. Statistical analyses were performed utilizing one-way analysis of variance (ANOVA) and Student’s t-test (unpaired t-test), with a significance level established at p < 0.05.
3 Results3.1 Proteomics analysisThrough TMT quantitative analysis, a total of 152 proteins exhibiting high expression levels were identified in the wild-type strain, while 102 highly expressed proteins were identified in the vaccine strain (see Supplementary Material 2). From the highly expressed proteins in the wild-type strain, seven target proteins were selected for the prediction of B-cell epitopes (refer to Table 1).
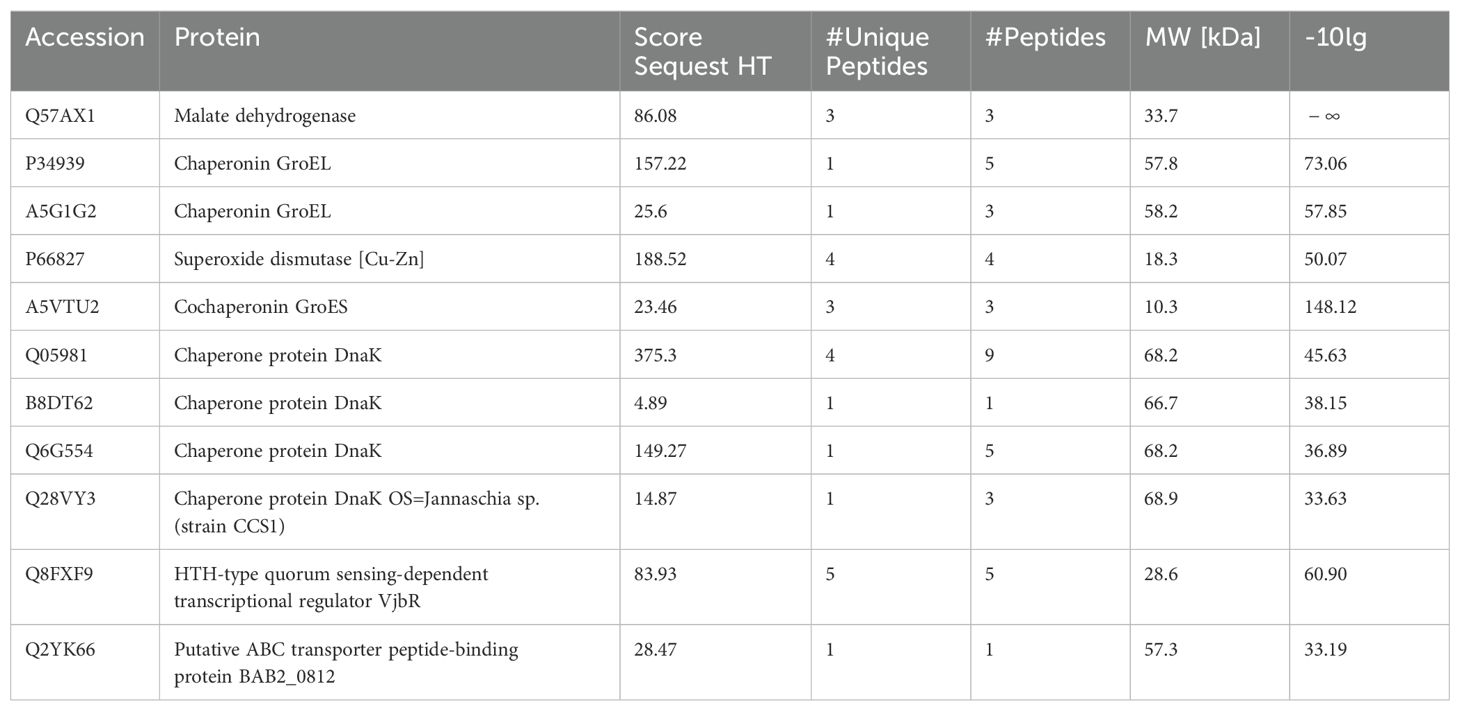
Table 1. Protein information selected based on TMT proteomics results.
3.2 Epitope predictionBased on the results obtained from proteomics and a comprehensive review of the pertinent literature, specific proteins were identified as potential candidate targets. A total of 32 epitopes were predicted, as presented in Table 2. Subsequently, these epitopes were concatenated to create the amino acid sequence of the fusion protein using a linker.
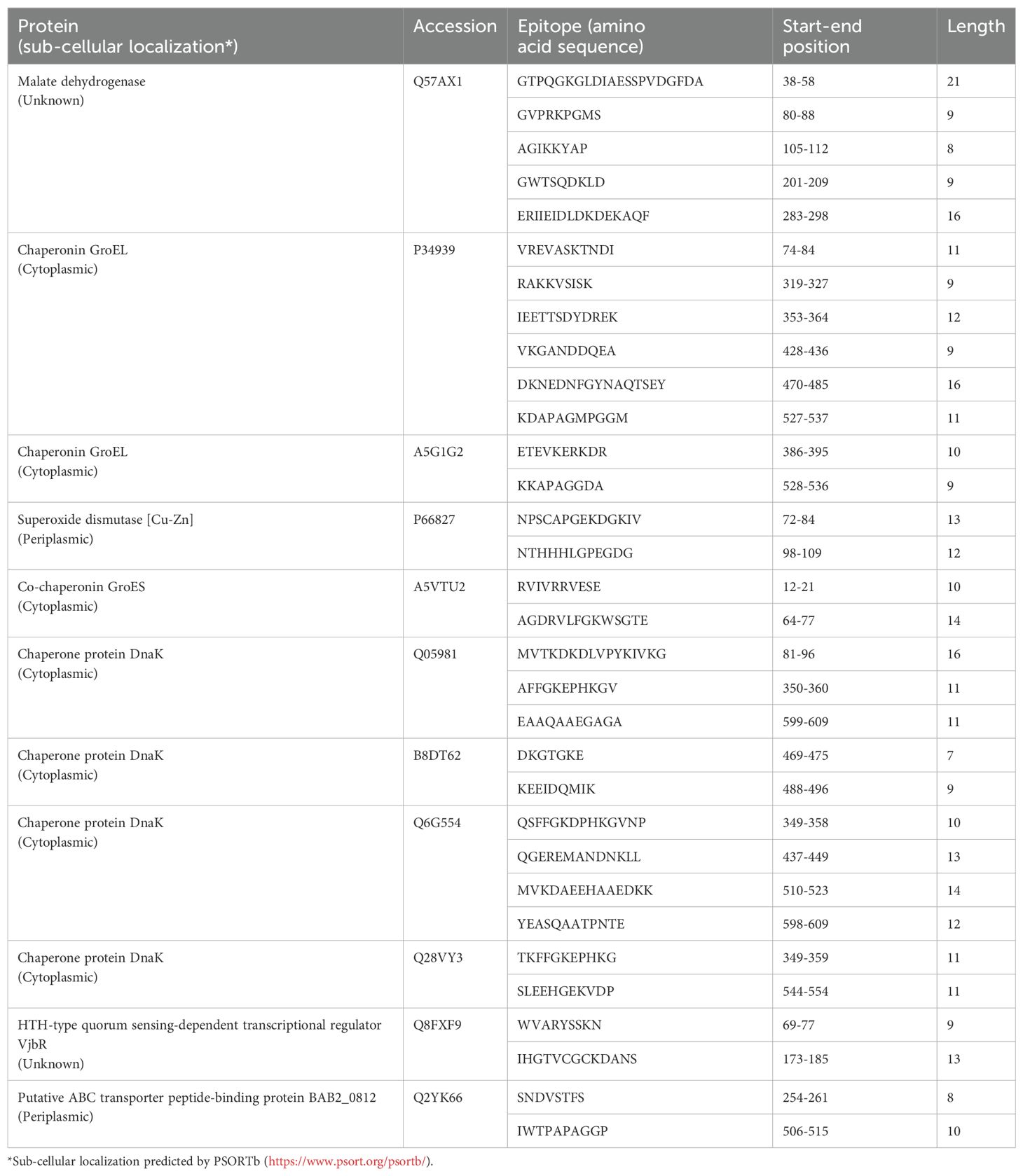
Table 2. Predicted candidate epitope information.
3.3 Fusion protein preparationFollowing prokaryotic expression, the target protein was identified in the supernatant. Subsequent purification of the fusion protein resulted in a purity level of 90.1%. The findings are illustrated in Figure 1. The prediction from VaxiJen indicated an antigenicity score of 1.052, suggesting that it is a probable antigen.
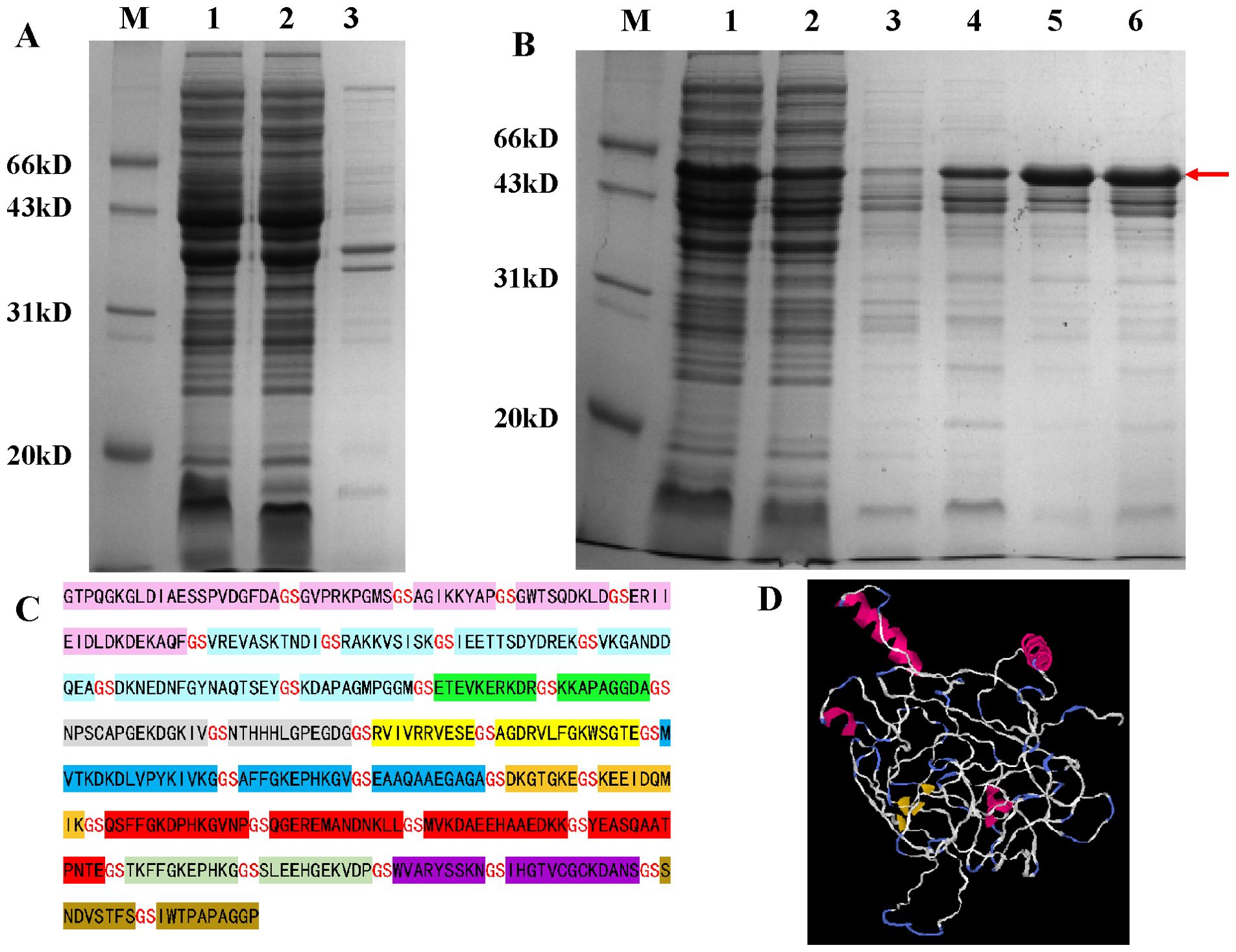
Figure 1. 12% SDS-PAGE analysis of fusion protein prokaryotic expression results. (A) Detection of fusion protein expression. M: marker; Lane 1: whole-cell lysate after sonication; Lane 2: supernatant after sonication; Lane 3: pellet after sonication. (B) Results of large-scale expression and purification of the fusion protein. M: marker; Lane 1: crude protein sample; Lane 2: flow-through fraction; Lane 3: elution with 15 mM imidazole; Lane 4: elution with 60 mM imidazole; Lanes 5 and 6: elution with 300 mM imidazole; (C) Amino acid sequence of the fusion protein; (D) 3D structural models of fusion proteins predicted by I-TASSER.
3.4 Results of iELISAThe analysis of the ROC curve revealed that the area under the diagnostic curve (AUC) for the fusion protein was 0.9576 (95% CI, 0.9337–0.9814), while the AUC for LPS was 0.9999 (95% CI, 0.9995–1.000). These findings indicated that both antigens possess excellent diagnostic value. Utilizing the Youden index, the cutoff value for the diagnosis using the fusion protein was established at 0.1970. At this threshold, the sensitivity and specificity of the diagnostic method were found to be 0.9300 (95% CI, 0.8611–0.9714) and 0.8542 (95% CI, 0.7674–0.9179), respectively. Conversely, the cutoff value for the diagnosis using LPS was determined to be 0.1953, with a sensitivity of 0.9900 (95% CI, 0.9455–0.9997) and a specificity of 1.000 (95% CI, 0.9623–1.000). The detailed results are illustrated in Table 3, Figure 2, and the Supplementary Material.
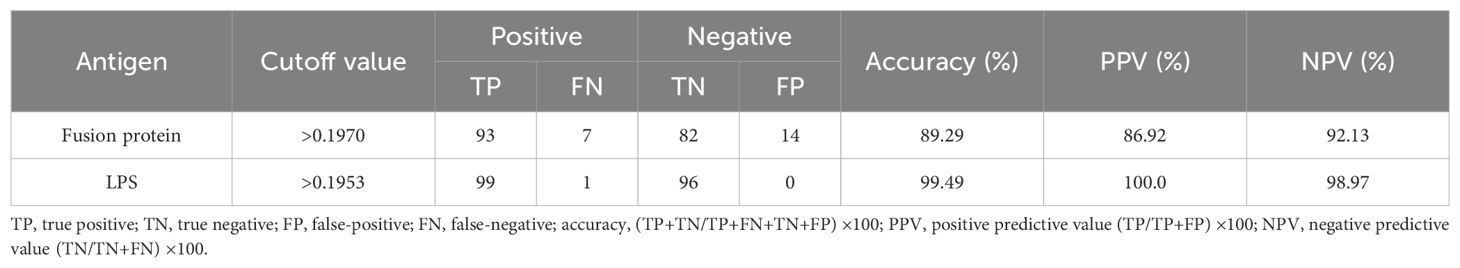
Table 3. Positive and negative predictive values of the test calculated for different cutoff values.
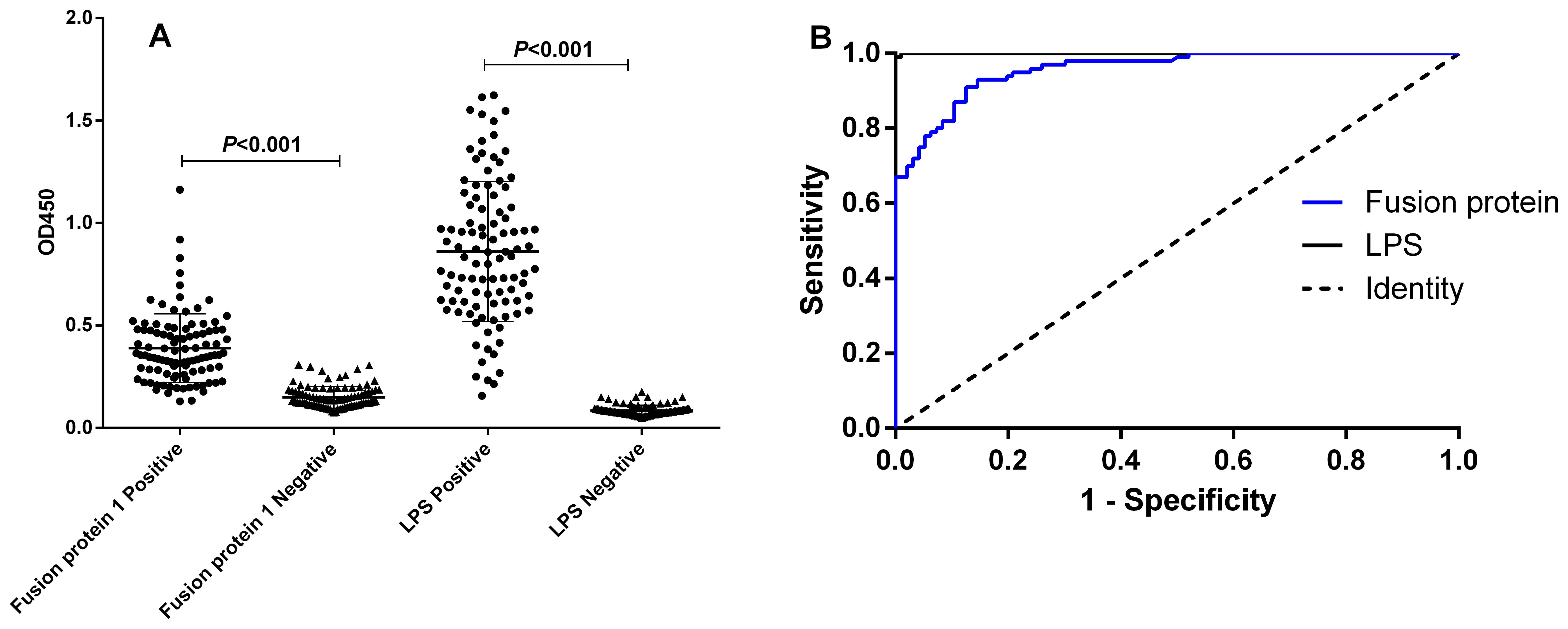
Figure 2. I-ELISA analysis of human serum samples. (A) Dot plot of human serum samples. (B) ROC analysis of human sera.
3.5 Cross-reactivity assessmentUtilizing the iELISA and the established cutoff values, cross-reactivity was identified in 5 out of 40 serum samples obtained from febrile patients who did not have brucellosis when the fusion protein was evaluated. These samples comprised 3 instances of Escherichia coli infection, 1 instance of Pseudomonas putida infection, and 1 instance of Streptococcus dysgalactiae infection. In contrast, cross-reactivity with LPS was detected in 14 cases, which included 8 instances of Escherichia coli infection, 3 instances of Staphylococcus aureus infection, and 1 instance each of Enterococcus faecium, Klebsiella pneumoniae, Moraxella osloensis, Pseudomonas putida, and Streptococcus dysgalactiae infection. The detailed results are presented in the Supplementary Material.
4 DiscussionIndirect ELISA is endorsed by the World Organization for Animal Health (OIE) for a variety of applications, including the assessment of population freedom from infection, the determination of individual animal freedom from infection, contributions to eradication strategies, confirmation of criminal suspects or clinical cases, and the surveillance of herd or flock prevalence of infection in animals (18). However, traditional iELISAs utilized for the diagnosis of brucellosis typically employ LPS as the antigen. LPS is commonly found in gram-negative bacteria and has been demonstrated to cross-react with Escherichia coli O157:H7 and Yersinia enterocolitica O:9 when used as a diagnostic antigen (8, 9). Consequently, the identification of more specific diagnostic antigens may enhance the efficacy of iELISA applications.
Proteomics encompasses the comparative analysis of the complete set of proteins expressed by a microorganism, thereby facilitating the classification and identification of pertinent proteins that can be characterized as protein molecules (19, 20). These molecules may serve as potential targets for novel drug development or as molecular markers for the early diagnosis of diseases. In the present study, we selected both wild and vaccine strains of Brucella for proteomic analysis. Drawing upon the results obtained and a comprehensive review of the literature, several proteins exhibiting significant differences were identified as candidate targets.
ABC transporters are integral to the biosynthetic pathways of extracellular polysaccharides. The deletion of the ABC transporter ATPase gene has been shown to diminish the virulence of wild-type strains and enhance the resistance of mice to challenges posed by these strains, thereby underscoring its significant immunological role in wild-type strains (21). Research has established that this transporter can be utilized for immunoenzymatic assays, specifically iELISA, in the diagnosis of bovine brucellosis (22). Heat shock proteins are recognized as major antigens that play a critical role during Brucella abortus infection; notably, DnaK, GroEL, and GroES are three heat shock proteins identified in Brucella. Immunization of animals with these recombinant proteins has the potential to elicit a Th1 immune response and generate protective antibodies, thereby highlighting their applicability in the serological diagnosis of brucellosis (23–28). Malate dehydrogenase (MDH), a pivotal enzyme in the tricarboxylic acid cycle, is capable of inducing Th2-related immune responses, and recombinant MDH (rMDH) has been employed in the diagnosis of bovine brucellosis (29–31). Additionally, Brucella Cu-Zn superoxide dismutase (Cu-Zn SOD), a periplasmic protein, has been shown to confer protection against Brucella abortus infection when mice are immunized with recombinant Cu-Zn SOD protein (32, 33). Furthermore, the transcriptional regulator VjbR is essential for the interaction with host cells during Brucella infection and is critical for the virulence of the intracellular facultative pathogen Brucella (34). Animal studies have confirmed its significant immunoprotective effect against brucellosis (35, 36).
Bioinformatics technologies have been extensively utilized in the identification of disease diagnostic antigens and the development of vaccines. A variety of techniques have been employed to generate predictions for antigen epitopes. Among the most commonly used methods for predicting linear epitopes are Bepipred, ABCpred, COBEpro, and SVMTriP (14–17). Bepipred integrates hidden Markov models with propensity scale methods to predict linear B-cell epitopes. ABCpred, which is based on a neural network framework, achieves predictions with an accuracy of approximately 65.93%. COBEpro employs a two-step approach to predict linear B-cell epitopes, initially predicting short peptides using a mechanical model, followed by scoring each amino acid residue. SVMTriP, a leading method in this domain, utilizes a support vector machine model that combines tri-peptide similarity with propensity scores. When applied to non-redundant B-cell linear epitopes sourced from the Immune Epitope Database (IEDB), SVMTriP demonstrated a sensitivity of 80.1% and a precision of 55.2% through fivefold cross-validation, resulting in an area under the curve (AUC) value of 0.702. However, the accuracy of individual prediction tools is frequently suboptimal. Consequently, this study selected four prediction tools to enhance overall accuracy. We integrated immunological information parameter predictions with B-cell epitope predictions and compared the B-cell epitopes identified by the four methods. Overlapping B-cell epitopes were subsequently selected as candidate B-cell epitopes for the construction of multiepitope fusion proteins.
In this study, a total of 32 epitopes were predicted, and the constructed fusion protein, utilized as a diagnostic antigen, was compared to LPS. Although the fusion protein demonstrated slightly inferior performance, it exhibited reduced cross-reactivity, thereby highlighting its advantages in the diagnosis of brucellosis. The approach of developing multiepitope fusion proteins not only preserves the immunogenicity of individual antigenic epitopes but also significantly diminishes the cross-reactivity of the diagnostic antigen, thereby enhancing specificity and improving diagnostic accuracy. It is important to note that the sera selected for testing in the evaluation of the constructed iELISA, particularly the 96 negative samples, did not allow for the determination of whether these samples were from individuals infected with other pathogens or from healthy individuals. The evaluation indices, including sensitivity, specificity, and false-positive rates, are subject to change with variations in sample selection. In contrast, when assessing cross-reactivity, the selected sera were sourced from patients with confirmed infections from other pathogens, all of whom presented with fever symptoms necessitating differential diagnosis from brucellosis. Consequently, these sera are particularly relevant for evaluating the validity of the iELISA. LPS is known to be present in gram-negative bacteria, and its cross-reactivity with Escherichia coli varies. In this study, the purity of the fusion protein expressed in prokaryotic systems was only 90.1%, with some impurities originating from E. coli, which may contribute to its cross-reactivity with E. coli infections. Future efforts to enhance the specificity of the fusion protein may involve improvements in its purification process. Furthermore, the advancement of this technology presents new perspectives and methodologies for the diagnosis of brucellosis, potentially addressing the limitations of current serological diagnostic techniques. However, the assessment of the advantages and disadvantages of the multiepitope fusion protein compared to LPS based solely on the testing of this limited sample size is not reliable. While LPS demonstrates an advantage concerning false positivity, fusion proteins exhibit a benefit in terms of reduced cross-reactivity; this discrepancy may be influenced by the selection of serum samples.
Currently, there is no vaccine available for human brucellosis, which renders the findings of this study inconclusive in determining whether this protein can effectively differentiate between sera from naturally infected individuals and those who have been immunized with a vaccine. Additional research is necessary to obtain sera from both immunized and naturally infected animals for further testing.
5 ConclusionIn summary, a novel multiepitope fusion protein, developed utilizing bioinformatics and TMT proteomics technology, has demonstrated significant potential as a diagnostic antigen for brucellosis, exhibiting high sensitivity and specificity. This innovative approach marks a considerable advancement in the field of brucellosis diagnosis, providing a more accurate and reliable alternative to existing methodologies. However, this study is not without its limitations. The sample size, comprising 100 positive and 96 negative serum samples, is relatively small, which may hinder the ability to make robust claims regarding diagnostic efficacy, particularly concerning the generalizability of results to diverse populations or various geographic regions where brucellosis is endemic. Additionally, it is crucial to consider the subcellular localization of candidate proteins, as this factor may significantly influence the immunogenicity of the fusion protein. Furthermore, the validity of the constructed iELISA method necessitates future comparisons with commercially available kits, and further validation through larger-scale clinical trials is essential to confirm the diagnostic performance of this novel antigen.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statementThe studies involving humans were approved by Ethics Committee of Xuzhou Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributionsQW: Methodology, Writing – original draft. YY: Methodology, Writing – review & editing. LG: Writing – review & editing. YX: Writing – review & editing. MY: Methodology, Writing – review & editing. DY: Conceptualization, Funding acquisition, Project administration, Writing – original draft.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Xuzhou Science and Technology Bureau (Grant number KC23306), the Medical Research Program of Jiangsu Commission of Health (Grant number Z2023080), Postgraduate Research & Practice Innovation Program of Jiangsu Province (Grant number KYCX23-2963), and QingLan Project of Jiangsu Province (2024). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
AcknowledgmentsWe thank the China Animal Health and Epidemiology Center for the gift of LPS, and the Xuzhou Center for Disease Control and Prevention for the gift of human brucellosis sera (positive and negative).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI was used in the preparation of this manuscript. Specifically, ChatGPT was primarily used for fine-tuning and proofreading the manuscript, particularly focusing on grammar, sentence structure, and the use of specialized terminology.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1509534/full#supplementary-material
Supplementary Material 1 | Sheet 1, OD450 of positive sera; sheet 2, OD450 of negative sera; sheet 3, data for Cross-Reactivity Assessment, sheet 3, 152 highly expressed proteins in Brucella wild strain.
Supplementary Material 2 | Results of TMT-Proteomics quality control analysis.
AbbreviationsTMT, Tandem Mass Tag; iELISA, indirect Enzyme-Linked Immunosorbent Assay; SAT, standard agglutination test; RBPT, rose bengal plate agglutination test; LPS, lipopolysaccharide; TSB, Tryptic Soy Broth; OD, optical density; AUC, area under the curve; ROC, receiver operating characteristic curve; ANOVA, One-way analysis of variance; MDH, Malate dehydrogenase; Cu-Zn SOD, Cu-Zn superoxide dismutase.
References2. Lai S, Zhou H, Xiong W, Gilbert M, Huang Z, Yu J, et al. Changing epidemiology of human brucellosis, China, 1955-2014. Emerg Infect Dis. (2017) 23:184–94. doi: 10.3201/eid2302.151710
PubMed Abstract | Crossref Full Text | Google Scholar
8. Bonfini B, Chiarenza G, Paci V, Sacchini F, Salini R, Vesco G, et al. Cross-reactivity in serological tests for brucellosis: a comparison of immune response of Escherichia coli O157:H7 and Yersinia enterocolitica O:9 vs Brucella spp. Vet Ital. (2018) 54:107–14. doi: 10.12834/VetIt.1176.6539.2
PubMed Abstract | Crossref Full Text | Google Scholar
9. Chart H, Okubadejo OA, Rowe B. The serological relationship between Escherichia coli O157 and Yersinia enterocolitica O9 using sera from patients with brucellosis. Epidemiol Infect. (1992) 108:77–85. doi: 10.1017/S0950268800049529
PubMed Abstract | Crossref Full Text | Google Scholar
10. Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J BioMed Inform. (2015) 53:405–14. doi: 10.1016/j.jbi.2014.11.003
PubMed Abstract | Crossref Full Text | Google Scholar
11. Kaewpongsri S, Sukasem C, Srichunrusami C, Pasomsub E, Zwang J, Pairoj W, et al. An integrated bioinformatics approach to the characterization of influenza A/H5N1 viral sequences by microarray data: Implication for monitoring H5N1 emerging strains and designing appropriate influenza vaccines. Mol Cell Probes. (2010) 24:387–95. doi: 10.1016/j.mcp.2010.08.006
PubMed Abstract | Crossref Full Text | Google Scholar
12. Nan J, Brostromer E, Liu XY, Kristensen O, Su XD. Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance. PloS One. (2009) 4:e7245. doi: 10.1371/journal.pone.0007245
PubMed Abstract | Crossref Full Text | Google Scholar
13. Deng H, Xue B, Wang M, Tong Y, Tan C, Wan M, et al. TMT-Based Quantitative Proteomics Analyses Reveal the Antibacterial Mechanisms of Anthocyanins from Aronia melanocarpa against Escherichia coli O157:H7. J Agric Food Chem. (2022) 70:8032–42. doi: 10.1021/acs.jafc.2c02742
PubMed Abstract | Crossref Full Text | Google Scholar
14. Sweredoski MJ, Baldi P. COBEpro: a novel system for predicting continuous B-cell epitopes. Protein engineering design selection: PEDS. (2009) 22:113–20. doi: 10.1093/protein/gzn075
PubMed Abstract | Crossref Full Text | Google Scholar
18. Khairullah AR, Kurniawan SC, Puspitasari Y, Aryaloka S, Silaen OSM, Yanestria SM, et al. Brucellosis: Unveiling the complexities of a pervasive zoonotic disease and its global impacts. Open Vet J. (2024) 14:1081–97. doi: 10.5455/OVJ.2024.v14.i5.1
PubMed Abstract | Crossref Full Text | Google Scholar
19. Jean Beltran PM, Federspiel JD, Sheng X, Cristea IM. Proteomics and integrative omic approaches for understanding host-pathogen interactions and infectious diseases. Mol Syst Biol. (2017) 13:922. doi: 10.15252/msb.20167062
PubMed Abstract | Crossref Full Text | Google Scholar
21. Zhang M, Han X, Liu H, Tian M, Ding C, Song J, et al. Inactivation of the ABC transporter ATPase gene in Brucella abortus strain 2308 attenuated the virulence of the bacteria. Vet Microbiol. (2013) 164:322–9. doi: 10.1016/j.vetmic.2013.02.017
PubMed Abstract | Crossref Full Text | Google Scholar
22. Faria AR, Dorneles EMS, Pires SDF, Andrade HM, Lage AP. Immunoproteomics of Brucella abortus reveals potential of recombinant antigens for discriminating vaccinated from naturally infected cattle. Microb Pathog. (2020) 147:104345. doi: 10.1016/j.micpath.2020.104345
PubMed Abstract | Crossref Full Text | Google Scholar
23. Kim JY, Sung SR, Lee K, Lee HK, Kang SI, Lee JJ, et al. Immunoproteomics of Brucella abortus RB51 as candidate antigens in serological diagnosis of brucellosis. Vet Immunol Immunopathol. (2014) 160:218–24. doi: 10.1016/j.vetimm.2014.05.009
PubMed Abstract | Crossref Full Text | Google Scholar
24. Al Dahouk S, Nöckler K, Scholz HC, Tomaso H, Bogumil R, Neubauer H. Immunoproteomic characterization of Brucella abortus 1119-3 preparations used for the serodiagnosis of Brucella infections. J Immunol Methods. (2006) 309:34–47. doi: 10.1016/j.jim.2005.11.003
PubMed Abstract | Crossref Full Text | Google Scholar
25. Oliveira SC, Harms JS, Banai M, Splitter GA. Recombinant Brucella abortus proteins that induce proliferation and gamma-interferon secretion by CD4+ T cells from Brucella-vaccinated mice and delayed-type hypersensitivity in sensitized Guinea pigs. Cell Immunol. (1996) 172:262–8. doi: 10.1006/cimm.1996.0241
PubMed Abstract | Crossref Full Text | Google Scholar
26. Delpino MV, Estein SM, Fossati CA, Baldi PC, Cassataro J. Vaccination with Brucella recombinant DnaK and SurA proteins induces protection against Brucella abortus infection in BALB/c mice. Vaccine. (2007) 25:6721–9. doi: 10.1016/j.vaccine.2007.07.002
PubMed Abstract | Crossref Full Text | Google Scholar
27. Stevens MG, Olsen SC, Pugh GW, Mayfield JE. Role of immune responses to a GroEL heat shock protein in preventing brucellosis in mice vaccinated with Brucella abortus strain RB51. Comp Immunol Microbiol Infect Dis. (1997) 20:147–53. doi: 10.1016/S0147-9571(96)00036-7
PubMed Abstract | Crossref Full Text | Google Scholar
28. Baloglu S, Toth TE, Schurig GG, Sriranganathan N, Boyle SM. Humoral immune response of BALB/c mice to a vaccinia virus recombinant expressing Brucella abortus GroEL does not correlate with protection against a B. abortus challenge. Vet Microbiol. (2000) 76:193–9. doi: 10.1016/S0378-1135(00)00231-5
PubMed Abstract | Crossref Full Text | Google Scholar
29. Andrade RS, Faria AR, Andrade HM, de Sousa Bueno Filho JS, Mansur HS, Mansur AAP, et al. Use of recombinant malate dehydrogenase (MDH) and superoxide dismutase (SOD) [CuZn] as antigens in indirect ELISA for diagnosis of bovine brucellosis. J Microbiol Methods. (2024) 217-218:106874. doi: 10.1016/j.mimet.2023.106874
PubMed Abstract | Crossref Full Text | Google Scholar
30. Im YB, Shim S, Park WB, Kim S, Yoo HS. Th2-related immune responses by the Brucella abortus cellular antigens, malate dehydrogenase, elongation factor, and arginase. Microb Pathog. (2017) 110:7–13. doi: 10.1016/j.micpath.2017.06.019
PubMed Abstract | Crossref Full Text | Google Scholar
31. Reyes AW, Simborio HL, Hop HT, Arayan LT, Kim S. Molecular cloning, purification and immunogenicity of recombinant Brucella abortus 544 malate dehydrogenase protein. J Vet Sci. (2016) 17:119–22. doi: 10.4142/jvs.2016.17.1.119
PubMed Abstract | Crossref Full Text | Google Scholar
32. Escalona E, Sáez D, Oñate A. Immunogenicity of a multi-epitope DNA vaccine encoding epitopes from cu-zn superoxide dismutase and open reading frames of brucella abortus in mice. Front Immunol. (2017) 8:125. doi: 10.3389/fimmu.2017.00125
PubMed Abstract | Crossref Full Text | Google Scholar
33. Liu X, Zhou M, Yang Y, Wu J, Peng Q. Overexpression of Cu-Zn SOD in Brucella abortus suppresses bacterial intracellular replication via down-regulation of Sar1 activity. Oncotarget. (2018) 9:9596–607. doi: 10.18632/oncotarget.24073
PubMed Abstract | Crossref Full Text | Google Scholar
34. Altamirano-Silva P, Meza-Torres J, Zúñiga-Pereira AM, Zamora-Jaen S, Pietrosemoli N, Cantos G, et al. Phenotypes controlled by the Brucella abortus two component system BvrR/BvrS are differentially impacted by BvrR phosphorylation. Front Microbiol. (2023) 14:1148233. doi: 10.3389/fmicb.2023.1148233
PubMed Abstract | Crossref Full Text | Google Scholar
35. Arenas-Gamboa AM, Rice-Ficht AC, Fan Y, Kahl-McDonagh MM, Ficht TA. Extended safety and efficacy studies of the attenuated Brucella vaccine candidates 16 M(Delta)vjbR and S19(Delta)vjbR in the immunocompromised IRF-1-/- mouse model. Clin Vaccine Immunol. (2012) 19:249–60. doi: 10.1128/CVI.05321-11
PubMed Abstract | Crossref Full Text | Google Scholar
36. Arenas-Gamboa AM, Ficht TA, Kahl-McDonagh MM, Gomez G, Rice-Ficht AC. The Brucella abortus S19 DeltavjbR live vaccine candidate is safer than S19 and confers protection against wild-type challenge in BALB/c mice whe
留言 (0)