The prevalence and fatality rate of cancer is consistently rising, resulting in cancer being the second most prominent cause of mortality globally. This alarming trend poses a significant risk to human well-being while also placing substantial healthcare and economic strain on society (1). Cancer is a pathological condition characterized by dysregulation across several biological levels, known as omics, and fundamental traits of cancer cells include sustained proliferation and mitosis. These traits are facilitated by the stimulation of growth signals that exert influence on the development and advancement of cancer (2).
B-cell receptor-associated protein 31 (BCAP31) was first identified and characterized by the research team led by Michael Reth in the year 1994 (3). The nomenclature of this entity was derived from its co-purification with immunoglobulin D, a component of B-cell receptors, as well as its manifestation of an observable molecular weight of 31 kDa on electrophoretic gels under denaturing conditions. BAP31, a transmembrane protein, has been promptly recognized as having widespread expression and mostly localizing in the Endoplasmic Reticulum (ER). Its primary roles in this cellular compartment include acting as a specific transmembrane protein chaperone as well as modulating apoptosis (4, 5).
Previous studies have shown an elevation of BCAP31 in several types of cancer, including cervical cancer (6), colorectal cancer (7), hepatocellular carcinoma (8), non-small lung cancer (9), gastric intestinal-type adenocarcinoma (10), as well as malignant melanoma (11). This elevation has been associated with a poor prognosis. It has been observed that low expression regarding BCAP31 corresponds with poor prognosis in colorectal cancer and hepatocellular carcinoma (12, 13). Furthermore, the act of eliminating it in U2OS osteosarcoma cells resulted in increased tumorigenicity both in-vitro as well as in a xenograft model. This effect was related to its role in suppressing autophagy (14). It was indicated that BCAP31 possesses the ability to function as a prognostic indicator for several types of malignancies.
Furthermore, it is noteworthy to mention that much research has shown an unexpected immunotherapy impact of BCAP31 on many types of malignancies. In the first stages of the study, it was discovered that the use of an immunotoxin that consists of Pseudomonas exotoxin A conjugated to a fragment of an anti-BAP31 antibody showed efficacy in inducing cell death in MDA-231 breast cancer cells when tested in a controlled laboratory setting (15). In a recent study, it was shown that vaccine immunotherapy utilizing BAP31-encoding plasmid resulted in a robust immunogenic reaction. This reaction effectively inhibited the development of tumors in a mouse model specifically designed to mimic malignant melanoma (11). Moreover, it has been demonstrated that the production of an intracellular antibody fragment against BAP31, known as an intrabody, effectively hinders the interaction with p27Kip1 and consequently inhibits the growth of mouse gastric cancer tumor xenografts. This inhibition was observed without any observable adverse effects (10). Additionally, the administration of the BAP31 antibody through interperitoneal injections demonstrated similar efficacy in limiting the development with regard to mouse hepatocellular carcinoma xenografts (8). Moreover, there is an increasing inclination towards using microRNAs for medicinal purposes (16). Prior research has demonstrated miR-362’s effectiveness in suppressing cancer development in mouse tumor xenografts via its targeting of BAP31 following transfection (17).
Nevertheless, most studies that have looked into how BCAP31 functions in malignancies thus far have only looked at a specific type of malignancy. There has not been a pan-cancer investigation into the relationship between BCAP31 and various tumors. As a result, we examined BCAP31 expression levels and their associations with prognosis in various malignancies using a number of databases, including UCSC, CCLE, as well as cBioPortal. Additionally, we examined potential relationships between BCAP31 expression and the Tumor Mutational Burden (TMB), copy number mutation, as well as immune infiltration in 33 distinct cancer types. We also performed enrichment analysis as well as co-expression studies with respect to immune-related genes with BCAP31 to investigate the biological roles of BCAP31 in malignancies. By influencing tumor-infiltrating immune cells and TMB, the outcomes we discovered demonstrate that BCAP31 can be utilized as a prognostic factor for several malignancies as well as can be a key player in tumor immunity. This investigation can shed light on BCAP31’s function in tumor immunotherapy.
Materials and methodsData collectionUtilizing the UCSC Xena platform, the RNA-sequencing samples derived from the Genotype-Tissue Expression (GTEx) as well as TCGA datasets were acquired (18). The CCLE database served as the source of information regarding every tumor cell line. Subsequently, the expression levels of these cell lines across 21 different tissues were examined and assessed based on their tissue origins. The data pertaining to the cohort undergoing immunotherapy was obtained by accessing the Supplementary Materials of a previously published study (19–21).
Cox regression analysis and survival analysisThrough the R software, Cox regression analysis was performed to investigate the relationship between BCAP31 expression as well as the disease-specific survival (DSS), overall survival (OS) and disease-free interval (DFI), in addition to the progression-free interval (PFI) of patients for various cancer types. The research utilized the The Cancer Genome Atlas (TCGA) datasets. Patients were divided into two groups based on BCAP31 expression levels using the median value as the threshold. The R packages “survival” as well as “survminer” were used to create the survival curves. We also looked at the relationship between BCAP31 expression as well as OS in a pan-cancer cohort using the R package “forestplot”.
Analysis of CNA in BCAP31The identification of copy number variations (CNVs) of BCAP31 in pan-cancer patients was conducted via the cBioPortal for Cancer Genomics platform, accessible at http://www.cbioportal.org/.
Tumor mutant burdenThe mutation data was acquired from the UCSC Xena. Subsequently, it was then processed using the Pearl programming language for extraction. Subsequently, we conducted an examination and integration of mutant data using the “maftools” software (22), followed by an analysis of the disparities in TMB between the BCAP31 high-risk as well as low-risk cohorts. esophageal squamous cell carcinoma (ESCA) samples were stratified into high and low groups based on the median expression level of BCAP31. Logistic regression was then employed to investigate the association between tumor mutation burden and ESCA cohort.
Gene set variation analysisThe use of GSVA was performed in order to examine the probable biological mechanism associated with BCAP31 across various types of cancer. The Molecular Signatures Database (Msigdb) provided the gene set for the Hallmark pathway (23). The R language “GSVA” package (24) was employed to determine the Hallmark pathway scores for every type of malignancy.
Immune feature analysisThe StromalScore, ImmunoScore, ESTIMATEScore, and TumorPurity scores have been generated using the ESTIMATE (25). Additionally, we used the approach proposed by Zeng et al. (26) to evaluate the TME’s biological mechanisms, including those connected to the immune system. Utilizing the Immune Cell Abundance Identifier (ImmuCellAI) as well as Tumor Immune Estimation Resource 2.0 (TIMER2.0) databases, the relationship between BCAP31 and immune cell infiltration was examined. Additionally, spearman test is utilized to examine the relationship between BCAP31 expression and immune-associated genes, for instance, Major Histocompatibility (MHC), chemokine-receptor, chemokine, in addition to immune-activating genes.
Drug sensitivity analysisNext, we investigated the correlation between BCAP31 expression as well as drug sensitivity using data obtained from the GDSC2 database. Following this, the relationship between medicines as well as the level of BCAP31 expression was investigated using spearman correlation analysis. The drugs exhibiting the highest six positive correlations were then presented.
Cell cultureKYSE-150 cell was obtained from Wuhan Punosai Life Technology Co., Ltd. KYSE-150 cell was grown in Roswell Park Memorial Institute-1640 (RPMI-1640) medium supplemented with 10% FBS and 1% streptomycin and penicillin. During cultivation at 37°C and 5.0% CO2, the KYSE-150 cell was frequently examined for mycoplasma contamination.
Human specimensThe study comprised a total of 5 tissue samples of ESCA and 5 corresponding paracancerous samples, 5 samples of lung adenocarcinoma (LUAD) and 5 corresponding paracancerous samples, and 5 samples of gastric adenocarcinoma (GA) and 5 corresponding paracancerous samples. All samples were collected from individuals who underwent surgery at the Affiliated People’s Hospital of Jiangsu University between April 2023 and September 2023. All procedures were conducted in accordance with relevant rules and regulations. The study and experimental procedures were approved by the Ethics Committee of the Affiliated People’s Hospital of Jiangsu University (Ethics Number: K-20230079-W), and informed consent was obtained from the patients.
Knockdown assay, cell transfectionTo generate BCAP31-knockdown KYSE-150 cells, small interfering RNA (siRNA) targeting BCAP31 (si-BCAP31) and a negative control siRNA (si-NC) were synthesized by Shanghai Jima Biotechnology Co., Ltd. Three knockdown sequences were constructed and transfected into KYSE-150 cells using Polyplus jetPRIME reagent (Polyplus, Illkirch, France) via liposome transfection. Transfection efficiency was verified by Western blot 48 hours post-transfection. All experiments were performed in triplicate, including both the BCAP31 knockdown group (si-BCAP31) and the negative control group (si-NC), to ensure statistical reliability.
Western blot analysisThe KYSE-150 cells were washed three times with ice-cold PBS and then lysed in RIPA buffer for 30 minutes at 4°C. Protein concentrations in the supernatants were determined using a bicinchoninic acid (BCA) protein assay kit after the lysates were centrifuged. Protein concentrations were then adjusted using lysis buffer. The samples were mixed with protein loading buffer and heated at 100°C for 10 minutes. Equal amounts of protein from all groups were separated on 12.5% SDS-PAGE and then transferred to PVDF membranes. The membranes were blocked at room temperature for 2 hours with 5% non-fat milk, followed by overnight incubation at 4°C with primary antibodies against BCAP31 and β-actin, each at a 1:1000 dilution. After three washes with TBST, the membranes were incubated with HRP-conjugated goat anti-rabbit or anti-mouse IgG for 2 hours at room temperature. Protein bands were then detected and quantified using ImageJ software.
MTT assayKYSE-150 cells were seeded into 96-well plates at a density of 1×104 cells per well in 100 μL of cell growth medium. Each group was organized into six replicate wells and incubated at 37°C in an incubator with 5% CO2. After the incubation period, 10 μL of MTT solution was added to each well, followed by an additional 4 hours of incubation. After horizontal centrifugation, the supernatant was carefully removed, and 150 μL of dimethyl sulfoxide (DMSO) was added to each well. The plates were then gently agitated for 10 minutes, and the optical density at 490 nm was measured using a microplate reader.
Colony formation assayEach group of KYSE-150 cells (si-BCAP31 and si-NC) was seeded in 6-well plates (500 cells per well) and cultured for 2 weeks to allow colony formation. Colonies were fixed with paraformaldehyde and stained with 0.4% crystal violet, making them visible for counting. Colony formation was assessed in triplicate wells for each group, and the entire experiment was independently repeated three times to confirm reproducibility.
Transwell assayThe Transwell migration and invasion assays were conducted using 24-well plates with Transwell inserts. Cells were counted and resuspended in serum-free RPMI-1640 medium at a concentration of 5×104 cells/mL. A 200 μL aliquot of the cell suspension was added to the upper chamber of the Transwell, while the lower chamber was filled with RPMI-1640 medium supplemented with 10% FBS as a chemoattractant. Both the BCAP31 knockdown group (si-BCAP31) and the negative control group (si-NC) were set up with triplicate wells for each experimental condition. Following 24 hours of incubation at 37°C in 5% CO2, migrated cells were fixed, stained with 0.1% crystal violet, and quantified. Images were captured from six randomly selected fields per well using an inverted microscope, and the experiment was independently repeated three times for consistency.
Wound healing assayThe studies were conducted according to the manufacturer’s instructions. Briefly, cells were seeded onto 24-well culture plates at a density of 2.0×105 cells per well and incubated at 37°C with 5% CO2 until they formed a monolayer. Once the monolayer was established, a scratch was made using a 1000 μL pipette tip, followed by two rinses with PBS. The medium was then replaced with RPMI-1640, and the cells were further incubated at 37°C with 5% CO2.
ImmunohistochemistryTo reduce endogenous peroxidase activity, paraffin-embedded ESCA tissues were dewaxed and incubated in 3% hydrogen peroxide at room temperature for 30 minutes. Antigen retrieval was performed by steam-heating the tissues in citrate buffer for 30 minutes. The tissues were then treated with 5% bovine serum albumin to block non-specific binding. Afterward, the BCAP31 antibody (1:200) was added, and the tissues were incubated overnight at 4°C. The following day, a secondary antibody was applied, and the tissues were incubated at 37°C for 30 minutes. Subsequently, the StreptAvidin Biotin Complex was added and incubated for 30 minutes at 37°C. After resin sealing, the tissue sections were treated with a diaminobenzidine substrate and counterstained with hematoxylin for microscopic examination.
Statistics analysisThe Statistical Product and Service Solutions (SPSS) software was used to perform the statistical calculations for the molecular biology verification. A two-tailed student’s t-test was the statistical analysis employed in the present investigation to evaluate and compare the differences between the two groups. To ascertain the statistical significance across numerous groups, the One-factor Analysis of Variance was employed. The data are presented in the form of means ± Standard Deviation (SD). Any difference with a P<0.05 was considered statistically significant.
ResultsThe expression profiles of BCAP31BCAP31 expression in different tumors as well as normal tissues was determined by analyzing mRNA data obtained from the UCSC Xena and CCLE databases. Based on the analysis of the UCSC Xena dataset, it was observed that BCAP31 exhibited differential expression across various tumor types. Specifically, high expression levels were detected in certain tumors, while low expression levels were observed in others. Notably, this pattern was not observed in mesothelioma (MESO), ovarian Serous cystadenocarcinoma (OV), sarcoma (SARC), pheochromocytoma and paraganglioma (PCPG), as well as uveal melanoma (UVM) (Figure 1A). These findings were generally consistent with the results obtained from the TCGA dataset (Figure 1B). The expression pattern of BCAP31 in GTEx cohorts was shown in Figure 1C, revealing a progressive rise in BCAP31 expression from the left to the right. Additional analysis of several cancer cell lines revealed that adipose tissue had the greatest level of gene expression, while muscle tissue exhibited the lowest level of gene expression. Additionally, an investigation was conducted on the BCAP31 protein expression via the CCLE database. The BCAP31 expression in cancer cells exhibited a five-fold rise, as seen in Figure 1D.
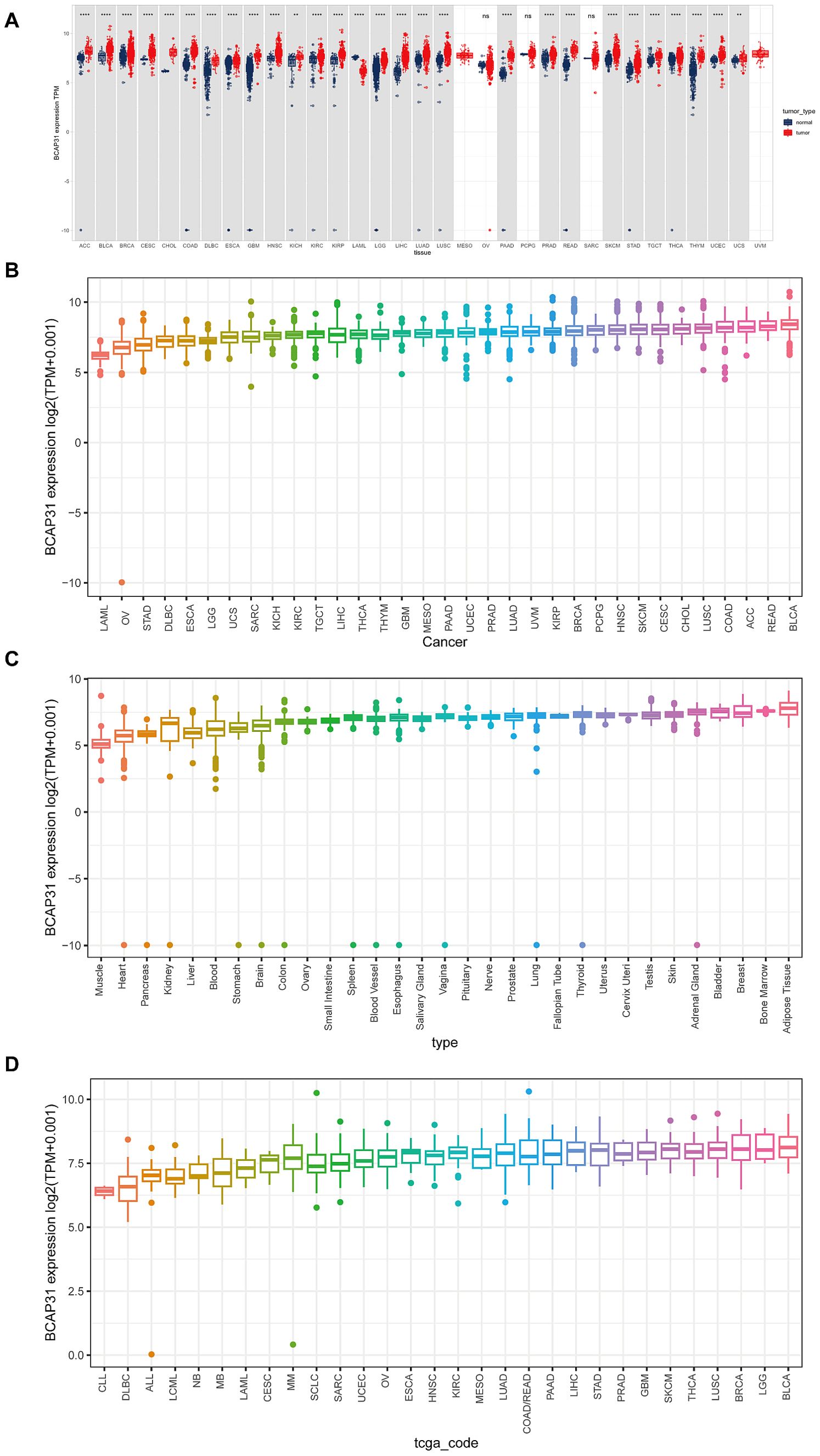
Figure 1. Differential expression of BCAP31 gene across various datasets. (A) Differential expression of the BCAP31 gene between tumor (red) and normal (blue) tissues across multiple tissue types. The y-axis represents log2(TPM + 0.001) values, with each box representing the distribution of expression levels for each cancer type. **p < 0.01, ****p < 0.0001, ns = not significant. (B) Expression boxplot of BCAP31 gene across different cancer types in the TCGA dataset. The y-axis shows log2(TPM + 0.001) values, with each box representing the distribution of expression levels for each cancer type. (C) Expression boxplot of BCAP31 gene across various tissue types in the GTEx dataset. The y-axis shows log2(TPM + 0.001) values, illustrating the expression distribution across normal tissues from different organs. (D) Expression boxplot of BCAP31 gene across different cancer cell lines in the CCLE dataset. The y-axis shows log2(TPM + 0.001) values, with each box representing the distribution of expression levels in specific cell lines.
Expression of BCAP31 in paired samples of 12 tumors in TCGA databaseAn analysis was conducted to examine BCAP31 expression across 33 distinct cancer types using TCGA-matched data. Significant differences in BCAP31 expression were observed in cholangiocarcinoma (CHOL), esca, bladder urothelial carcinoma (BLCA), kidney renal clear cell carcinoma (KIRC), adrenocortical carcinoma (ACC), colon adenocarcinoma (COAD), lung squamous cell carcinoma (LUSC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), uterine corpus endometrial carcinoma (UCEC), head and neck squamous cell carcinoma (HNSC), and rectum adenocarcinoma (READ) (Figures 2A–L). The corresponding paired tissue sample numbers for these analyses were 19, 9, 26, 13, 72, 32, 50, 50, 33, 7, 43, and 6, respectively.
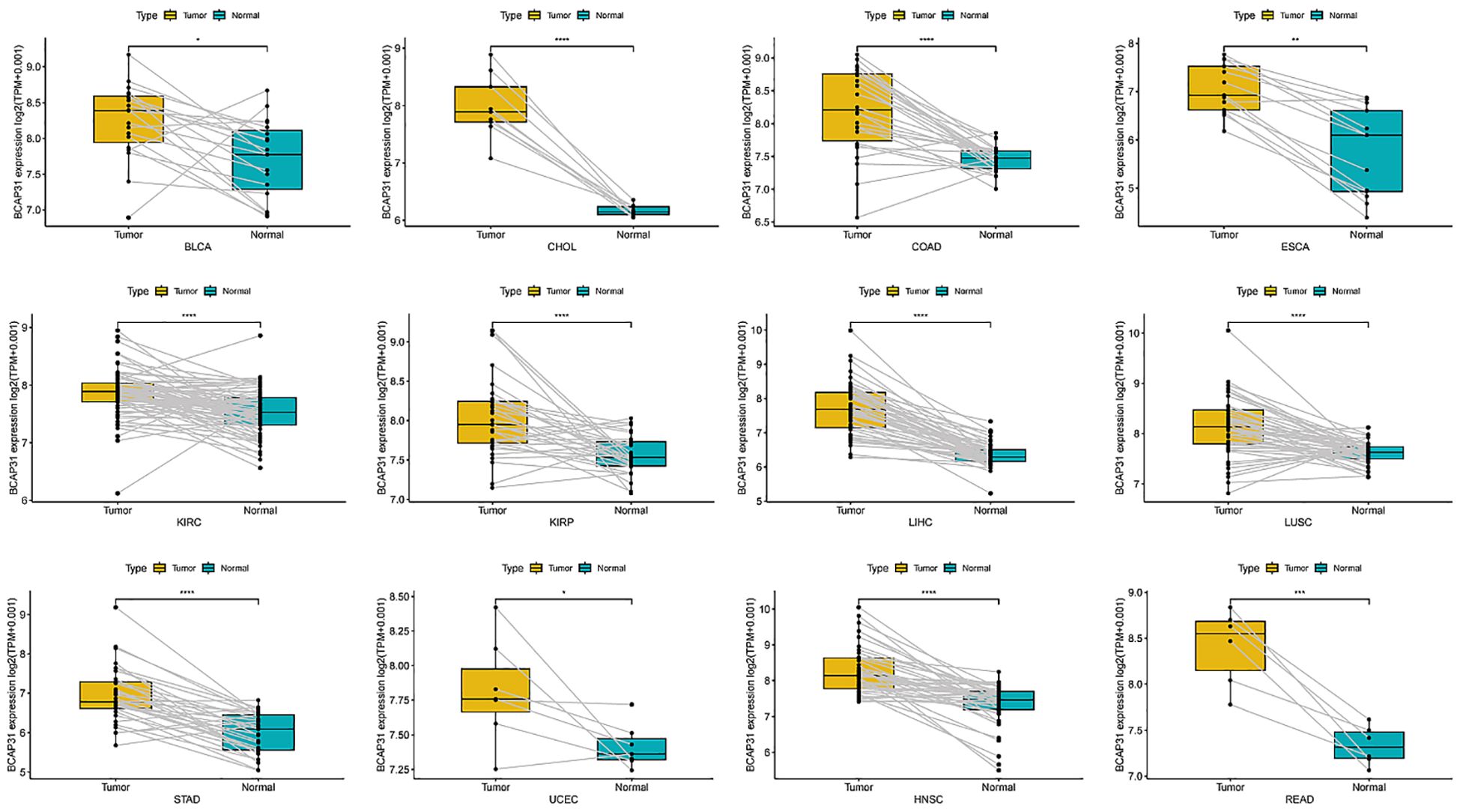
Figure 2. Differential expression of BCAP31 gene in paired tumor and adjacent normal tissues across various cancer types. Boxplots depict the log2(TPM + 0.001) expression values of BCAP31 in tumor (yellow) and matched adjacent normal (cyan) tissues. BLCA (Bladder Urothelial Carcinoma); CHOL (Cholangiocarcinoma); COAD (Colon Adenocarcinoma); ESCA (Esophageal Carcinoma); KIRC (Kidney Renal Clear Cell Carcinoma); KIRP (Kidney Renal Papillary Cell Carcinoma); LIHC (Liver Hepatocellular Carcinoma); LUSC (Lung Squamous Cell Carcinoma); STAD (Stomach Adenocarcinoma); UCEC (Uterine Corpus Endometrial Carcinoma); HNSC (Head and Neck Squamous Cell Carcinoma); READ (Rectum Adenocarcinoma). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Paired data points are connected by lines to show individual sample comparisons.
The correlation between BCAP31 expression and clinicopathologyWe investigated the relationship between BCAP31 expression and tumor stage across several cancer types, ranging from stage I to stage IV. Our results indicated a significant correlation between BCAP31 expression and tumor stage in specific cancers, including BLCA, breast invasive carcinoma (BRCA), HNSC, OV, READ, thyroid carcinoma (THCA), KIRP, and kidney chromophobe (KICH). Notably, in some cancer types, BCAP31 expression varied significantly between earlier and later stages, suggesting a potential role for BCAP31 in tumor progression (Figure 3).
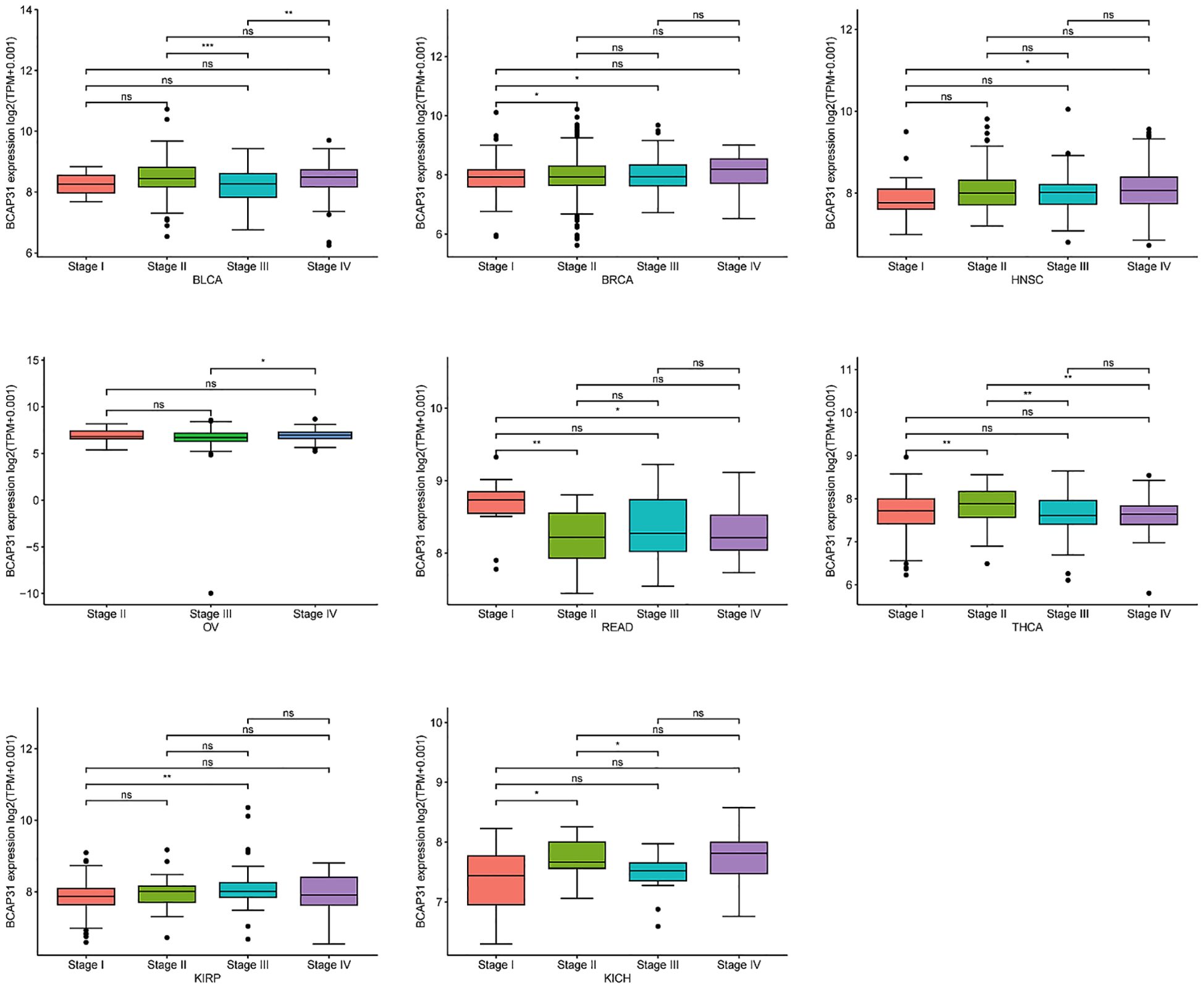
Figure 3. The expression of BCAP31 across different cancer stages in pan-cancer analysis. Boxplots show the log2(TPM + 0.001) expression levels of BCAP31 in various stages (Stage I, II, III, and IV) across different cancer types. BLCA (Bladder Urothelial Carcinoma); BRCA (Breast Invasive Carcinoma); HNSC (Head and Neck Squamous Cell Carcinoma); OV (Ovarian Serous Cystadenocarcinoma); READ (Rectum Adenocarcinoma); THCA (Thyroid Carcinoma); KIRP (Kidney Renal Papillary Cell Carcinoma); KICH (Kidney Chromophobe). *p < 0.05, **p < 0.01, ***p < 0.001, and “ns” for non-significant comparisons.
Prognostic value of BCAP31In order to examine the relationship between the BCAP31 expression level and prognosis, we used OS, DFI, DSS, as well as PFI as indicators to assess the prognostic implications of BCAP31 in several cancer types. The BCAP31 expression was demonstrated to be substantially linked with poor OS in many cancer types, including brain lower grade glioma (BLGG) (p < 0.001), HNSC (p < 0.001), BRCA (p = 0.001), glioblastoma multiforme (GBM) (p < 0.011), acute myeloid leukemia (LAML) (p = 0.022), ESCA (p = 0.032), and LUAD (p = 0.035) (Figure 4A). Pertaining to DSS, it was shown that greater expression with respect to BCAP31 was linked with shorter DSS in LGG (p < 0.001), HNSC (p < 0.001), GBM (p = 0.016), and BRCA (p = 0.047). In contrast, the downregulation of BCAP31 was shown to be associated with extended DSS in patients with STAD (p = 0.044), as seen in Figure 4B. Furthermore, the examination of DFI data demonstrated correlations between elevated BCAP31 expression as well as unfavorable prognosis in individuals diagnosed with COAD (p = 0.003), KIRC (p = 0.034), and CHOL (p = 0.044). Conversely, in patients with OV (p = 0.002), THCA (p = 0.003), SARC (p = 0.015), as well as STAD (p = 0.018), BCAP31 expression displayed a contrasting association with prognosis (Figure 4C). Lastly, the findings revealed that the overexpression of BCAP31 was associated with a significantly shorter PFI in patients with LGG (p < 0.001), HNSC (p = 0.002), ESCA (p = 0.028), and COAD (p = 0.039). On the contrary, the downregulation of BCAP31 was associated with a significant PFI in SARC (p = 0.012), OV (p = 0.015), and STAD (p < 0.021), as seen in Figure 4D.
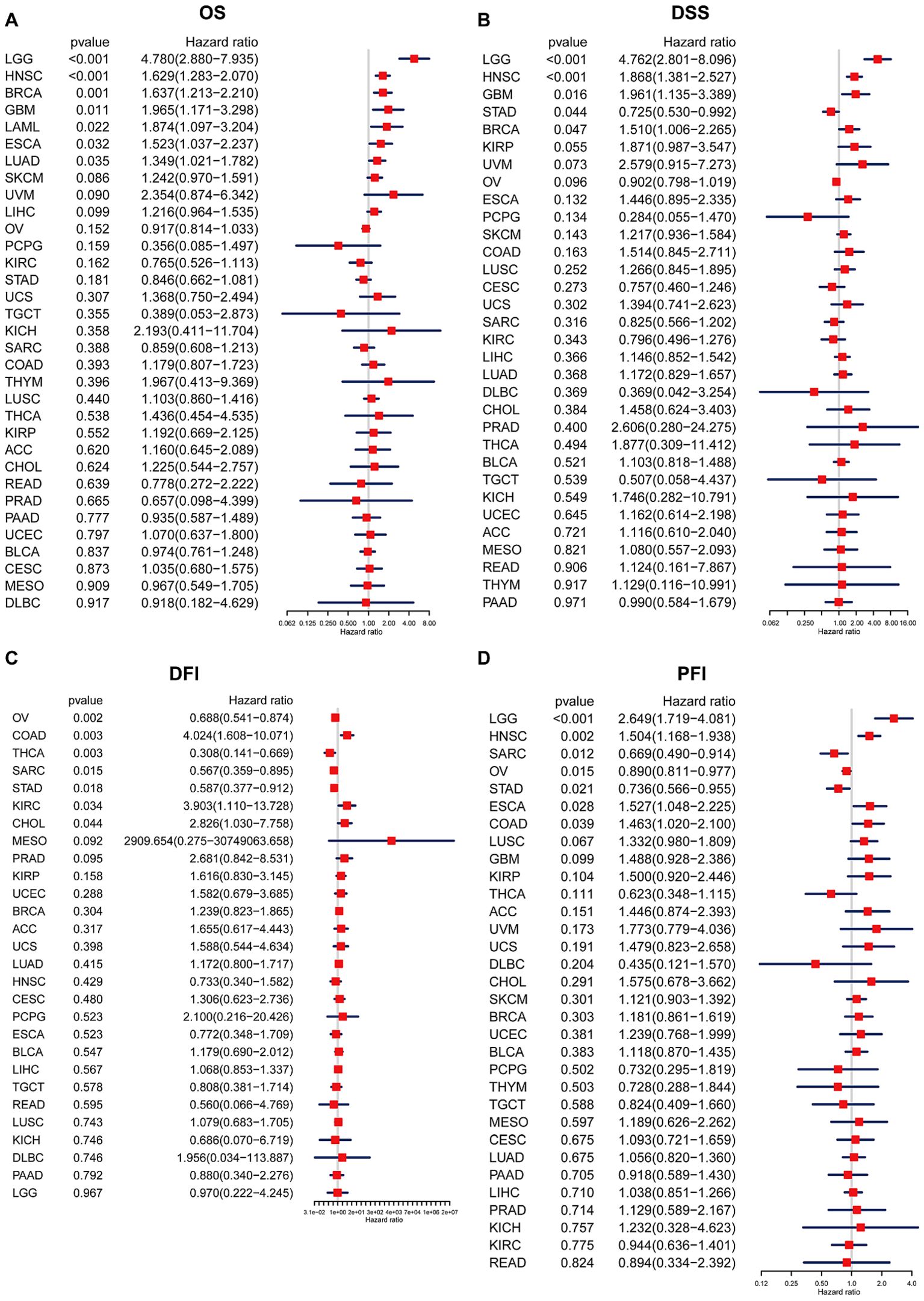
Figure 4. Association between BCAP31 expression levels and clinical outcomes in various cancer types. Forest plots illustrate the hazard ratios (HR) and p-values for the association of BCAP31 expression with four clinical outcomes. (A) OS (Overall Survival); (B) DSS (Disease-Specific Survival); (C) DFI (Disease-Free Interval); (D) PFI (Progression-Free Interval). The hazard ratio (HR) and its 95% confidence interval are presented for different cancer types. An HR > 1 indicates that higher BCAP31 expression is associated with an increased risk, while an HR < 1 suggests a protective effect. Red squares represent the HR values, with the horizontal blue lines indicating the 95% confidence intervals. Significant associations are highlighted based on the p-values provided.
Survival analysisSubsequently, a more thorough analysis of the relationship between BCAP31 expression as well as cancer prognosis was carried out utilizing the Kaplan-Meier plot. The outcomes regarding the correlation study showed a substantial relationship between BCAP31 expression as well as the prognosis of various cancer kinds. The findings indicated that the upregulation of BCAP31 expression was associated with an unfavorable prognosis in twelve distinct cancer types, namely ESCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), LIHC, BRCA, COAD, HNSC, GBM, LAML, KIRP, LGG, LUAD, and SKCM. Furthermore, the findings showed that survival rates were substantially lower for those in the high BCAP31 group than those in the low BCAP31 group (Figure 5).
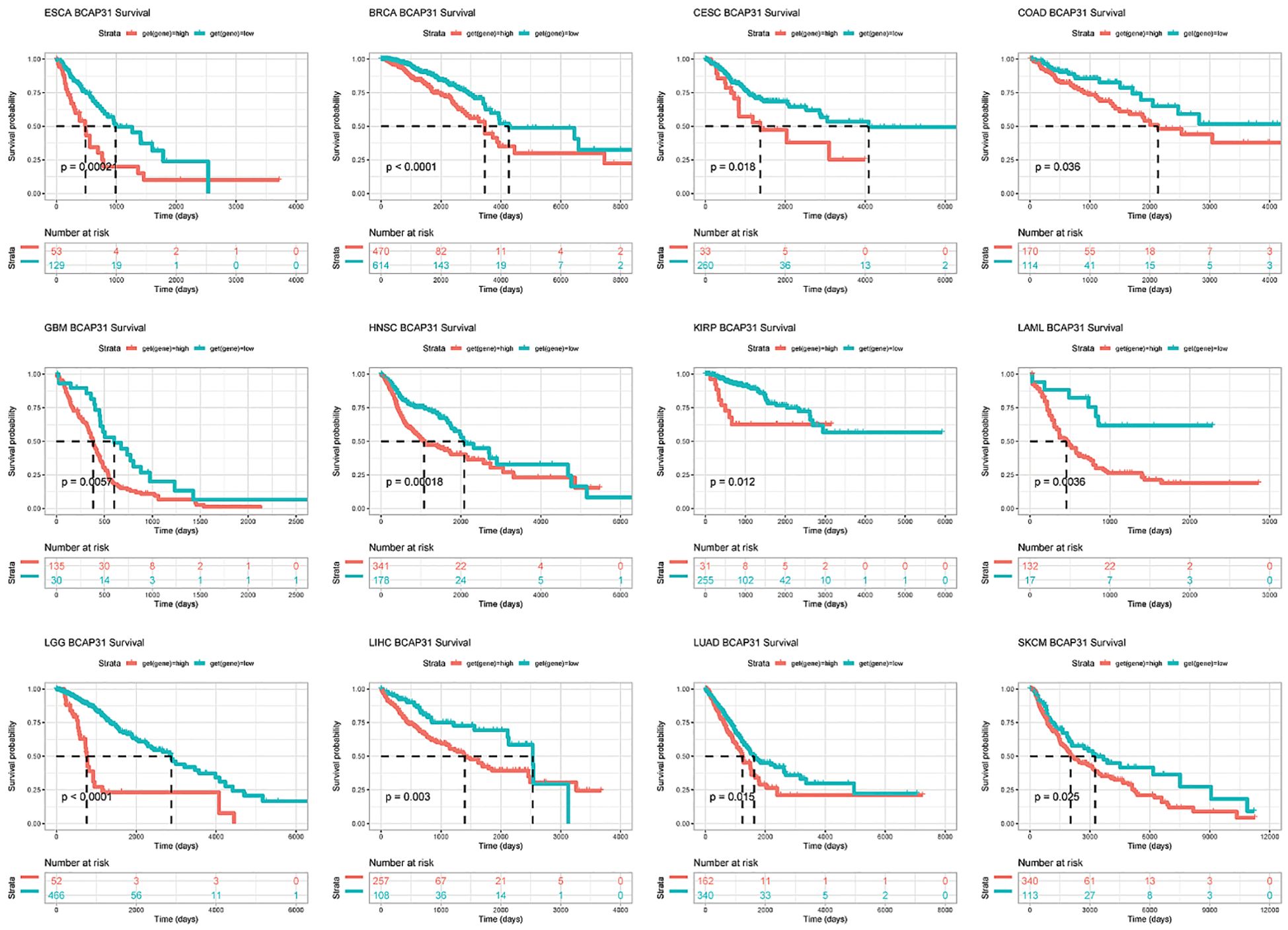
Figure 5. Kaplan-Meier survival curves for BCAP31 expression in various cancer types. Kaplan-Meier survival analysis was performed to assess the impact of high (red) versus low (blue) BCAP31 expression levels on overall survival across different cancer types. ESCA (Esophageal Carcinoma), p = 0.0002; BRCA (Breast Invasive Carcinoma), p < 0.0001; CESC (Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma), p = 0.018; ;COAD (Colon Adenocarcinoma), p = 0.036; GBM (Glioblastoma Multiforme), p = 0.005; HNSC (Head and Neck Squamous Cell Carcinoma), p = 0.0018; KIRP (Kidney Renal Papillary Cell Carcinoma), p = 0.012; LAML (Acute Myeloid Leukemia), p = 0.0036; LGG (Brain Lower Grade Glioma), p < 0.0001; LIHC (Liver Hepatocellular Carcinoma), p = 0.003; LUAD (Lung Adenocarcinoma), p = 0.015; SKCM (Skin Cutaneous Melanoma), p = 0.025. The x-axis represents time in days, and the y-axis represents survival probability. The Number at risk table below each plot indicates the number of patients remaining in each expression group at different time points, providing insight into survival trends based on BCAP31 expression levels.
Gene mutation analysis and tumor mutant burden analysisFollowing this, we performed an investigation into the correlation between BCAP31 as well as CNA in pan-cancer. The research’s findings show a substantial positive correlation between BCAP31 as well as CNA in a number of cancer types, which includes SARC, CESC, HNSC, thymoma (THYM), ESCA, STAD, LIHC, LUAD, CHOL, BLCA, OV, READ, BRCA, LUSC, UVM, pancreatic adenocarcinoma (PAAD), KIRC, as well as prostate adenocarcinoma (PRAD). Conversely, a negative correlation was observed in LAML and PCPG (Figure 6A), which aligns with the general BCAP31 expression pattern observed in various cancer types. Additionally, a relationship was found to be positive between BCAP31 expression as well as CNV in ESCA, as seen in Figure 6B. The gene mutation data for each kind of tumor was acquired from the UCSC Xena database. Subsequently, the R package “maftools” was used to generate visual representations of gene mutations within high and low expression categories. In the context of ESCA, the analysis revealed that the top ten genes that possessed the highest rates of mutation were TP53, DNAH5, FLG, CNTNAP5, TTN, CSMD3, MUC4, RYR2, MUC16, and SYNE1 in the BCAP31 high group. Conversely, the BCAP31 low group exhibited TP53, TIN, CSMD3, SYNE1, MUC16, FLG, MUC4, DNAH5, PCLO, and SPTA1 as the top ten genes (Figures 6C, D). Furthermore, the genes that exhibited variations in expression levels between the ESCA groups with high and low expression were identified as SDK1, CHD3, DCDC1, CNTNAP5, FCRL3, and NETO1, as seen in Figure 6E.
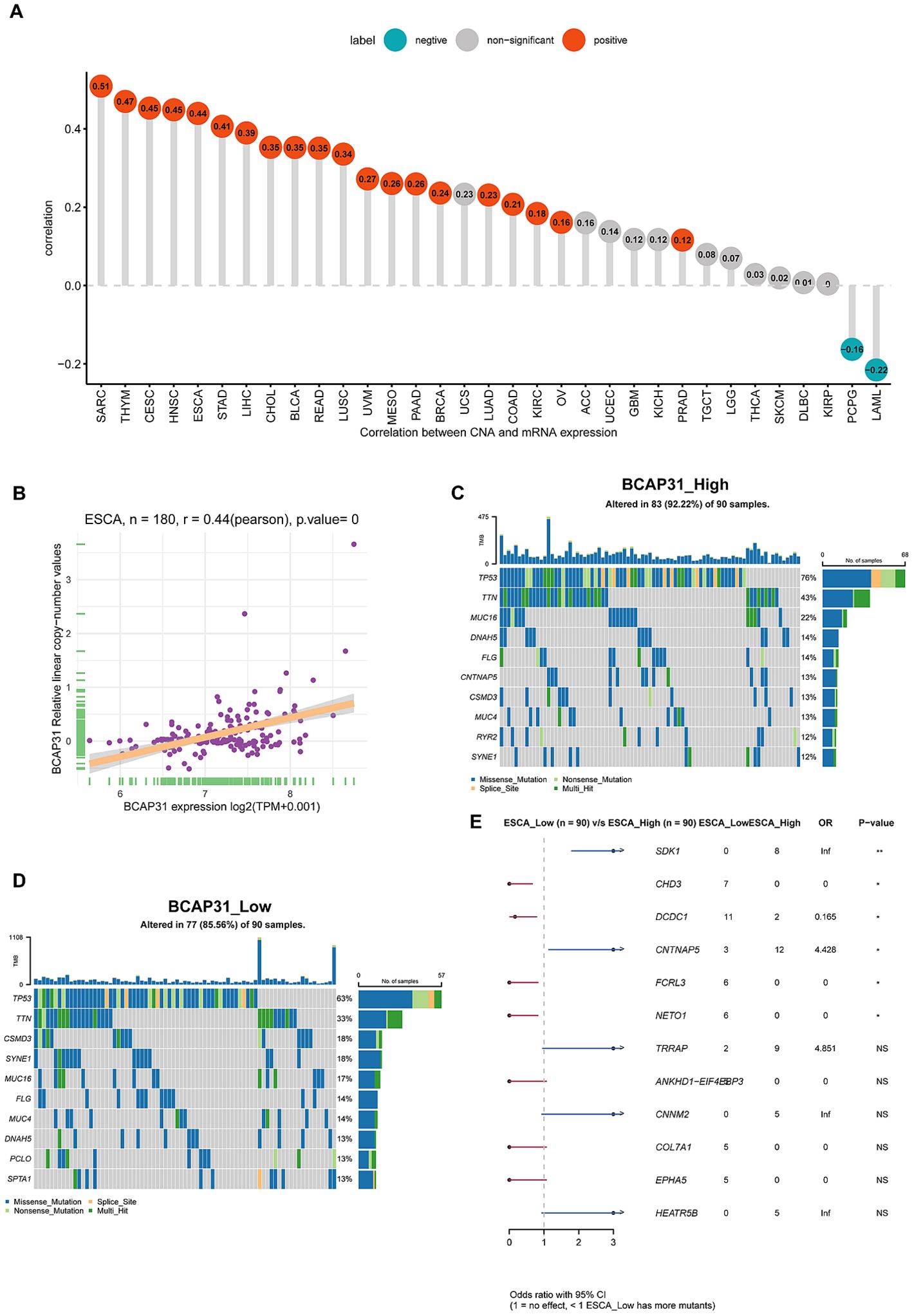
Figure 6. Correlations and genetic variations of BCAP31 in ESCA and pan-cancer analysis. (A) Bar chart showing the correlation between BCAP31 gene expression and copy number alterations (CNA) across various cancers, with green bars indicating negative correlations and red bars indicating positive correlations; (B) Scatter plot showing the correlation between BCAP31 expression and CNA specifically in ESCA (Esophageal Carcinoma), with a Pearson correlation coefficient of 0.44 (p < 0.001); (C, D) Mutation profiles of selected genes in high and low BCAP31 expression groups in ESCA, illustrating differences in mutation frequencies; (E) Forest plot displaying odds ratios and p-values for the comparison of gene mutation frequencies between high and low BCAP31 expression groups in ESCA. *p < 0.05, **p < 0.01. ns for non-significant comparisons.
GSVA analysisTo examine the possible involvement of BCAP31 in cancer, utilizing the GSVA was crucial to assess the associations between its gene expression and several biological signature pathways. In the context of GSVA, our findings indicate a significant association between BCAP31 and many pathways connected to proliferation and immunological response. These pathways include KRAS signaling up, PI3K AKT MTOR signaling, mTORC1 signaling route, TGF-β signaling, and IL-2 signaling pathway, as seen in Figure 7A. Furthermore, within the TCGA-ESCA cohort, there exists a positive correlation between the BCAP31 gene expression as well as the scores associated with various pathways. These pathways include G2M checkpoint, mTORC1 signaling, E2F targets, spermatogenesis, mitotic spindle, MYC targets v1, DNA repair, MYC targets v2, glycolysis, cholesterol, and homeostasis. Conversely, BCAP31 gene expression negatively correlates with the scores of the following pathways: UV response DN, myogenesis, TGF-beta signaling, inflammatory response, coagulation, androgen response, interferon gamma response, apoptosis, TNFA signaling via NFKB, KRAS signaling up, complement, allograft rejection, IL6 JAK STAT3 signaling, and IL2 STAT5 signaling (Figure 7B). The activation of tumor-related pathways, including those associated with the immune system, may contribute to worse patient outcomes in individuals with malignancies.
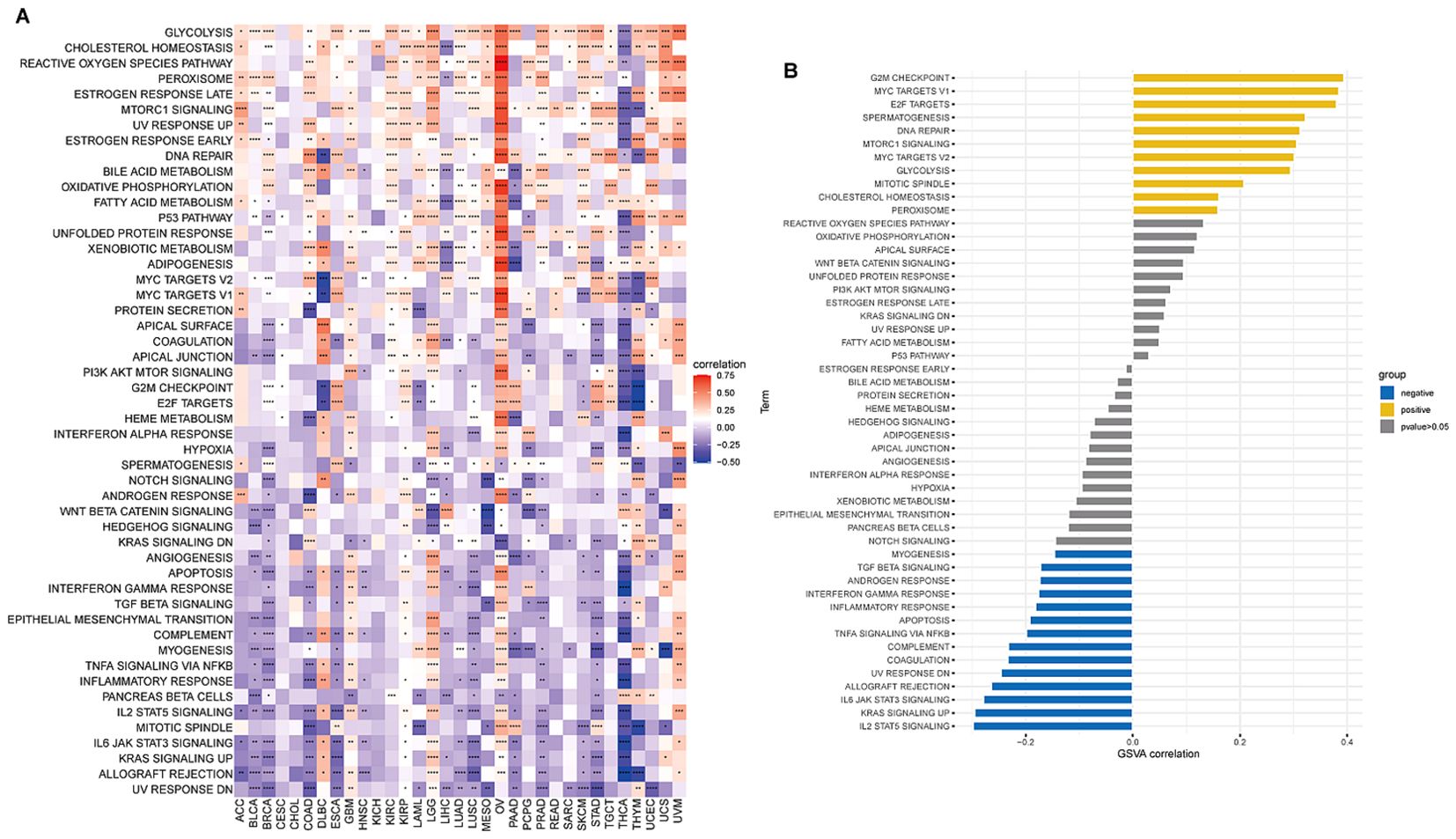
Figure 7. Results of GSVA (Gene Set Variation Analysis) for BCAP31 in pan-cancer and ESCA. (A) Heatmap showing GSVA enrichment of BCAP31 across various cancers, with red indicating a positive correlation and blue indicating a negative correlation. The color gradient ranges from purple (-0.5) to white (0) to red (0.75), representing the strength and direction of correlation between BCAP31 expression and pathway enrichment across different cancer types. (B) Bar plot displaying GSVA enrichment analysis results of BCAP31 specifically in ESCA (Esophageal Carcinoma), with pathways grouped by positive (yellow) and negative (blue) correlations, and gray indicating non-significant results (p-value > 0.05). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
ESTIMATE analysisThe behavior of cancer cells is significantly influenced by the surrounding cells within the tumor microenvironment (TME). Our analysis examined the relationship between BCAP31 expression and various factors, including stromal score, ESTIMATE score, immune score, and tumor purity. The results showed a negative correlation between BCAP31 expression and tumor purity in patients with DLBC, UVM, LGG, and GBM (Figure 8A). A positive correlation was observed between BCAP31 expression and immune scores in DLBC, UVM, LGG, GBM, and OV. Moreover, there was a significant correlation between BCAP31 expression and stromal scores, except in cancers such as OV, KIRP, UCS, SARC, CESC, PCPG, LAML, MESO, KIRC, TGCT, UCEC, READ, HNSC, CHOL, KICH, and THYM. Similarly, BCAP31 expression showed a significant correlation with ESTIMATE scores across several cancer types, including DLBC, UVM, LGG, GBM, and OV. Additionally, we calculated TME-related scores, including stromal and immune scores, as well as tumor purity, as illustrated in Figures 8B–E. These findings suggest that BCAP31 plays a critical role in the disruption of the TME, affecting both stromal and immune components.

Figure 8. Correlation between BCAP31 expression and tumor microenvironment scores in pan-cancer analysis. (A) Heatmap showing the correlation between BCAP31 expression and TumorPurity, ImmuneScore, StromalScore, and ESTIMATEScore across various cancers, with a color gradient from purple (-0.25) to white (0) to red (0.25) indicating the correlation strength and direction. (B) Radar plot illustrating BCAP31’s correlation with TumorPurity in different cancers; (C) Radar plot of StromalScore correlations; (D) Radar plot of ImmuneScore correlations; (E) Radar plot of ESTIMATEScore correlations. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TME analysisThis particular research’s outcomes illustrated a correlation that is positive between BCAP31 as well as the TME in several cancer types, including OV, LGG, UVM, PAAD, GBM, COAD, KIRP, KICH, TGCT, UCEC, and PCPG. However, an inverse correlation was seen between BCAP31 and TME in HNSC, BLCA, SARC, and SKCM (Figure 9A). Furthermore, it is crucial to highlight the differences in the TME between the high and low expression cohorts of BCAP31 in ESCA. In particular, the analysis reveals that DNA replication, mismatch repair, DNA damage response, base excision repair, and nucleotide excision repair signatures are significantly associated with the elevated expression observed in the BCAP31 high-expression group (Figure 9B).
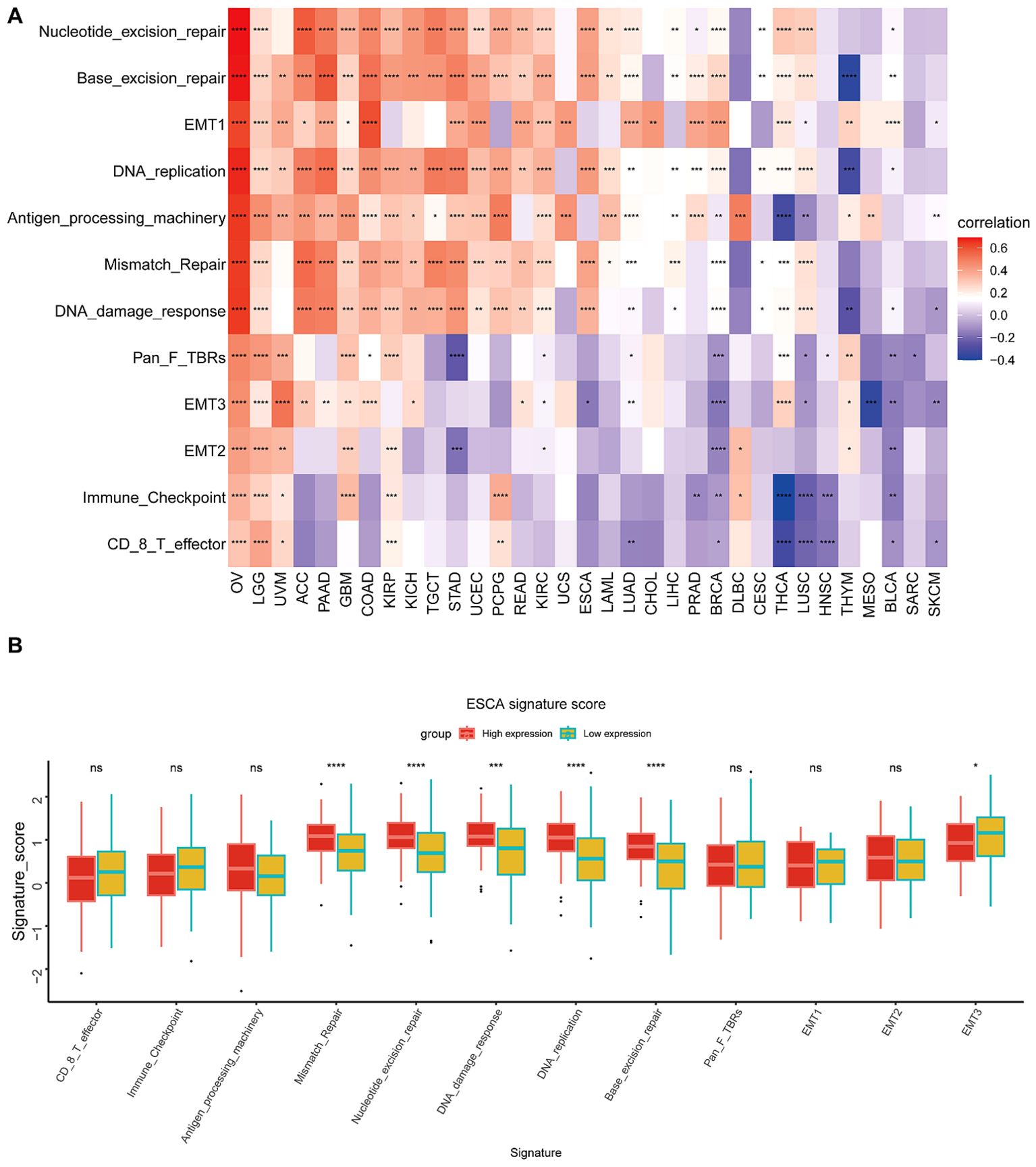
Figure 9. Tumor Microenvironment (TME) analysis of BCAP31 expression. (A) Heatmap showing the correlation between BCAP31 expression and various TME-related signatures across multiple cancer types. The color gradient ranges from purple (-0.4) to white (0) to red (0.6), representing the strength and direction of correlation between BCAP31 expression and TME components. (B) Boxplot showing the differences in TME signature scores between high and low BCAP31 expression groups in ESCA (Esophageal Carcinoma), with high expression in red and low expression in green. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TIMER2.0 analysisThe algorithms of TIMER2 were employed to assess the possible correlation between differences in the infiltration of different types of immune cells as well as the BCAP31 gene expression. In many types of tumors, BCAP31 expression shows a significant positive correlation with certain immune cells, such as myeloid cells, macrophages, neutrophils, and NK cells. Conversely, BCAP31 expression exhibits a significant negative correlation with other immune cells, including B cells, CD8+ T cells, regulatory T cells (Tregs), and follicular helper T cells. There is also a positive correlation between some tumors and cancer-associated fibroblasts (CAFs) (Figure 10).

Figure 10. Immune infiltration analysis. Correlation analysis between BCAP31 expression and immune cell infiltration across various cancer types, using data from the TIMER2 database. Red squares indicate a positive correlation, green squares indicate a negative correlation, and white squares indicate no significant correlation. The color gradient ranges from green (-0.4) to white (0) to red (0.2), representing the strength and direction of correlation. Black-bordered squares represent significant correlations with p < 0.05, while crossed squares (box with an “X”) represent non-significant correlations with p ≥ 0.05.
ImmueCellAI analysisEmerging studies have shown that tumor-infiltrating immunocytes might possess a significant influence on the OS outcomes of individuals. In this perspective, an examination was conducted on the relationships between the levels with regards to BCAP31 expression as well as the abundance of 25 different subtypes of immune cells that infiltrate at a pan-cancer level. The analysis was performed utilizing the ImmueCellAI database. Based on an analysis of data obtained from the ImmuCellAI database, results that we have obtained indicate a substantial positive correlation between BACP31 as well as various immune cell types, including Neutrophil, DC, Tem, macrophage, monocytes, nTreg, Th17, and NKT. In contrast, a correlation that is negative was discovered between BACP31 as well as Tgd, Tc, Tr1, CD8 T, NK, iTreg, Tcm, B cell, Tfh, and CD4 T (Figure 11A). These results suggest that BCAP31 may exert an immunosuppressive effect. In addition, we investigated the relationship between BCAP31 as well as immune cells in the TCGA-ESCA cohort. Our findings revealed a strong positive connection between BCAP31 and immune cell subsets such as Tem, neutrophil, monocyte, and DC. Conversely, BCAP31 displayed a negative correlation with B cell, Tr1, Tfh, Tcm, Tc, NK, iTreg, CD8 T, and CD4 T subsets (Figure 11B).
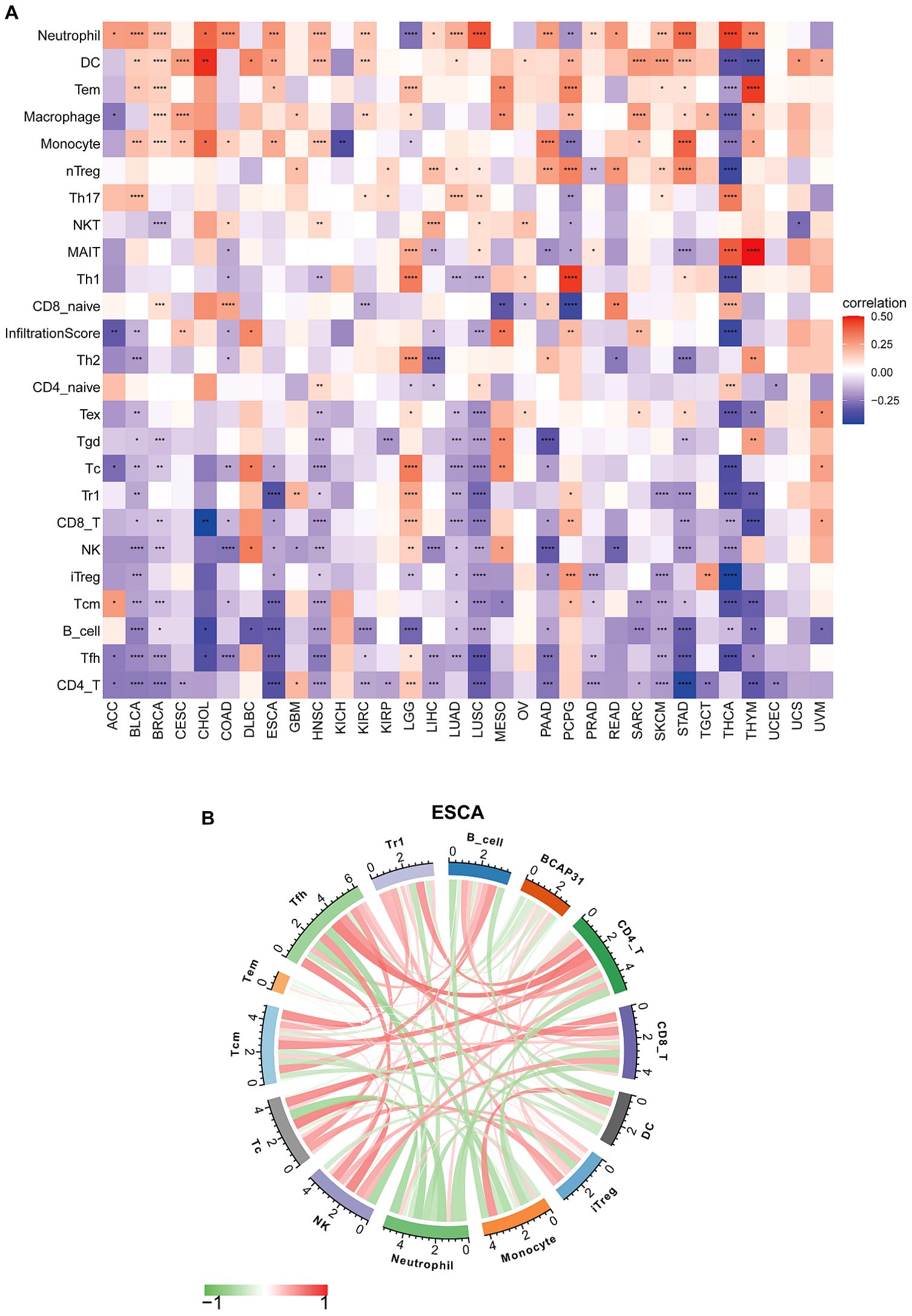
Figure 11. Immune cell infiltration analysis of BCAP31 expression. (A) Heatmap showing the correlation between BCAP31 expression and immune cell infiltration levels across various cancers using the ImmuCellAI database. The color gradient ranges from purple (-0.25) to white (0) to red (0.50), representing the correlation strength and direction. (B) Circos plot displaying immune cell infiltration analysis results specifically for BCAP31 in ESCA (Esophageal Carcinoma), with the color gradient ranging from green (-1) to red (1) indicating correlation values. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Immune-related genes analysis co-expression analysisTo attempt to get a deeper understanding of the regulatory mechanism behind tumor infiltration associated with BCAP31, this work examined the relationships between BCAP31 and four specific categories of immunomodulators: MHC genes, chemokine-receptor genes, chemokine genes, as well as immune-activating genes. These associations were analyzed using datasets obtained from TCGA. In addition, we performed gene co-expression studies in order to examine the associations between the BCAP31 expression as well as immune-related genes in a sample of 33 malignancies. The heatmap generated from the analysis revealed a significant co-expression between BCAP31 and almost all genes associated with the immune system. Furthermore, it has also been validated that the BCAP31 expression indicated a correlation that is positive with immunomodulators in OV, PCPG, LGG, KIRP, DLBC, GBM, UCEC, and UVM. Conversely, BCAP31 indicated a negative correlation with immunomodulators in ESCA, THCA, LUSC, CHOL, HNSC, BRCA, and STAD (Figures 12A–D).
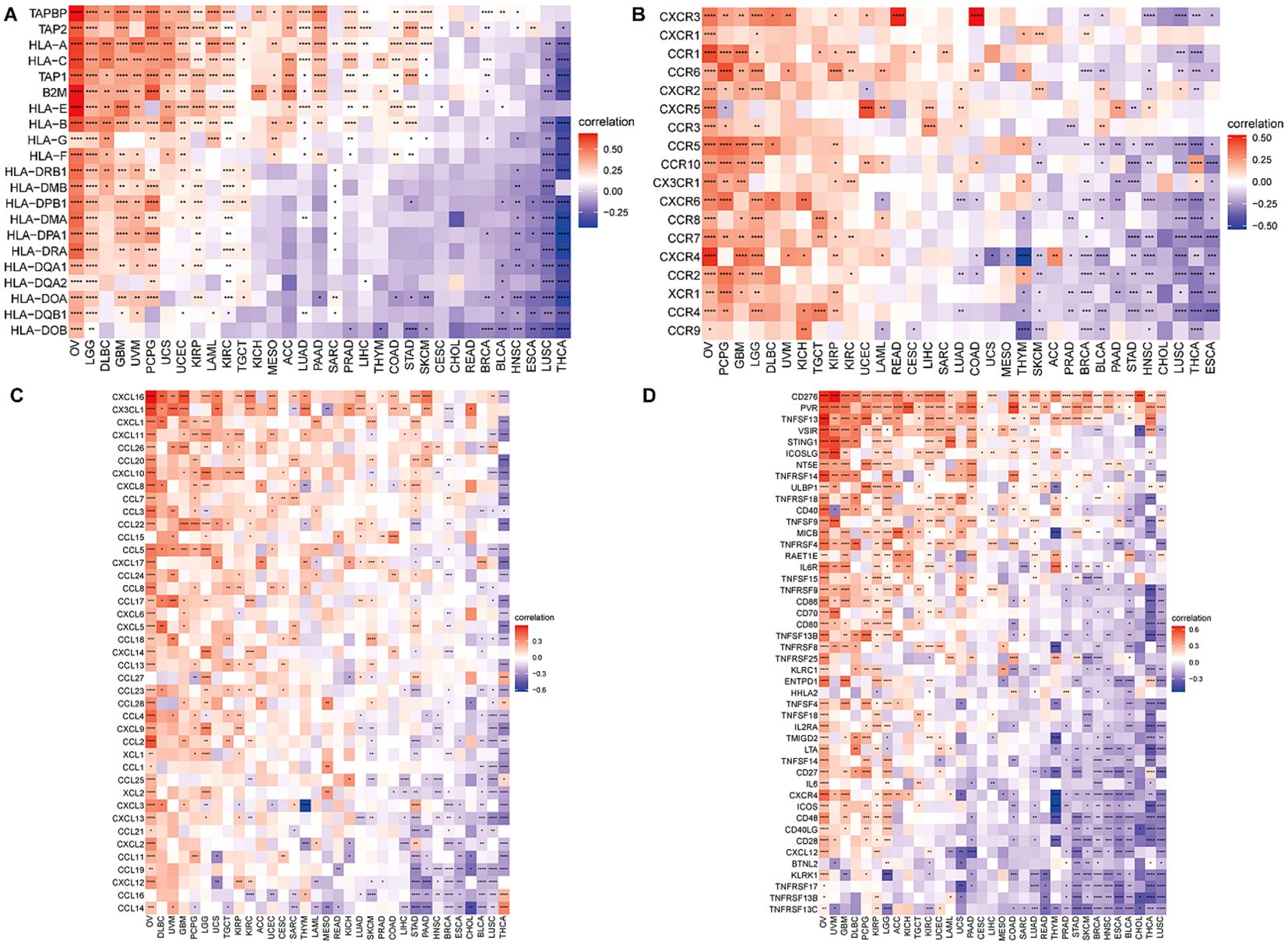
Figure 12. Correlation between BCAP31 and immune-related genes. (A) Heatmap showing the correlation between BCAP31 expression and MHC genes across various cancers, with a color gradient from purple (-0.25) to white (0) to red (0.50). (B) Correlation between BCAP31 and chemokine receptor genes, with the color gradient ranging from purple (-0.50) to white (0) to red (0.50). (C) Correlation between BCAP31 and chemokine genes, with a color gradient from purple (-0.60) to white (0) to red (0.30). (D) Correlation between BCAP31 and immune-activating genes, with the color gradient ranging from purple (-0.30) to white (0) to red (0.60). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Drug sensitivity analysisA comprehensive set of 198 pharmaceutical compounds has been discovered to have an association with BCAP31. The present study demonstrated the six medicines that had the most robust positive connection. The medicines shown to have a positive connection with BCAP31 include 5-Fluorouracil (R = 0.21), ABT737 (R = 0.25), Afuresertib (R = 0.2), AGI-5198 (R = 0.23), Alisertib (R = 0.29), and AGl-6780 (R = 0.25) (Figures 13A–F).
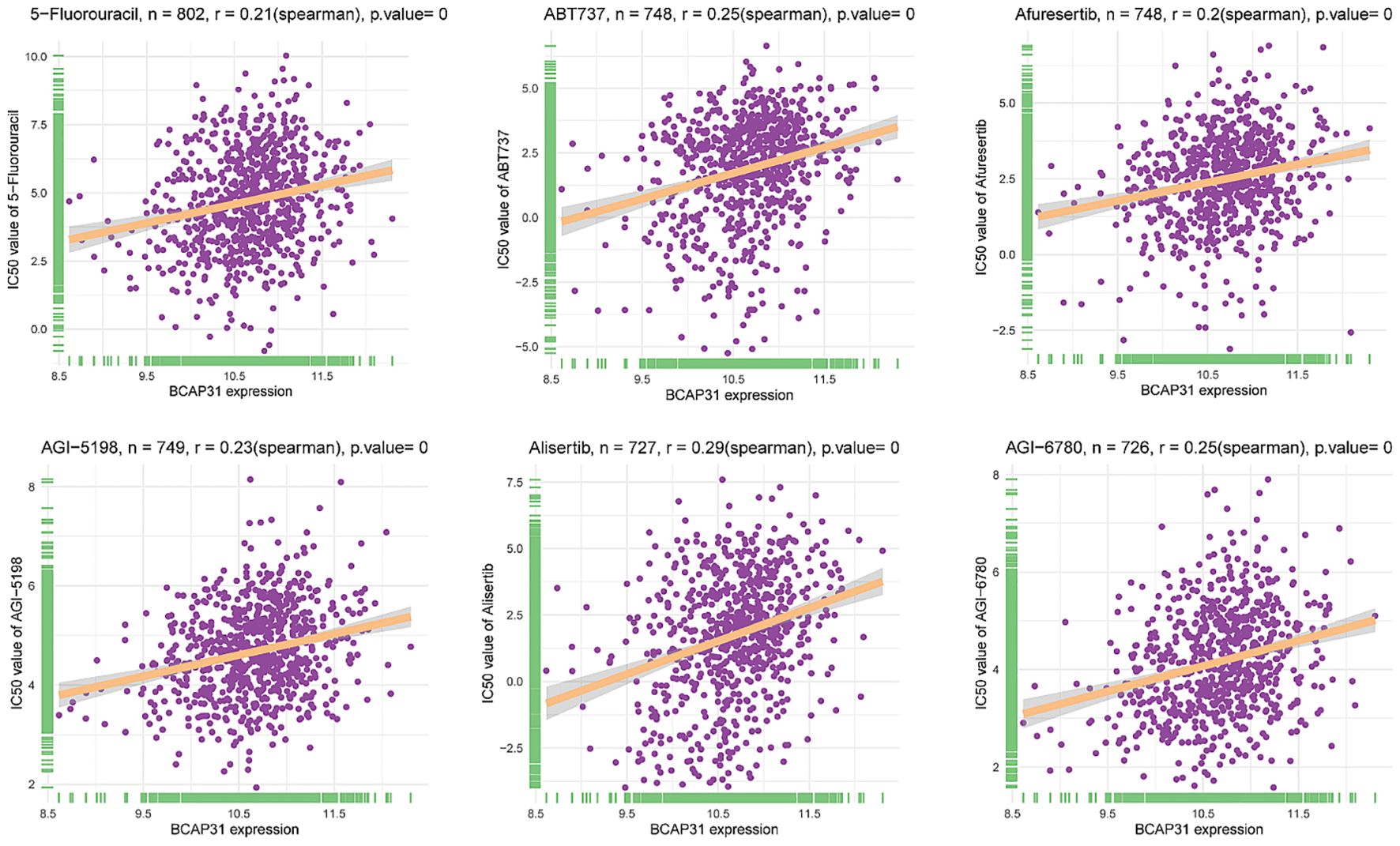
Figure 13. Correlation analysis between BCAP31 expression and drug sensitivity (IC50 values) in various compounds. Scatter plots illustrate the correlation between BCAP31 expression levels and IC50 values (drug sensitivity) for different compounds: 5-Fluorouracil (n = 802, r = 0.21, p < 0.001); ABT737 (n = 748, r = 0.25, p < 0.001); Afuresertib (n = 748, r = 0.20, p < 0.001); AGI-5198 (n = 749, r = 0.23, p < 0.001); Alisertib (n = 727, r = 0.29, p < 0.001); AGI-6780 (n = 726, r = 0.25, p < 0.001). Each plot shows a positive correlation between BCAP31 expression and drug IC50, suggesting that higher BCAP31 expression may be associated with increased resistance to these compounds. The Spearman correlation coefficient and p-value are indicated for each analysis.
Immunotherapy analysisPreliminary evidence suggests that BCAP31 has a significant impact on the immune cell infiltration in the TMB. Meanwhile, the efficacy of immunotherapy is directly linked with immunological checkpoint expressions on the surface of immune cells. To gain in-depth knowledge with respect to the possible correlation between the expression level of BCAP31 and the prognosis of tumor patients undergoing immunotherapy, we analyzed it in order to examine the variances in therapeutic response and prognosis across various subgroups of BCAP31 expression. This analysis was performed on three cohorts of patients who were receiving immunotherapy for melanoma (Figure 14A) and advanced renal cell carcinoma (Figures 14B, C). In accordance with our anticipated findings, a substantial correlation was discovered between elevated levels regarding BCAP31 expression. It diminished therapeutic response as well as worse outcomes among individuals with tumors who underwent immunotherapy.
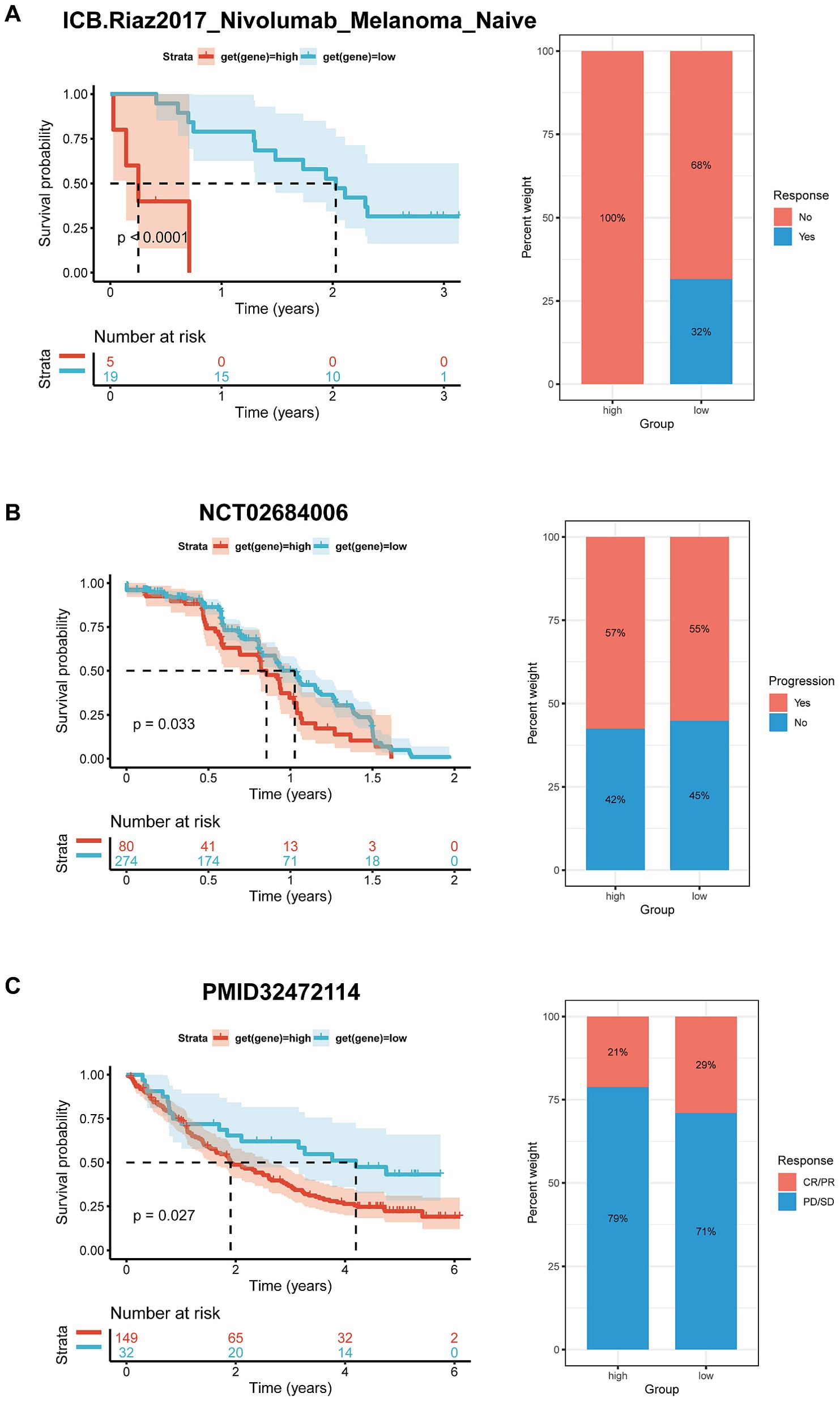
Figure 14. Correlation between BCAP31 expression and immunotherapy response. (A) Survival analysis and treatment response proportion in high and low BCAP31 expression groups in the ICB Riaz2017 Nivolumab Melanoma dataset, showing significant survival differences (p < 0.0001) and response rates with 100% non-responders in the high group vs. 68% in the low group. (B) Survival analysis and treatment response proportion in high and low expression groups in the NCT02684006 dataset, with a significant difference in survival (p = 0.033) and response rates (57% progression in high vs. 55% in low). (C) Survival analysis and treatment response proportion in high and low expression groups, showing significant survival differences (p = 0.027) and response rates with 21% responders in the high group vs. 29% in the low group. The left panels display Kaplan-Meier survival curves, and the right panels show bar charts representing the response proportions for each group.
Function assayWB analysis showed that BCAP31 was increased in four paired tissues, which was similar to the abovementioned bioinformatic result (Figure 15A). To explore the functions of BCAP31 in ESCA, we utilized a siRNA to reduce the BCAP31 expression levels in KYSE-150 cells. WB analysis showed that this siRNA could downregulate the expression of BCAP31 in the cell lines with high efficiency (Figure 15B). As a functional assay, transwell, as well as wound healing assays, were utilized to assess the impact of BCAP31 on the migration along with the metastatic capacity of KYSE-150 cells. Here, the findings demonstrated that migration as well as invasion with respect to BCAP31-downregulated KYSE-150 cells was sig
留言 (0)