Lung cancer is the most common cancer, and also the leading cause of cancer death in China (1). Non-small cell lung cancer (NSCLC) accounts for 75-80% of the total lung cancer cases, including adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma, large cell carcinoma, mucoepidermoid carcinoma, sarcomatoid carcinoma (2). Distant metastasis is the leading cause of most NSCLC deaths. Nearly one-third of patients with NSCLC have metastases at diagnosis, NSCLC often metastasizes to the brain, bone, pleural, liver and other organs (3–7).
The process of cancer metastasis and migration is a continuous process, where cancer cells can directly invade the surrounding organs/tissues, or invade the lymphatic system and vasculature and spread to other organs (8). Due to the complexity of the vascular and lymphatic systems, the site of metastasis is always unpredictable. One study showed that during tumor initiation, genomic instability causes various genetic mutations and leads to primary tumor cellular heterogeneity, thus increasing the possibility of tumor cell transformation, which is responsible for tumor spread and metastasis, such as immune evasion, epithelial-mesenchymal transition (EMT), and angiogenesis (9). Other studies have shown that the metastatic capacity of tumor cells is acquired through oncogene mutations in the initiation stage of early tumors and is stored in some subsets of tumor cells, known as “cancer stem cells” (10). Thus, somatic mutations seem to play a significant role in the tumor metastasis process in different cancer metastasis model studies.
With the rapid development of modern molecular biology techniques, more and more NSCLC driver mutations have been discovered, for example, epidermal growth factor receptor gene (EGFR), ananaplastic lymphoma kinase (ALK), rearrangements of the c-ros oncogene 1(ROS1), kirsten rat sarcoma viral oncogene homolog (KRAS), etc. Molecular genotyping has become critical in metastatic NSCLC, and the development of mutation-targeted therapies has revolutionized the treatment of NSCLC (11–14). With the continuous development of targeted therapy and immunotherapy, the overall prognosis of lung cancer is also constantly improving. To date, there is limited data on predicting or evaluating biological susceptibility factors at metastatic sites based on molecular profiles in NSCLC patients. A comprehensive analysis by Wu et al. found that both EGFR mutation and ALK mutations were associated with distant metastasis of NSCLC, however, no significant association was found between KRAS mutation and NSCLC metastasis (15). Several studies of ALK + NSCLC therapy have also shown that ALK + NSCLC has high rates of CNS metastasis even during first-line treatment with ALK TKI (16–19).
Based on these previous results, we propose that the biological predisposition to metastatic sites may differ between subsets of NSCLC molecules. There are no population-based data on the correlation of common gene mutation types with metastasis in NSCLC, especially the relationship between different gene mutation types and metastatic sites. This study focused on the analysis of the correlation between common gene mutation types and metastatic sites in NSCLC patients. These findings may provide a rationale for clinicians to develop immediate and accurate treatment plans for patients with lung cancer.
Materials and methodsPatientsA total of 1586 lung cancer patients who were treated in our hospital from December 2018 to August 2023 were enrolled in the study. This study was conducted with the approval of the Ethics Committee of Liaocheng People’s Hospital (NO. 2024086), and we guaranteed confidentiality throughout the study. Patient data were collected from our institutional cancer registry database and from patient follow-up visits to our outpatient office. The information collected from the records included patient characteristics, pathological diagnosis, metastasis site and follow-up data. Cancer staging was performed according to the Tumor node metastasis (TNM) classification of the International Union Against Cancer, 8th edition and histological typing was based on the World Health Organization classification.
Reagents and gene mutation detectionDNA and RNA were extracted separately from paraffin-embedded lung cancer tissues using an FFPE DNA/RNA extraction kit (AmoyDx, Xiamen, China). Sample processing and DNA/RNA extraction were performed according to the manufacturer’s protocol. The EGFR, ALK, ROS1, rearranged during Transfection (RET), mesenchymal-epithelial transition factor (MET), v-raf murine sarcoma viral oncogene homolog B (BRAF), human epidermal growth factor receptor 2 (HER2), KRAS, neuroblastoma RAS viral oncogene homolog (NRAS), and phosphatidylinositol 3-kinase (PIK3CA) gene mutations were detected using the human lung cancer multi-gene mutation detection kit (fluorescence PCR method) (AmoyDx, Xiamen, China). Detailed Gene test sites as listed in Supplementary Material, Table 1. Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS-PCR) was conducted on a cobas Z480 platform (Roche, Switzerland), and the amplification program was set up as follows: Phase 1: 5 min at 42°C and 5 min at 95°C; Phase 2: 25 s at 95°C, 20 s at 64°C, 20 s at 72°C for 10 cycles; Phase 3: 25 s at 93°C, 35 s at 60°C, 20 s at 72°C for 36 cycles; signal collection: The FAM and VIC signals were collected at 60°C in phase 3.The experiment process was performed according to the product instructions.
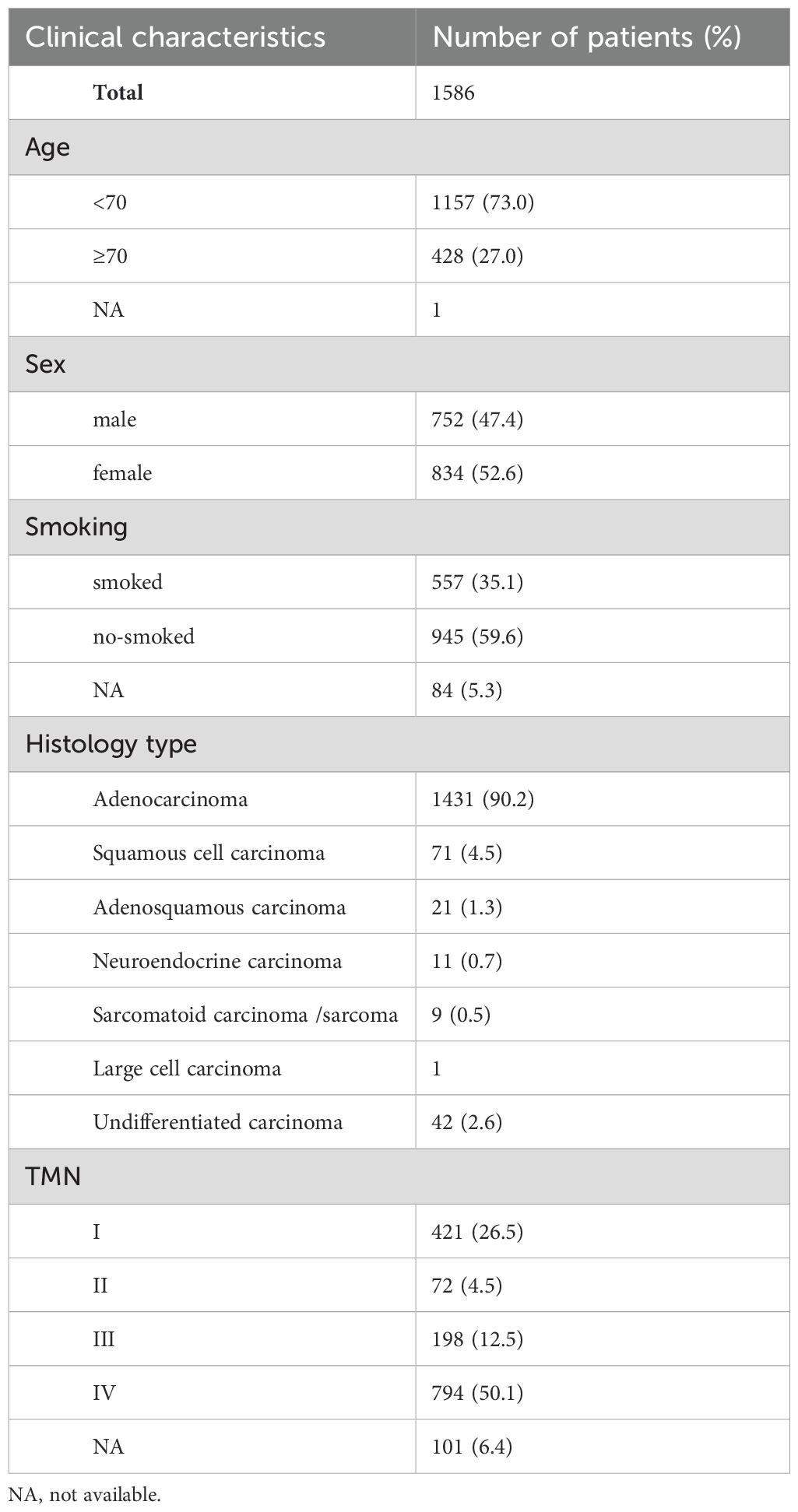
Table 1. Basic clinical information of 1586 cases of patients with non-small cell lung cancer.
Statistical analysisStatistical analysis performed by IBM SPSS statistics 27.0. Enumeration data expressed as numbers, the association between different groups were analyzed by Chi-square test. When the chi-square test was not met, Fisher’s exact probability test was used. Statistical significance was set at P value less than 0.05.
ResultsPatient characteristicsThe clinicopathological characteristics of the 1586 patients are summarized in Table 1. Tissue types included 684 surgical resection specimens, 667 bronchoscopic biopsy or crude needle biopsy specimens and 194 pleural effusion or pericardial effusion specimens. Among the 1586 patients, 752(47.4%) were males and 834 (52.6%) were females, with an average age of 62.8 years (range = 23–92 years). Among the 1586 lung cancer cases, 84 had an unknown smoking history, 557 (35.1%) were smokers, and 945 (59.6%) were non-smokers. Tumor staging was performed according to the 8th edition of Lung cancer staging criteria for IASLC (international association for the study of lung cancer); 421 cases (36.5%), 72 cases (4.5%), 198 cases (12.5%) and 794 cases (50.1%) were in stages I, II, III, and IV, respectively, and 101 cases staging were unknown. The pathological types of these patients were mostly adenocarcinoma (1431 cases, 90.2%), squamous cell carcinoma (71 cases, 4.5%), adenosquamous carcinoma (21 cases, 1.3%), neuroendocrine carcinoma (11 cases, 0.7%), sarcomatoid carcinoma and sarcoma (8 cases, 0.5%), large cell carcinoma (1 cases, 0.06%), and undifferentiated carcinoma (42 case, 2.6%).
Molecular characteristicsAs shown in Figure 1, among 1586 patients with non-small cell lung cancer, 1198 (75.5%) had genetic mutations detected, and the most common genetic mutation was EGFR (872,55.0%). The frequency of mutations in the other genes were KRAS (120,7.6%), ALK (62,3.9%), HER2 (46,2.9%), RET (24,1.5%), MET (22,1.4%), BRAF (17,1.1%), ROS1(16,1.0%), NRAS (5,0.3%) and PIK3CA (4,0.3%) (Figure 1). As shown in Figure 1, in patients with the EGFR mutation, the most common type of EGFR mutation was the L858R point mutation (421 cases, 26.5%). The deletion of exon 19 was detected in 367 (23.1%) patients, T790M mutation in 13 (0.8%) patients (19Del+T790M 7 cases, L858R+T790M 5 cases, L858R+G719X+T790M 1 case), and other rare EGFR mutations were detected in 59 (3.7%) patients (exon 20 insertion 29 cases, G719X 19 cases, L861Q 10 cases, S768I 1 case). Two types of EGFR mutations were detected in 12 (0.8%) patients (L858R+19Del 6 cases, G719X+S768I 4 cases, 19Del+20Ins 1 case, and L858R+S768I 1 case) (Figure 1). Two genetic mutations were detected in 10 patients. Combined mutations in EGFR and other genes were detected in 6 (0.4%) patients (L858R+MET, L858R+RET, L858R+KRAS, L858R+ PIK3CA, 20Ins+PIK3CA, 19Del+PIK3CA) (Figure 2). Additional combinations of genes were detected in four patients (KRAS+PIK3CA, KRAS+ROS1, HER2+PIK3CA, RET+BRAF).
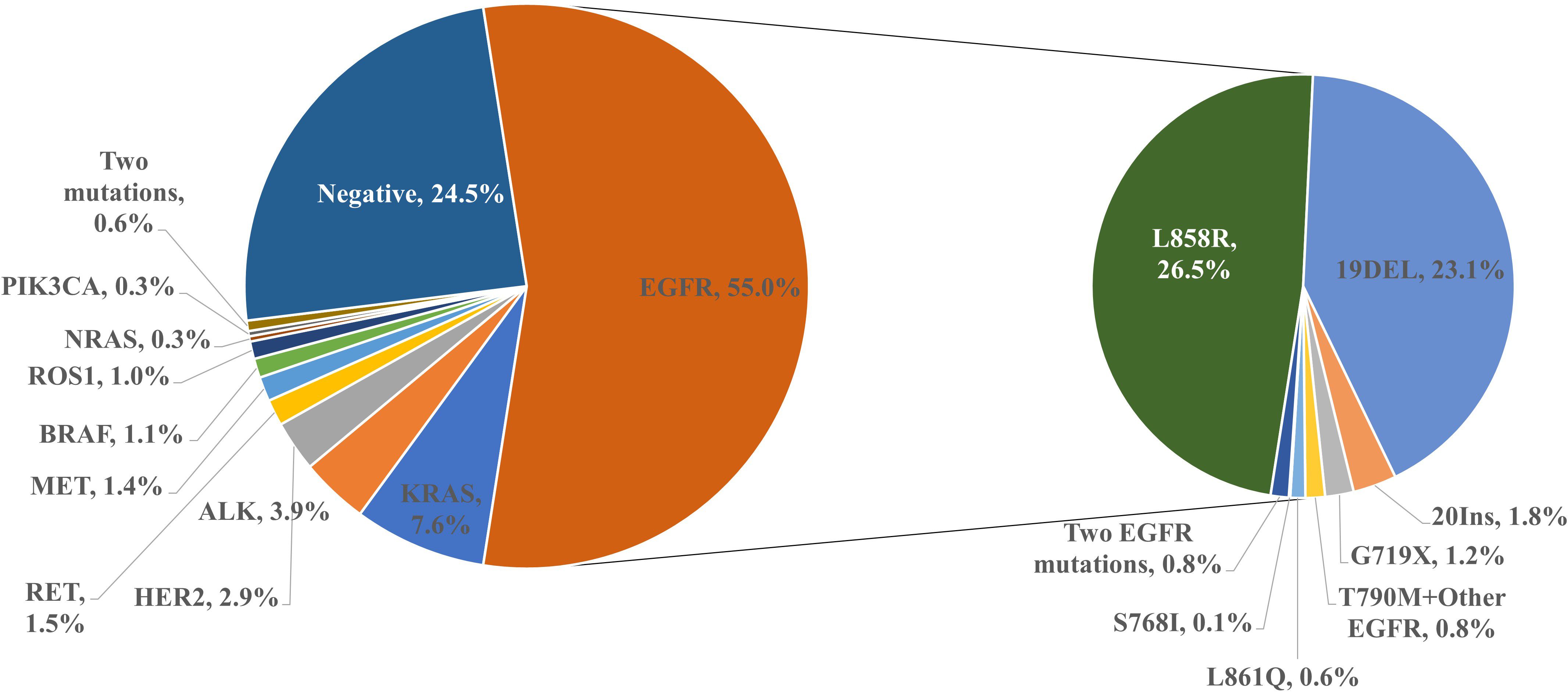
Figure 1. Frequency of detection of gene mutation in non-small cell lung cancer.
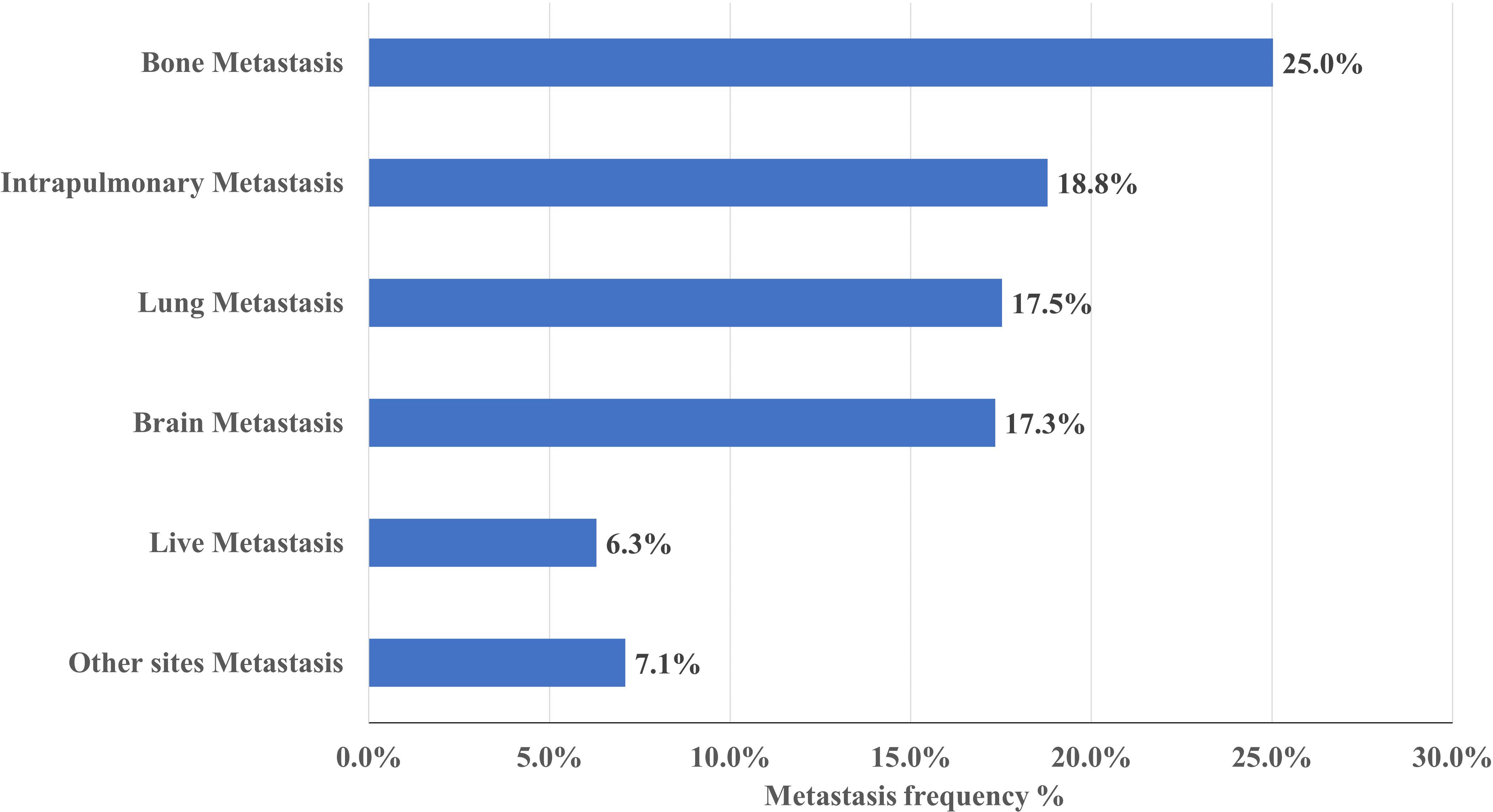
Figure 2. Frequency of metastases in NSCLC. Bone is the most common site of metastasis, occurring in approximately 25% of NSCLC.
Metastasis siteAs shown in Figure 2, of the 1586 NSCLC cases, 796(50.2%) patients had metastasis, 397(25.0%) patients had bone metastasis, 298(18.8%) patients had intrapulmonary metastasis, 278(17.5%) patients had lung metastasis, 275(17.3%) patients had brain metastasis, 100(6.3%) patients had liver metastasis and 112 (7.1%) patients had other metastatic sites.
Analysis of molecular features with metastatic sitesAs shown in Tables 2, 3, 431 (49.1%) patients in the EGFR mutation group developed metastasis, lung metastasis (166 cases, 18.9%, P = 0.004), brain metastasis (166 cases, 18.9%, P = 0.001) and bone metastasis (238 cases, 27.1%, P =0.004)were more common in the EGFR mutation group than in the wild-type group. The EGFR 19Del mutation group had more common lung metastasis (88 cases, 24%, P < 0.001), bone metastasis (94 cases, 25.6%, P = 0.048) and pleural metastasis (85 cases, 23.2%, P = 0.027). The EGFR L858R mutation group was more prone to metastasize to brain (89, 21.1%, P = 0.001) and bone (121 case, 28.7%, P = 0.002). In addition, no difference in the EGFR rare mutation group and 19Del/L858R+T790M mutation group compared with wild-type group metastasis.
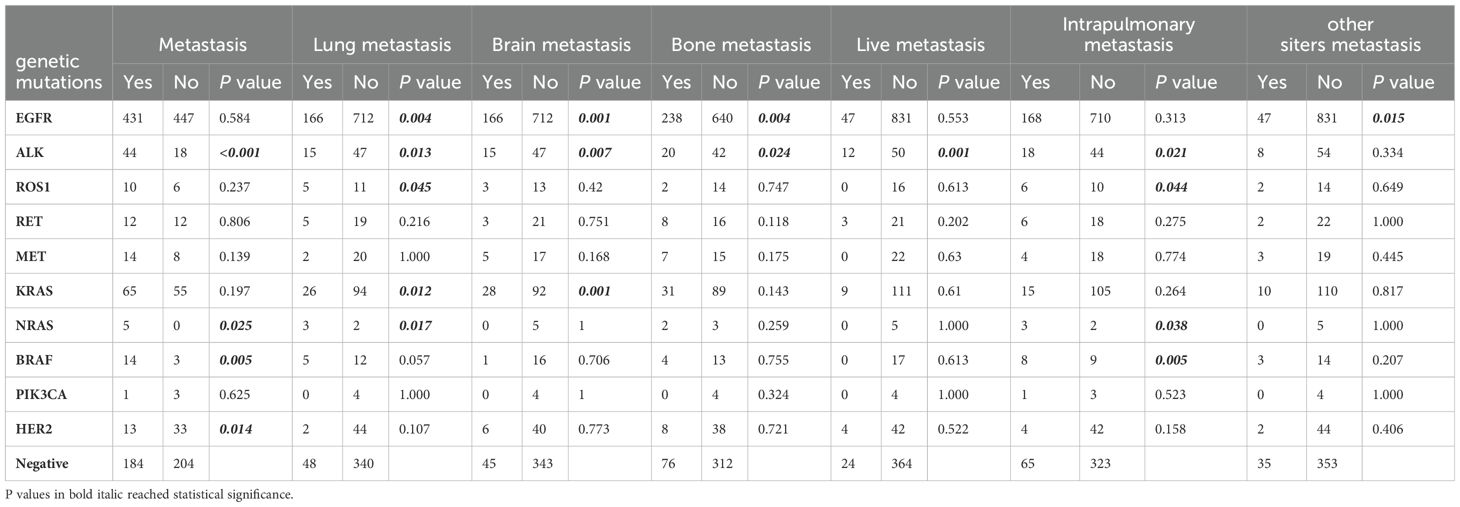
Table 2. Analysis of the association between metastases and genetic mutations.
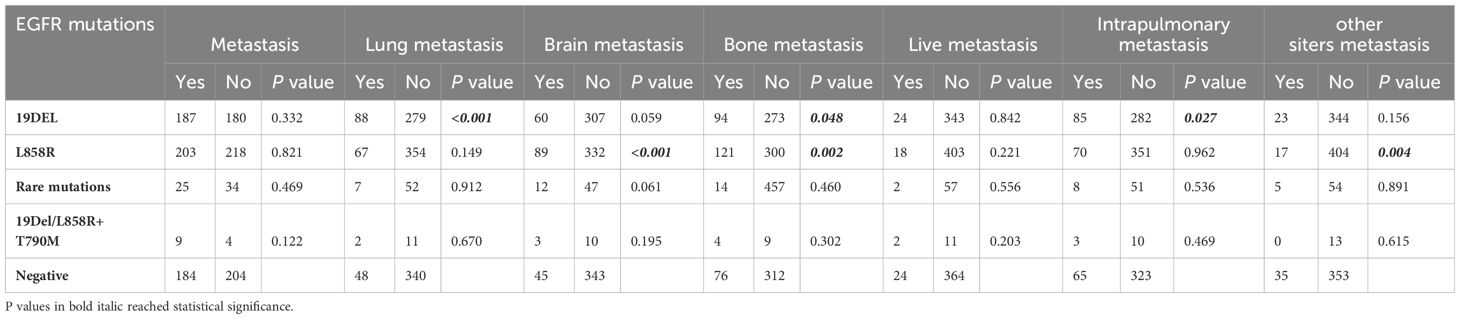
Table 3. Analysis of the association between metastases and EGFR mutations.
As shown in Table 2, the ALK mutation group (44 cases, 71.0%, P = 0.001) was more prone to metastasis than the wild-type group. Most metastatic sites in the ALK mutation group were lung (15 cases, 24.2%, P = 0.013), brain (15 cases, 24.2%, P = 0.007), bone (20 cases, 32.3%, P = 0.024), liver (12 cases, 19.4%, P = 0.001), and pleura (18 cases, 29.0%, P = 0.021). The BRAF mutation group (14 cases, 82.4%, P = 0.005) was more prone to metastasis than the wild-type group. Pleural metastasis was common in the BRAF mutation group (8 cases, 47.1%, P = 0.005). The NRAS mutation group (5 cases, 100%, P = 0.025) was more prone to metastasis than the wild-type group. Lung metastasis (3 cases, 60.0%, P = 0.017) and plural metastasis (3 cases, 60.0%, P = 0.038) frequently occurred in the NRAS mutation group. In the KRAS mutation group, 120 cases (54.2%) developed tumor metastasis, and the frequent sites were lung metastasis (26 cases, 21.7%, P = 0.012) and brain metastasis (27 cases, 23.3%, P = 0.001). In the ROS1 mutation group, 16 patients (62.5%) developed tumor metastasis, and lung metastasis (5 cases, 31.3%, P = 0.045) and pleural metastasis (6 cases, 37.5%, P = 0.044) often occurred. Compared with the wild-type group, the HER2 mutation group had less metastasis (13 cases, 28.3%, P = 0.014). In addition, there was no difference in the RET mutant group, MET mutant group and PIK3CA mutant group compared with wild-type group metastasis.
DiscussionThis study demonstrates different results for common metastatic sites in NSCLC patients with common genetic mutations. First, as EGFR was the most common mutation in this study, we studied the common sites of metastasis in 872 NSCLC patients with EGFR mutations. In terms of single organ metastasis, EGFR mutant NSCLC prefer to transfer to bone, brain, and lung, with L858R mutation patients mostly developing bone and brain metastasis, 19Del mutation patients mostly developing intrapulmonary, pleural, and bone metastasis. This is in consistent with previous studies on EGFR mutant NSCLC metastasis. Although a retrospective study found no metastasis site differences in EGFR mutation versus wild-type NSCLC patients. However, this study has only involved 105 patients, and there may be a bias (20). The study of Chen et al. showed that EGFR 19Del is usually associated with intrapulmonary and bone metastasis, with higher metastasis rate of bone, brain, and liver in patients with concurrent T790M mutation (21). Kuijpers et al. showed that the classical EGFR mutation (19Del/L858R) was associated with pleural and bone metastasis (22). These results are generally in agreement with our study, but we did not find differences in metastatic sites in patients with coexisting T790M mutation and wild-type patients. In addition, EGFR mutation showed no association with liver metastasis in NSCLC patients, which is consistent with studies in the literature (23).
ALK rearrangement is one of the most common targeted mutations in NSCLC, ALK mutations in 3.9% of NSCLC were detected in this study, metastasis in 71% of ALK positive NSCLC patients, the metastasis rate was much higher than that in the wild-type group (71.0% vs. 47.4%, P < 0.001). The most common site of metastasis was bone (32.3% vs. 19.6%, P = 0.024), followed by the pleura (29% vs. 16.8%, P = 0.021), lung (24.2% vs. 12.4%, P = 0.013), brain (24.2% vs. 11.6%, P = 0.007) and the liver (19.4% vs. 6.2%, P = 0.001). In previous studies, ALK positive NSCLC often developed metastasis, with brain metastasis being the most common, with a baseline cohort metastasis rate of approximately 24% – 29%, followed by bone metastasis of approximately 27% (24–26). However, the results of pleural metastasis and lung metastasis are inconsistent, and ALK+NSCLC Meta-analysis by Dexter et al. showed that patients with ALK rearrangement NSCLC are more likely to develop pleural metastasis, but are less likely to develop lung metastasis (12). However, the study of Gang Chen et al. showed that in 97 ALK-positive NSCLC, the incidence of brain metastases was significantly higher in ALK-positive patients both at baseline and during treatment than in the negative group, but the pleural effusion was opposite (25). Di Ma et al. analyzed 52 patients with ALK-positive advanced NSCLC at the Cancer Hospital of Chinese Academy of Medical Sciences, and the incidence of pleural metastasis was 36.5% (27). In the review by Chang et al., metastases of central nervous system (CNS), Bone, pleura, and liver often occurred in ALK positive NSCLC, which was basically consistent with our study (28). Moreover, mechanistic studies of metastasis in ALK-positive NSCLC were unclear. A novel circular RNA F-circEA-2a mainly located in the cytoplasm generated by the EML4-ALK fusion gene was found, and although F-circEA-2a does not affect the proliferation of NSCLC cells, it can significantly affect the migration and invasion of cancer cells (29). In addition, epithelial-mesenchymal transformation (epithelial-mesenchymal transition, EMT) is closely associated with cancer cell invasion and metastasis, and EML 4-ALK expression also increases the expression of EMT-inducible transcription factors, snail and slug, indicating that EMT is ongoing (30). All these studies showed that EML 4-ALK fusion is associated with invasion and metastasis of NSCLC.
ROS1 are also important drivers of NSCLC and are found in 1-2% of NSCLC patients (28, 31, 32). In our study, the ROS1 mutation rate was 1.0% (16/1586), in single organ metastasis, ROS1-positive NSCLC was more transferred to the pleura (37.5% vs.16.8%, P = 0.044) and lung (31.3% vs.12.4%, P = 0.045) compared with wild type. The brain metastasis rate of ROS 1-positive NSCLC was 18.75% (3/16, P = 0.42), but there was no statistical difference due to the small number of cases. The largest prospective trial of ROS1 TKI in East Asia (including Taiwan, Japan, Korea, China) reported a CNS transfer rate of 18.1% in ROS 1-positive NSCLC at baseline, which is consistent with our study (33). Jun et al. found that the ROS 1 cells expressing the CD74-ROS 1 fusion were highly invasive in vitro and metastatic in vivo. Expression of CD74-ROS 1 results in the phosphorylation of the extended synaptotagin-like protein E-Syt 1 (34). Another study further confirmed cellular EMT in CD74-ROS 1 or CD74-ROS1 G2032R mutation by constructing a G2032R mutant A549-CD74 ROS1 crizotinib resistant cell line. This promotes migration and increases the expression of matrix metalloproteinase (MMP) -9 and Twist1 transcription factors (35). In a study from Shanghai Chest Hospital, showing that the specific ROS 1 fusion variant CD74-ROS 1 may increase the preference for CNS metastasis, among 19 CD74-ROS1 patients, 31.6% had CNS metastases compare to no (0%) CNS metastasis among the 17 non-CD74-ROS1 patients (36).
KRAS is an important driver gene of NSCLC, the KRAS mutation rate in the Chinese NSCLC population is approximately 12.1% (37). Studies of KRAS and NSLCL metastasis are rare, and the results are inconsistent. As Chan KH et al. showed that in single-organ metastasis, KRAS/NRAS mutated NSCLC tended to transfer to the brain and bone, however, the sample size of this study was relatively small, with only 29 cases of KRAS/NRAS mutated NSCLC (38). However, KRAS was not considered to be associated with distant metastasis of NSCLC in a meta-analysis study by Wu Y et al. (15). In this study, KRAS mutation rate was 7.6% (120/1586), and in single organ metastasis, KRAS mutant NSCLC tended to transfer to brain (23.3% vs.11.6%, P = 0.001) and lung (21.7% vs.12.4%, P = 0.012) compared with wild type. The study population of Chan KH et al. was western group, and the population included in Wu Y et al. et al. meta-analysis was admixed population containing 10249 Western patients and 26319 Asian patients, while our study was single-center Chinese population, which may have ethnic and geographical differences, leading to incomplete consistent study results.
In this study, the frequency of HER 2 mutations in NSCLC was approximately 2.9% (46/1586), which is consistent with previous reports. This study showed that patients with HER2-mutated NSCLC had less metastasis compared with wild type (28.3% vs. 47.4%, P = 0.014), however, Le Y reported that HER2 positivity was associated with interpulmonary metastasis, but the intrapulmonary metastasis rate in patients with HER2 mutant NSCLC was only 4.3% in this study (39).
In addition, we also analyzed the association of rare mutations such as BRAF, NRAS, RET, MET, and PIK3CA with metastasis in NSCLC patients. When stratified by mutation profiles, patients with BRAF mutation or NRAS mutation NSCLC were more susceptible to metastasis (BRAF, 82.4% vs. 47.4%, P = 0.005; NRAS, 100% vs. 47.4%, P = 0.025). Among these, NSCLC patients with BRAF mutations had more pleural metastasis (47.1% vs.16.8%, P = 0.005), and NRAS mutations had lung (60.0% vs.12.4%, P = 0.025) and pleural metastasis (60.0% vs.16.8%, P = 0.038). However, a negative association between BRAF and distant metastasis of NSCLC was previously reported by Wu Y et al. (15). The study of Chan KH et al. showed that NRAS mutant NSCLC preferred to transfer to the brain and bone (38). In addition, in terms of total metastasis rate and single organ metastasis, the RET mutation, MET mutation and PIK3CA mutation groups were not different compared with the wild-type group, and the sample size of the cohort with rare mutation was small, which makes the analysis and interpretation difficult.
This study has several strengths. First, this is a comprehensive study to explore the correlation between NSCLC gene profile and metastasis. We not only investigated the relationship of common gene mutations such as EGFR, ALK, ROS1 and NSCLC metastasis, but also extended the gene spectrum to rare gene mutations such as KRAS, HER2, BRAF, RET, MET, and PIK3CA. Secondly, the large number of NSCLC patients included in this study makes the study results more credible. To the best of our knowledge, this is the first study of the correlation between gene mutation profile and metastasis in Chinese NSCLC patients based on real-world data.
However, this study also has some limitations. First, this was a single-center retrospective study, which may limit the generalizability of the results. Second, due to the low rate of NSCLC rare gene mutations, the small sample size of our rare gene mutation cohort makes analysis and interpretation difficult. However, the mechanism of different gene transfer preferences was unclear and further studies were essential, which may be necessary to find new ways to limit the development of metastasis.
ConclusionsThis retrospective study further suggested a correlation between gene profiles and metastatic sites in NSCLC patients. First, ALK mutations, BRAF mutations, and NRAS mutations were all positively associated with NSCLC metastasis, while HER2 mutations in NSCLC had less metastasis. Secondly, in terms of single organ metastasis, EGFR mutation was associated with lung, brain, and bone metastasis of NSCLC, KRAS mutation was associated with lung and brain metastasis of NSCLC, and ROS1 mutation was associated with lung and pleural metastasis of NSCLC, but the total metastasis rate was not different compared with wild type. Furthermore, no association was found between RET, MET, PIK3CA mutations and NSCLC metastasis. This is an important study using real-world data to predict metastasis in NSCLC patients, and these results need to be confirmed in larger studies.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statementThe studies involving humans were approved by Ethics Committee of Liaocheng People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from gifted from another research group. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributionsYL: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. ZZ: Methodology, Writing – review & editing. LW: Data curation, Visualization, Writing – review & editing. LH: Data curation, Visualization, Writing – review & editing. YD: Methodology, Writing – review & editing. XZ: Data curation, Visualization, Writing – review & editing. XL: Methodology, Writing – review & editing. JX: Conceptualization, Supervision, Writing – review & editing.
FundingThe author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1451576/full#supplementary-material
AbbreviationsALK, Anaplastic lymphoma kinase gene; ARMS-PCR, Amplification Refractory Mutation System-Polymerase Chain Reaction; BRAF V-raf, murine sarcoma viral oncogene homolog B; CNS, Central nervous system; EGFR, Epidermal growth factor receptor gene; EMT, Epithelial-mesenchymal transition; HER2, Human epidermal growth factor receptor 2; KRAS, Kirsten rat sarcoma viral oncogene homolog; MET, Mesenchymal-epithelial transition factor; NRAS, Neuroblastoma RAS viral oncogene homolog; NSCLC, Non-small cell lung cancer; PCR, Polymerase chain reaction; PIK3CA, Phosphatidylinositol 3-kinase; RET, Rearranged during Transfection; ROS1, Rearrangements of the c-ros oncogene 1; TNM, Tumor node metastasis.
References1. Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. (2022) 135:584–90. doi: 10.1097/cm9.0000000000002108
PubMed Abstract | Crossref Full Text | Google Scholar
3. Khasraw M, Yalamanchili P, Santhanagopal A, Wu C, Salas M, Meng J, et al. Clinical management of patients with non-small cell lung cancer, brain metastases, and actionable genomic alterations: A systematic literature review. Adv Ther. (2024) 41:1815–42. doi: 10.1007/s12325-024-02799-9
PubMed Abstract | Crossref Full Text | Google Scholar
4. Durbin L, Murali B, Li S, Zhao L, Hawthorne S, Kanas G, et al. Treatment patterns in non-small-cell lung cancer in China: Results from the CancerMPact survey 2020. Cancer Treat Res Commun. (2021) 29:100462. doi: 10.1016/j.ctarc.2021.100462
PubMed Abstract | Crossref Full Text | Google Scholar
5. Ganti AK, Klein AB, Cotarla I, Seal B, Chou E. Update of incidence, prevalence, survival, and initial treatment in patients with non–small cell lung cancer in the US. JAMA Oncol. (2021) 7:1824. doi: 10.1001/jamaoncol.2021.4932
PubMed Abstract | Crossref Full Text | Google Scholar
6. Waqar SN, Morgensztern D, Govindan R. Systemic treatment of brain metastases. Hematology/Oncology Clinics North America. (2017) 31:157–76. doi: 10.1016/j.hoc.2016.08.007
Crossref Full Text | Google Scholar
7. Zhou H, Watson M, Bernadt CT, Lin S, Lin CY, Ritter JH, et al. AI-guided histopathology predicts brain metastasis in lung cancer patients. J Pathol. (2024) 263:89–98. doi: 10.1002/path.6263
PubMed Abstract | Crossref Full Text | Google Scholar
11. Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies. Oncotarget. (2017) 8:57680–92. doi: 10.18632/oncotarget.17016
PubMed Abstract | Crossref Full Text | Google Scholar
12. Mendoza DP, Stowell J, Muzikansky A, Shepard J-AO, Shaw AT, Digumarthy SR. Computed tomography imaging characteristics of non–small-cell lung cancer with anaplastic lymphoma kinase rearrangements: A systematic review and meta-analysis. Clin Lung Cancer. (2019) 20:339–49. doi: 10.1016/j.cllc.2019.05.006
PubMed Abstract | Crossref Full Text | Google Scholar
14. Del Re M, Luculli GI, Petrini I, Sbrana A, Scotti V, Perez D, et al. Clinical utility of Next Generation Sequencing of plasma cell-free DNA for the molecular profiling of patients with NSCLC at diagnosis and disease progression. Trans Oncol. (2024) 41:101869. doi: 10.1016/j.tranon.2023.101869
Crossref Full Text | Google Scholar
15. Wu Y, Ni H, Yang D, Niu Y, Chen K, Xu J, et al. Driver and novel genes correlated with metastasis of non-small cell lung cancer: A comprehensive analysis. Pathol - Res Pract. (2021) 224:153551. doi: 10.1016/j.prp.2021.153551
PubMed Abstract | Crossref Full Text | Google Scholar
16. Camidge DR, Kim HR, Ahn MJ, Yang CH, Popat S. Brigatinib versus crizotinib in ALK-positive non–small-cell lung cancer. New Engl J Med. (2018) 379:2027–39 doi: 10.1056/NEJMoa1810171
PubMed Abstract | Crossref Full Text | Google Scholar
17. Gadgeel S, Peters S, Mok T, Shaw AT, Kim DW, Ou SI, et al. Alectinib versus crizotinib in treatment-nave anaplastic lymphoma kinase-positive (ALK+) non-small-cell lung cancer: CNS efficacy results from the ALEX study. Ann Oncol. (2018) 29:2214–22. doi: 10.1093/annonc/mdy405
PubMed Abstract | Crossref Full Text | Google Scholar
18. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. New Engl J Med. (2017) 377:829–38. doi: 10.1056/NEJMoa1704795
PubMed Abstract | Crossref Full Text | Google Scholar
19. Zhou C, Lu Y, Kim SW, Reungwetwattana T, Zhou J, Zhang Y, et al. Primary results of ALESIA: Phase III, randomised open-label study of alectinib (ALC) vs crizotinib (CRZ) in Asian patients (pts) with treatment-nave ALK+ advanced non-small-cell lung cancer (NSCLC). Ann Oncol. (2018) 29:174. doi: 10.1093/annonc/mdy483.001
Crossref Full Text | Google Scholar
20. Beypinar I, Demir H, Araz M, Uysal M. The relationship between EGFR mutation and metastasis pattern in lung adenocarcinoma. J Oncological Sci. (2019) 5:65–9. doi: 10.1016/j.jons.2019.08.002
Crossref Full Text | Google Scholar
21. Chen Y, Deng J, Liu Y, Wang H, Zhao S, He Y, et al. Analysis of metastases in non-small cell lung cancer patients with epidermal growth factor receptor mutation. Ann Trans Med. (2021) 9:206–6. doi: 10.21037/atm-20-2925
Crossref Full Text | Google Scholar
22. Kuijpers CCHJ, Hendriks LEL, Derks JL, Dingemans AMC, van Lindert ASR, van den Heuvel MM, et al. Association of molecular status and metastatic organs at diagnosis in patients with stage IV non-squamous non-small cell lung cancer. Lung Cancer. (2018) 121:76–81. doi: 10.1016/j.lungcan.2018.05.006
PubMed Abstract | Crossref Full Text | Google Scholar
23. Jiang T, Cheng R, Zhang G, Su C, Zhao C, Li X, et al. Characterization of liver metastasis and its effect on targeted therapy in EGFR-mutant NSCLC: A multicenter study. Clin Lung Cancer. (2017) 18:631–639.e632. doi: 10.1016/j.cllc.2017.04.015
PubMed Abstract | Crossref Full Text | Google Scholar
24. Mendoza DP, Lin JJ, Rooney MM, Chen T, Sequist LV, Shaw AT, et al. Imaging features and metastatic patterns of advanced ALK-rearranged non–small cell lung cancer. Am J Roentgenology. (2020) 214:766–74. doi: 10.2214/ajr.19.21982
Crossref Full Text | Google Scholar
25. Chen G, Chen X, Zhang Y, Yan F, Fang W, Yang Y, et al. single-center, real-world study of clinicopathological characteristics and treatment in advanced ALK-positive non-small-cell lung cancer. Cancer Med. (2017) 6:953–61. doi: 10.1002/cam4.1059
PubMed Abstract | Crossref Full Text | Google Scholar
26. Nakamura T, Yoshida T, Takeyasu Y, Masuda K, Sinno Y, Matsumoto Y, et al. Distinct metastatic spread and progression patterns in patients treated with crizotinib for ROS1- and ALK-rearranged non-small cell lung cancer: a single-center retrospective study. Trans Lung Cancer Res. (2023) 12:1436–44. doi: 10.21037/tlcr-23-10
Crossref Full Text | Google Scholar
27. Ma D, Hao X, Wang Y, Xing P, Li J. Clinical effect of pemetrexed as the first-line treatment in Chinese patients with advanced anaplastic lymphoma kinase-positive non-small cell lung cancer. Thorac Cancer. (2016) 7:452–8. doi: 10.1111/1759-7714.12353
PubMed Abstract | Crossref Full Text | Google Scholar
28. Chang X, Liu Z, Man S, Roys A, Li Z, Zuo D, et al. Metastasis manners and the underlying mechanisms of ALK and ROS1 rearrangement lung cancer and current possible therapeutic strategies. RSC Adv. (2019) 9:17921–32. doi: 10.1039/c9ra02258a
PubMed Abstract | Crossref Full Text | Google Scholar
29. Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. (2018) 17:138. doi: 10.1186/s12943-018-0887-9
PubMed Abstract | Crossref Full Text | Google Scholar
30. Kogita A, Togashi Y, Hayashi H, Sogabe S, Nishio K. Hypoxia induces resistance to ALK inhibitors in the H3122 non-small cell lung cancer cell line with an ALK rearrangement via epithelial-mesenchymal transition. Int J Oncol. (2014) 45:1430–6. doi: 10.3892/ijo.2014.2574
PubMed Abstract | Crossref Full Text | Google Scholar
31. ten Berge DMHJ, Damhuis RAM, Aerts JGJV, Dingemans A-MC. Real-world treatment patterns and survival of patients with ROS1 rearranged stage IV non-squamous NSCLC in the Netherlands. Lung Cancer. (2023) 181:107253. doi: 10.1016/j.lungcan.2023.107253
PubMed Abstract | Crossref Full Text | Google Scholar
32. Zhang Q, Wu C, Ding W, Zhang Z, Qiu X, Mu D, et al. Prevalence of ROS1 fusion in Chinese patients with non-small cell lung cancer. Thorac Cancer. (2018) 10:47–53. doi: 10.1111/1759-7714.12899
PubMed Abstract | Crossref Full Text | Google Scholar
34. Jun HJ, Johnson H, Bronson RT, de Feraudy S, White F, Charest A. The oncogenic lung cancer fusion kinase CD74-ROS activates a novel invasiveness pathway through E-syt1 phosphorylation. Cancer Res. (2012) 72:3764–74. doi: 10.1158/0008-5472.can-11-3990
PubMed Abstract | Crossref Full Text | Google Scholar
35. Gou W, Zhou X, Liu Z, Wang L, Shen J, Xu X, et al. CD74-ROS1 G2032R mutation transcriptionally up-regulates Twist1 in non-small cell lung cancer cells leading to increased migration, invasion, and resistance to crizotinib. Cancer Lett. (2018) 422:19–28. doi: 10.1016/j.canlet.2018.02.032
PubMed Abstract | Crossref Full Text | Google Scholar
36. Li Z, Shen L, Ding D, Huang J, Zhang J, Chen Z, et al. Efficacy of crizotinib among different types of ROS1 fusion partners in patients with ROS1-rearranged non–small cell lung cancer. J Thorac Oncol. (2018) 13:987–95. doi: 10.1016/j.jtho.2018.04.016
PubMed Abstract | Crossref Full Text | Google Scholar
37. Chen H, Huang D, Lin G, Yang X, Zhuo M, Chi Y, et al. The prevalence and real-world therapeutic analysis of Chinese patients with KRAS-Mutant Non-Small Cell lung cancer. Cancer Med. (2022) 11:3581–92. doi: 10.1002/cam4.4739
留言 (0)