For decades, cancer has been the leading cause of human health and death. According to the latest statistical data, there were nearly 20 million new cancer cases and 9.7 million cancer-related deaths worldwide in 2022 (Bray et al., 2024). It is predicted that by 2050, the number of new cancer cases will reach 35 million (Bray et al., 2024). In 2022, there were approximately 4.82 million new cancer cases and 2.57 million new cancer-related deaths in China (Han et al., 2024). In the United States, it is expected that there will be approximately 2 million new cancer cases and 0.61 million cancer-related deaths by 2024 (Siegel et al., 2024). These data reflect that unremitting efforts in cancer prevention, diagnosis, and treatment are required to reduce the burden of cancer.
During this period, it was discovered that long non-coding RNAs (lncRNAs) in regulatory non-coding RNAs can extensively participate in the occurrence and development of cancer at various levels (Rajakumar et al., 2023) and can be developed into diagnostic, therapeutic, and prognostic biomarkers (Chi et al., 2019; Goyal et al., 2021; Najafi et al., 2022; Zhang et al., 2023). LncRNA is a type of non-coding RNA with length exceeding 200 nucleotides (Pierouli et al., 2022). LncRNA is believed to play a role in many cellular processes, including cell cycle (Hashemi et al., 2022; Yin et al., 2023; Zangouei et al., 2023), differentiation (Liu et al., 2022; Wang and Qi, 2021), apoptosis (Iwai et al., 2023; Wang et al., 2023), migration (Ye et al., 2021), and metabolism (Liu et al., 2021; Tan et al., 2021a).
The HOX gene is a member of the homeobox superfamily, and its gene sequence is highly conserved (Li et al., 2019a). There are 39 HOX genes in humans, divided into four clusters: HOXA, HOXB, HOXC, and HOXD. The HOX cluster also contains many non-coding RNAs, including some lncRNAs. The expression of these lncRNAs has exhibited abnormalities in different types of cancer and is associated with the occurrence and progression of cancer, such as HOXA cluster antisense RNA2 (Chen and He, 2021; Jiang et al., 2019; Wang et al., 2019; Zhang et al., 2022a; Zheng et al., 2019), HOXB cluster antisense RNA3 (Jiang et al., 2020; Wu et al., 2024; Xing et al., 2021a; Xu et al., 2021), and HOXC cluster antisense RNA3 (Su et al., 2022; Su et al., 2020; Yang et al., 2021; Zhang et al., 2022b; Zhao et al., 2021). In addition to the aforementioned lncRNAs, HOXC13 antisense RNA (HOXC13-AS, also known as HOXC-AS5) has attracted attention due to its abnormal expression patterns found in many malignant tumors. HOXC13-AS is an antisense lncRNA transcribed from chromosome 12. It is located on the reverse strand of chromosome 12 (Figure 1A) (https://www.genecards.org/cgi-bin/carddisp.pl?gene=HOXC13-AS&keywords=HOXC13-AS) (Stelzer et al., 2016), with coordinates 53935328–53939643, spans three exons, and has a length of 4,316 nucleotides (source https://www.ncbi.nlm.nih.gov/gene/100874366). In addition, HOXC13-AS overlaps with the HOXC13 gene on the sense strands. More interestingly, it shares genomic position 12q13.13 with HOX transcript antisense RNA (Figure 1B) [National Center for Biotechnology Information (NCBI) (Internet). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information (1988)–(cited 2025 January 10). Available from: https://www.ncbi.nlm.nih.gov/]. The oncogenic lncRNA HOX transcript antisense RNA is involved in the occurrence and development of various types of cancer (Fu et al., 2016; Guo et al., 2023; Shi et al., 2022; Wei et al., 2020; Zhang et al., 2024), indicating the potential role of HOXC13-AS in cancer. Some studies have found that the expression of HOXC13-AS is dysregulated in various types of malignant tumors, and upregulated HOXC13-AS expression is associated with clinical pathological features such as tumor node metastasis (TNM) staging, metastasis, clinical staging, and prognosis (Gao et al., 2019; Zhou J. F. et al., 2019). More studies have found that HOXC13-AS is involved in the occurrence and development of tumors by affecting processes such as cell proliferation, invasion, migration, and epithelial–mesenchymal transition (EMT) (Gao et al., 2019; Li et al., 2020; Li et al., 2019b; Liu et al., 2019; Wang et al., 2021). HOXC13-AS has exhibited potential application value as a tumor treatment, a prognostic biomarker, and a therapeutic target.
In this review, we systematically searched PubMed and Web of Science databases using the keywords “HOXC13-AS,” “HOXC13 antisense RNA,” and “HOXC-AS5” and screened for research literature related to human cancer. Based on the aforementioned search results, this review summarizes the expression levels of HOXC13-AS in different types of cancer and its correlation with clinical pathological characteristics, prognosis, and diagnostic value. This review also outlines the biological role and molecular regulatory mechanisms of HOXC13-AS in various types of cancer, aiming to lay a foundation for the clinical application of HOXC13-AS.
2 Expression of HOXC13-AS and its clinical significance as a potential biomarker in human cancerMany studies have revealed the abnormal expression of HOXC13-AS in various types of human tumors, and it has been established that the dysregulation of HOXC13-AS is associated with certain clinical pathological characteristics and the prognosis of patients (Table 1). This section provides an overview of the expression changes of HOXC13-AS in human malignancies and its correlation with some clinical pathological characteristics and discusses the potential of HOXC13-AS as a valuable biomarker for prognosis in different types of tumors.

Table 1. Expression of HOXC13-AS in tissue samples and its relationship with clinical characteristics and survival in human tumorsa.
2.1 Expression of HOXC13-AS in human cancerPrevious studies have revealed that HOXC13-AS is abnormally expressed in various types of human cancer, including hepatocellular carcinoma, nasopharyngeal carcinoma, breast cancer, head and neck squamous cell carcinoma (HNSCC), oral squamous cell carcinoma, glioma, cervical cancer, and intrahepatic cholangiocarcinoma (Table 1).
We extensively investigated the expression of HOXC13-AS in pan-cancer. The interactive BodyMap in GEPIA2 (http://gepia2.cancer-pku.cn/) (Tang et al., 2019) identified the expression profiles of HOXC13-AS RNA transcripts in both tumor and normal tissues. According to the interactive BodyMap, HOXC13-AS is expressed at higher levels in tumor tissues than in normal tissues, primarily in malignancies of the head and neck, lung, breast, cervix uteri, and bladder (Figure 2A). In tumor samples, HNSCC exhibited the highest expression of RNA transcripts (Figure 2B), whereas in normal tissues, the highest expression was observed in normal tissues adjacent to skin cutaneous melanoma (Figure 2C). Furthermore, the results demonstrated that HOXC13-AS was significantly upregulated in many types of malignancies, including HNSCC, cervical squamous cell carcinoma, and endocervical adenocarcinoma. Conversely, skin cutaneous melanoma exhibited a marked downregulation of HOXC13-AS (Figure 2D).
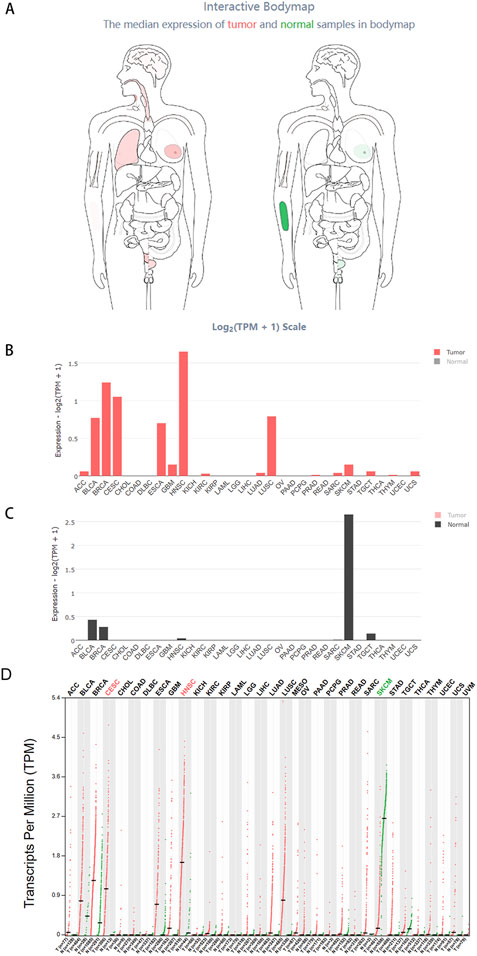
Figure 2. The comprehensive expression profile of HOXC13-AS in human tissues. Using data from GEPIA 2 (https://www.kmplot.com/analysis/): Interactive Bodymap, the analysis of HOXC13-AS gene expression including all identified isoforms depicted the median expression of HOXC13-AS in tumor and normal samples (A). The bar chart provides detailed gene expression profiles of tumor samples (B) and corresponding normal tissues (C). The dot plot represents the gene expression profiles of all tumor samples and paired normal tissues (D). ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
2.2 HOXC13-AS as a prognostic and diagnostic biomarkerHOXC13-AS, as a potential biomarker in human tumors, has been reported in many studies for its enormous clinical potential (Table 1). A study has found that the expression level of HOXC13-AS is correlated with the clinical characteristics of some types of cancer (Table 1). For example, in hepatocellular carcinoma (Zhou Q. et al., 2019), high expression of HOXC13-AS is closely associated with TNM staging and lymph node metastasis. In nasopharyngeal carcinoma (Gao et al., 2019), overexpression of HOXC13-AS is significantly correlated with local recurrence, distant metastasis, and clinical staging. However, the relationship between the expression of HOXC13-AS and patient prognosis was also discovered through research. Upregulation of the expression of HOXC13-AS in hepatocellular carcinoma (Zhou J. F. et al., 2019), nasopharyngeal carcinoma (Gao et al., 2019), glioma (Liu et al., 2019), and intrahepatic cholangiocarcinoma (Angenard et al., 2019) indicates poor prognosis for patients. In addition, HOXC13-AS is a new candidate diagnostic biomarker for patients with HNSCC (Xiong et al., 2020).
We analyzed the relationship between HOXC13-AS expression levels and prognosis using the Kaplan-Meier plots (https://www.kmplot.com/analysis/) (Győrffy, 2024). The expression level of HOXC13-AS is significantly related to the prognosis of patients with bladder cancer, esophageal adenocarcinoma, esophageal squamous cell carcinoma, HNSCC, kidney renal papillary cell carcinoma, ovarian cancer, pheochromocytoma and paraganglioma, sarcoma, stomach adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma (Figure 3). The high expression of HOXC13-AS indicates poorer overall survival in esophageal adenocarcinoma, esophageal squamous cell carcinoma, kidney renal papillary cell carcinoma, ovarian cancer, pheochromocytoma and paraganglioma, and sarcoma (Figures 3B, C, E–H), whereas its high expression indicates that overall survival is better in stomach adenocarcinoma and thyroid carcinoma, which can serve as favorable predictive factors (Figures 3I, J). These findings indicate that HOXC13-AS has different prognostic significance in various types of cancer, rendering it a potentially effective prognostic biomarker for a range of human malignancies.
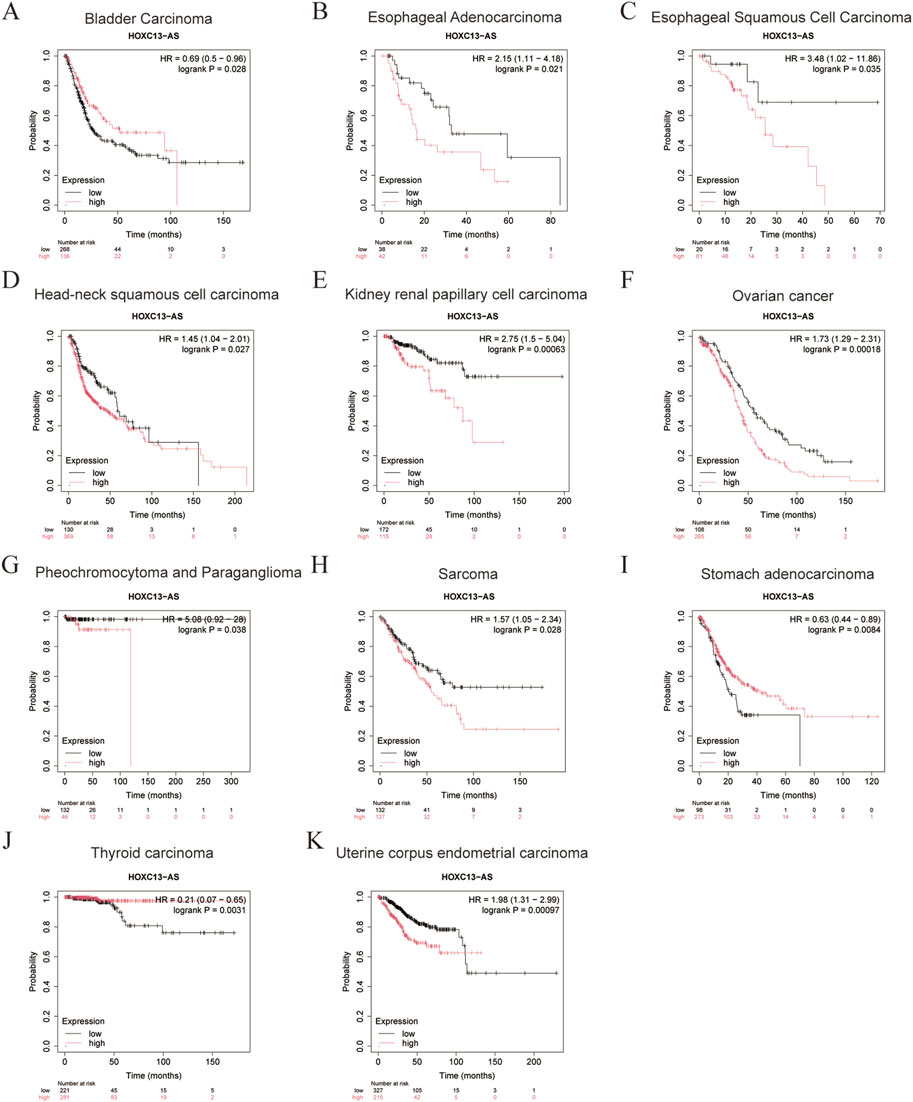
Figure 3. The relationship between HOXC13-AS expression and overall survival (OS) in bladder carcinoma (A), esophageal adenocarcinoma (B), esophageal squamous cell carcinoma (C), head-neck squamous cell carcinoma (D), kidney renal papillary cell carcinoma (E), ovarian cancer (F), pheochromocytoma and paraganglioma (G), sarcoma (H), stomach adenocarcinoma (I), thyroid carcinoma (J), uterine corpus endometrial carcinoma (K) from the Kaplan–Meier plots (https://www.kmplot.com/analysis/). HOXC13-AS: HOXC13 antisense RNA; HR: hazard rate.
3 The molecular mechanisms and role of HOXC13-AS in cancerHOXC13-AS participates in regulating biological functions through different molecular mechanisms in various types of cancer, thereby affecting the occurrence and development of cancer. These biological functions include cell proliferation, migration, invasion, EMT, and tumor growth (Table 2; Figure 4). Based on these findings, HOXC13-AS is expected to become a new target for human cancer treatment. Many studies have explored the regulatory mechanism of lncRNA HOXC13-AS in different types of cancer (Figure 5), including nasopharyngeal carcinoma (Gao et al., 2019), breast cancer (Li et al., 2019a), oral squamous cell carcinoma (Li et al., 2020), glioma (Liu et al., 2019), and cervical cancer (Wang et al., 2021). Among these regulatory mechanisms, the most common one is HOXC13-AS as a competing endogenous RNA (ceRNA) to bind with microRNA (miRNA). LncRNAs can be seen as a “sponge” to competitively bind miRNA through their specific sequence, which are complementary to the miRNA, thus reducing the regulatory effects of miRNA to downstream target genes (Ma et al., 2023). In this section, the molecular mechanisms underlying the regulatory role of HOXC13-AS in different types of malignant tumors are discussed.
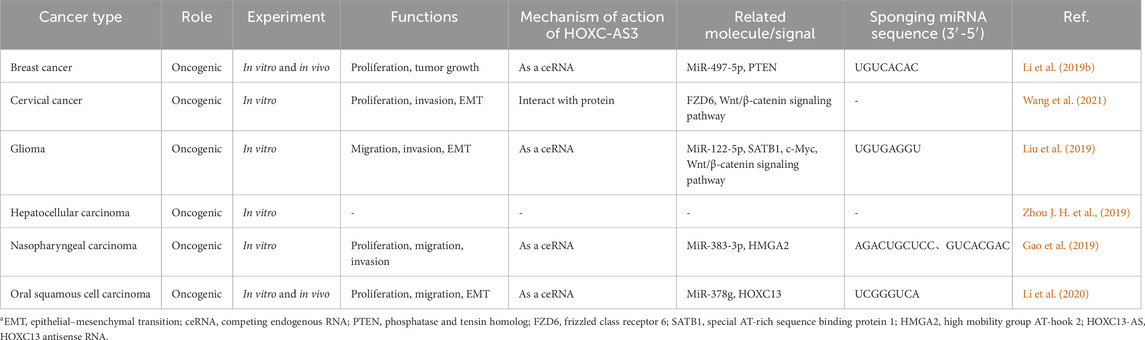
Table 2. The role and regulatory mechanism of HOXC13-AS in human malignanciesa.
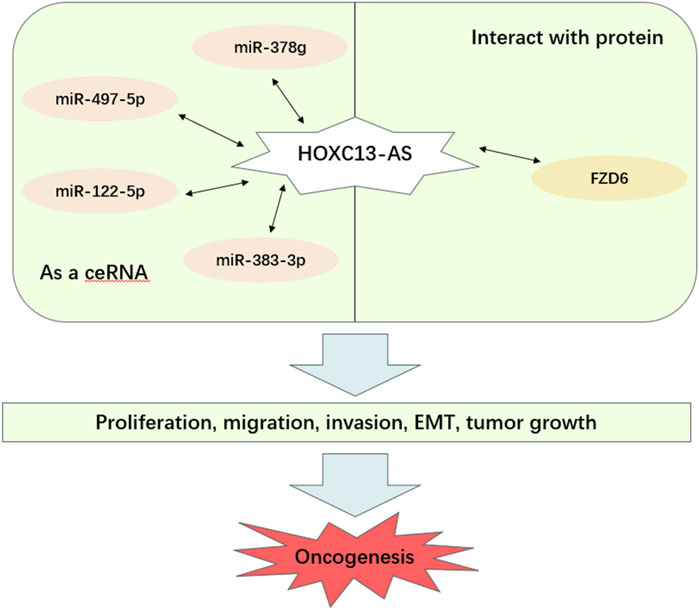
Figure 4. Main mechanisms of HOXC13-AS in cancer progression. HOXC13-AS, HOXC13 antisense RNA; FZD6, frizzled class receptor 6; ceRNA, competing endogenous RNA; EMT, epithelial mesenchymal transition.
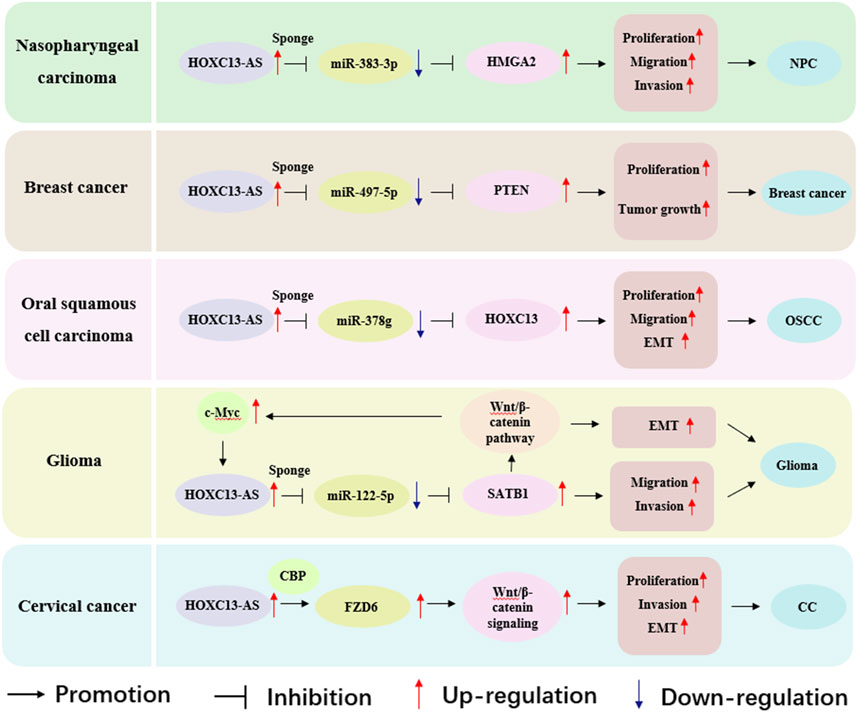
Figure 5. Role of HOXC13-AS in modulating molecular pathways across nasopharyngeal carcinoma, breast cancer, oral squamous cell carcinoma, glioma and cervical cancer. HOXC13-AS, HOXC13 antisense RNA; HMGA2, high mobility group AT-hook 2; NPC, nasopharyngeal carcinoma; PTEN, phosphatase and tensin homolog; EMT, epithelial mesenchymal transition; OSCC, oral squamous cell carcinoma; SATB1, special AT rich sequence binding protein 1; CBP, cAMP-response element binding protein-binding protein; FZD6, frizzled class receptor 6; CC, cervical cancer.
3.1 Nasopharyngeal carcinomaNasopharyngeal carcinoma (NPC) is a type of HNSCC that predominantly occurs in East Asia and Southeast Asia (Chen et al., 2019). The expression of HOXC13-AS was observed to increase in NPC tissues, and HOXC13-AS was highly expressed in five cancer cell lines (CNE 1, 6-10B, SUNE 2, HNE-1, and CNE 2), exhibiting a significant correlation with local-regional recurrence, distant metastasis, and clinical stage (Gao et al., 2019). In addition, high expression of HOXC13-AS is associated with poor patient prognosis and low overall survival (Gao et al., 2019). Functionally, in vitro experiments have indicated that knocking down HOXC13-AS inhibits cell proliferation, migration, and invasion (Gao et al., 2019). Mechanistically, HOXC13-AS acts through the HOXC13-AS–miR-383-3p–high mobility group AT-hook 2 (HMGA2) axis, where HOXC13-AS functions as a ceRNA and promotes NPC progression by competitively sponging with miR-383-3p to enhance the expression of HMGA2 (Gao et al., 2019). In summary, although HOXC13-AS exerts oncogenic function by regulating the miR-383-3p/HMGA2 axis, it also provides new ideas for the treatment of nasopharyngeal carcinoma patients to a certain extent.
3.2 Breast cancerBreast cancer is one of the most common malignant tumors in women and is also a major cause of cancer-related deaths in women worldwide (Wilkinson and Gathani, 2022). Real-time quantitative polymerase chain reaction, Cell Counting Kit-8 assay, and colony formation assay demonstrated that compared with adjacent normal tissues, the expression of HOXC13-AS in breast cancer tissues was significantly upregulated, and the upregulated HOXC13-AS promoted the growth of breast cancer cells (Li et al., 2019b). In the mechanism study, in vivo and in vitro experiments demonstrated that HOXC13-AS promotes breast cancer cell proliferation and tumor growth through the miR-497-5p/phosphatase and tensin homolog (PTEN) axis. Put differently, HOXC13-AS, which is significantly upregulated in breast cancer, can act as the “sponge” of miR-497-5p, reduce the expression of miR-497-5p, and further increase PTEN to promote cell proliferation (Li et al., 2019a). These findings suggest that HOXC13-AS may play a carcinogenic role in breast cancer, but it is expected to become a potential target for treatment.
3.3 Oral squamous cell carcinomaStudies have found that HOXC13-AS is upregulated in oral squamous cell carcinoma (OSCC) tissues and cell lines, leading to oncogenic functions (Li et al., 2020). HOXC13-AS upregulates the expression of HOXC13 in OSCC cells by sequestering miR-378g, promoting OSCC cell proliferation, migration, and EMT (Li et al., 2020). The expression of HOXC13 is positively correlated with HOXC13-AS and negatively correlated with miR-378g. Overall, HOXC13-AS, as a ceRNA, promotes OSCC development through the HOXC13-AS/miR-378g/HOXC13 axis (Li et al., 2020), providing a new approach for lncRNA targeted therapy of OSCC.
3.4 GliomaGliomas account for approximately 80% of all intracranial malignancies (Li et al., 2022). HOXC13-AS is considered an oncogene of glioma (Liu et al., 2019). The expression of HOXC13-AS is elevated in the tissues and cells of gliomas, and high levels of HOXC13-AS indicate poor prognosis in patients. Knockdown of HOXC13-AS can hinder the migration, invasion, and EMT process of glioma cells (Liu et al., 2019). Mechanistically, HOXC13-AS as a ceRNA indirectly regulates special AT-rich sequence binding protein 1 (SATB1) expression by sponging miR-122-5p, thereby affecting the EMT process, and the Wnt/β-catenin pathway is also involved in this process (Liu et al., 2019). Interestingly, the Wnt/β-catenin pathway target gene c-Myc can regulate the expression of HOXC13-AS at the transcriptional level by binding to the promoter region of HOXC13-AS, thereby forming a positive feedback loop (Liu et al., 2019). The discovery of the HOXC13-AS–miR-122-5p–SATB1–c-Myc feedback loop has improved the regulatory mechanism of HOXC13-AS in malignant tumors and provided potential therapeutic targets for gliomas (Liu et al., 2019).
3.5 Cervical cancerCervical cancer (CC) is the fourth most common cancer among women worldwide and the leading cause of cancer-related deaths among women in developing countries (Buskwofie et al., 2020). HOXC13-AS is highly expressed in CC tissues and cell lines and is positively correlated with frizzled class receptor 6 (FZD6) (Wang et al., 2021). HOXC13-AS upregulates FZD6 and activates Wnt/β-catenin signaling to promote CC proliferation, invasion, and EMT (Wang et al., 2021). HOXC13-AS upregulates FZD6 through interaction with cAMP-response element binding protein-binding protein (CBP), inducing histone H3 lysine 27 acetylation on the FZD6 promoter (Wang et al., 2021). Fat mass and obesity-associated protein improves the stability of HOXC13-AS by reducing N6-methyladenosine (Wang et al., 2021). In general, FZD6 is an oncogene in CC, and HOXC13-AS has great potential as a new target for CC therapy.
4 Conclusions and perspectivesNumerous lines of evidence have indicated that multiple lncRNAs are involved in the occurrence and development of human tumors through various complex mechanisms (Chen et al., 2022; Hashemi et al., 2022; McCabe and Rasmussen, 2021; Xing et al., 2021b), and HOXC13-AS is one of them, playing an important role in tumor research. Studies have found that HOXC13-AS is dysregulated in various types of human tumors (Angenard et al., 2019; Gao et al., 2019; Li et al., 2020; Li et al., 2019b; Liu et al., 2019; Wang et al., 2021; Xiong et al., 2020; Zhou Q. et al., 2019). The expression level of lncRNA HOXC13-AS is closely related to the clinical characteristics of tumors. For example, in hepatocellular carcinoma, HOXC13-AS levels are significantly correlated with TNM staging and lymph node metastasis, and high expression levels of HOXC13-AS indicate advanced liver cancer status in patients (Zhou J. F. et al., 2019). In nasopharyngeal carcinoma, the expression level of HOXC13-AS is correlated with local recurrence, distant metastasis, and clinical staging, further demonstrating the effect of HOXC13-AS on cancer-related features (Gao et al., 2019). Moreover, HOXC13-AS is involved in various biological processes. High expression of HOXC13-AS promotes cell proliferation, migration, invasion, EMT, and tumor growth, thereby accelerating tumor progression (Gao et al., 2019; Li et al., 2020; Li et al., 2019a; Liu et al., 2019; Wang et al., 2021).
HOXC13-AS has the potential to become a prognostic and diagnostic biomarker. It presents unique prognostic factors in different types of cancer and provides new ideas and insights for cancer treatment. According to relevant research reports, in nasopharyngeal carcinoma and glioma, patients with higher levels of HOXC13-AS have a lower overall survival rate than those with lower levels of HOXC13-AS (Gao et al., 2019; Liu et al., 2019). Similarly, in hepatocellular carcinoma and intrahepatic cholangiocarcinoma, the expression level of HOXC13-AS is negatively correlated with overall survival and disease-free survival (Angenard et al., 2019; Zhou Q. et al., 2019). In addition, HOXC13-AS is a new candidate diagnostic biomarker for patients with HNSCC (Xiong et al., 2020). Furthermore, it was found that the high expression of HOXC13-AS in different cancers represents different prognostic effects though a comprehensive analysis of Kaplan-Meier plots (Figure 3). It indicated poor prognosis in esophageal adenocarcinoma, esophageal squamous cell carcinoma, kidney renal papillary cell carcinoma, ovarian cancer, pheochromocytoma and paraganglioma, and sarcoma, while it indicated good prognosis in bladder carcinoma, stomach adenocarcinoma and thyroid carcinoma. This variability in prognosis might be caused by tissue-specific expression of lncRNA (Galamb et al., 2019; Sarropoulos et al., 2019).
The functions of lncRNA are diverse (Nemeth et al., 2024). It can directly interact with DNA (Arab et al., 2019; Luo et al., 2022), RNA (Hu et al., 2014; Shi et al., 2020; Tan et al., 2021b), and also proteins (Ferrè et al., 2016; Li et al., 2021; Zhang et al., 2018; Zhou J. F et al., 2019). In some types of cancer, the molecular mechanism of HOXC13-AS is achieved by interacting with microRNAs to exhibit sponge-like activity, which in turn regulates the expression of target genes to exert its regulatory effect. For example, in nasopharyngeal carcinoma, HOXC13-AS sponges miR-497-5p, forming a ceRNA network to enhance the expression of HMGA2, which means that it participates in tumor progression through the HOXC13-AS–miR-383-3p–HMGA2 axis (Gao et al., 2019). Similarly, in breast cancer, HOXC13-AS plays a role through the miR-497-5p/PTEN axis (Li et al., 2019b). In oral squamous cell carcinoma, HOXC13-AS affects tumor development through the miR-378g/HOXC13 axis (Li et al., 2020). In gliomas, HOXC13-AS exerts regulatory effects through the HOXC13-AS–miR-122-5p–SATB1–c-Myc feedback loop (Liu et al., 2019). In addition, HOXC13-AS can interact with proteins along with microRNAs. In cervical cancer, HOXC13-AS upregulates FZD6 by interacting with CBP, thereby activating Wnt/β-catenin signaling to accelerate tumor progression (Wang et al., 2021). In glioma, nasopharyngeal carcinoma, breast cancer and oral squamous cell carcinoma, HOXC13-AS is expected to become a potential target for treatment. Interestingly, HOXC13-AS plays a role in these tumors through the lncRNA-miRNA axis. And this axis is particularly important in clinical trials, as the two non-coding RNA molecules in the axis can regulate the effects of drugs on the body, reduce cell resistance, and provide new ideas for the development and application of targeted new drugs (Entezari et al., 2022; Ma et al., 2023). The discovery of these molecular mechanisms suggests that HOXC13-AS may become a potential target for cancer therapy, enhancing the sensitivity of cancer treatment.
Although there has been progress in the study of the role of HOXC13-AS in human tumors, there are still several issues that remain to be explored and resolved. Firstly, Figures 2B–D shows that HOXC13-AS is highly expressed in various cancers, but there is a lack of research on the upstream regulatory processes of HOXC13-AS. Existing research suggests that upregulation of lncRNA genes may be related to transcription factor activation (Wei et al., 2016), N6-methyladenosine demethylation (Yao et al., 2024), single nucleotide polymorphism (Yan et al., 2021), and other factors. Therefore, further exploration of the upstream mechanisms of HOXC13-AS is still needed. Secondly, there is a lack of existing research on the molecular mechanisms and related signaling pathways involved in the regulation of HOXC13-AS for different types of cancer, and a more comprehensive investigation is required. Therefore, there is an urgent need to search for downstream small molecules that can be paired with it to explore the corresponding molecular mechanisms and provide new targets and ideas for cancer treatment to meet the needs of developing and optimizing new treatment methods. Thirdly, more in vivo and in vitro experiments will be needed to validate the potential clinical application value of HOXC13-AS as a diagnostic and prognostic biomarker. It is expected to establish a multi–biomarker combined diagnostic or prognostic model in the near future, combining HOXC13-AS with other related biomarkers, comprehensively analyzing various indicators, and providing stronger evidence for clinical applications. During this process, due to the main regulatory role of lncRNA in the organism, its copy number is low and its expression is restricted in different developmental stages and cells, making detection difficult (Mattick et al., 2023). What’s even more complex is that most lncRNAs have different subtypes and their localization in cells varies, even if they are located the same, they still have different functions (Bridges et al., 2021). However, in current research on HOXC13-AS, there is a lack of studies on its subcellular localization, which is crucial for studying the function of HOXC13-AS. Therefore, further in-depth exploration is needed in this area.
In summary, HOXC13-AS plays a unique role in regulating tumor progression. A detailed and comprehensive understanding of the role of HOXC13-AS will facilitate the exploration of its clinical value as a prognostic and diagnostic biomarker and enhance its potential as a new therapeutic target. More in-depth scientific research on the diverse mechanisms of HOXC13-AS will be needed to conducted in the near future. Its plasticity in treatment resistance and drug development were looked forward to further clarify based on multi–center research cooperation, in order to promote the clinical application value of HOXC13-AS.
Author contributionsXG: Conceptualization, Writing–original draft. XH: Data curation, Formal Analysis, Software, Writing–review and editing. SZ: Validation, Visualization, Writing–review and editing. XZ: Data curation, Formal Analysis, Writing–review and editing. YW: Funding acquisition, Writing–original draft, Writing–review and editing. LL: Conceptualization, Investigation, Methodology, Supervision, Writing–original draft, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant no. 82103131), the Key R&D Program of Shandong Province (Major Science and Technology Innovation Project) (grant no. 2022CXGC010510), the Taishan Industrial Experts Program, China.
Conflict of interestThe authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAngenard, G., Merdrignac, A., Louis, C., Edeline, J., and Coulouarn, C. (2019). Expression of long non-coding RNA ANRIL predicts a poor prognosis in intrahepatic cholangiocarcinoma. Dig. Liver Dis. 51 (9), 1337–1343. doi:10.1016/j.dld.2019.03.019
PubMed Abstract | CrossRef Full Text | Google Scholar
Arab, K., Karaulanov, E., Musheev, M., Trnka, P., Schäfer, A., Grummt, I., et al. (2019). GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 51 (2), 217–223. doi:10.1038/s41588-018-0306-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74 (3), 229–263. doi:10.3322/caac.21834
PubMed Abstract | CrossRef Full Text | Google Scholar
Buskwofie, A., David-West, G., and Clare, C. A. (2020). A review of cervical cancer: incidence and disparities. J. Natl. Med. Assoc. 112 (2), 229–232. doi:10.1016/j.jnma.2020.03.002
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, R., and He, P. (2021). Long noncoding RNA HOXA-AS2 accelerates cervical cancer by the miR-509-3p/BTN3A1 axis. J. Pharm. Pharmacol. 73 (10), 1387–1396. doi:10.1093/jpp/rgab090
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, X., Song, J., Wang, X., Sun, D., Liu, Y., and Jiang, Y. (2022). LncRNA LINC00460: function and mechanism in human cancer. Thorac. Cancer 13 (1), 3–14. doi:10.1111/1759-7714.14238
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, Y. P., Chan, A. T. C., Le, Q. T., Blanchard, P., Sun, Y., and Ma, J. (2019). Nasopharyngeal carcinoma. Lancet 394 (10192), 64–80. doi:10.1016/s0140-6736(19)30956-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Chi, Y., Wang, D., Wang, J., Yu, W., and Yang, J. (2019). Long non-coding RNA in the pathogenesis of cancers. Cells 8 (9), 1015. doi:10.3390/cells8091015
PubMed Abstract | CrossRef Full Text | Google Scholar
Entezari, M., Taheriazam, A., Orouei, S., Fallah, S., Sanaei, A., Hejazi, E. S., et al. (2022). LncRNA-miRNA axis in tumor progression and therapy response: an emphasis on molecular interactions and therapeutic interventions. Biomed. Pharmacother. 154, 113609. doi:10.1016/j.biopha.2022.113609
PubMed Abstract | CrossRef Full Text | Google Scholar
Fu, W. M., Lu, Y. F., Hu, B. G., Liang, W. C., Zhu, X., Yang, H. D., et al. (2016). Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 7 (4), 4712–4723. doi:10.18632/oncotarget.6731
PubMed Abstract | CrossRef Full Text | Google Scholar
Galamb, O., Barták, B. K., Kalmár, A., Nagy, Z. B., Szigeti, K. A., Tulassay, Z., et al. (2019). Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 25 (34), 5026–5048. doi:10.3748/wjg.v25.i34.5026
PubMed Abstract | CrossRef Full Text | Google Scholar
Gao, C., Lu, W., Lou, W., Wang, L., and Xu, Q. (2019). Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J. Cell. Physiol. 234 (8), 12809–12820. doi:10.1002/jcp.27915
PubMed Abstract | CrossRef Full Text | Google Scholar
Goyal, B., Yadav, S. R. M., Awasthee, N., Gupta, S., Kunnumakkara, A. B., and Gupta, S. C. (2021). Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta Rev. Cancer 1875 (2), 188502. doi:10.1016/j.bbcan.2021.188502
PubMed Abstract | CrossRef Full Text | Google Scholar
Guo, Y., Liu, B., Huang, T., Qi, X., and Li, S. (2023). HOTAIR modulates hepatocellular carcinoma progression by activating FUT8/core-fucosylated Hsp90/MUC1/STAT3 feedback loop via JAK1/STAT3 cascade. Dig. Liver Dis. 55 (1), 113–122. doi:10.1016/j.dld.2022.04.009
PubMed Abstract | CrossRef Full Text | Google Scholar
Győrffy, B. (2024). Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 181 (3), 362–374. doi:10.1111/bph.16257
PubMed Abstract | CrossRef Full Text | Google Scholar
Han, B., Zheng, R., Zeng, H., Wang, S., Sun, K., Chen, R., et al. (2024). Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 4 (1), 47–53. doi:10.1016/j.jncc.2024.01.006
PubMed Abstract | CrossRef Full Text | Google Scholar
Hashemi, M., Moosavi, M. S., Abed, H. M., Dehghani, M., Aalipour, M., Heydari, E. A., et al. (2022). Long non-coding RNA (lncRNA) H19 in human cancer: from proliferation and metastasis to therapy. Pharmacol. Res. 184, 106418. doi:10.1016/j.phrs.2022.106418
PubMed Abstract | CrossRef Full Text | Google Scholar
Hu, G., Lou, Z., and Gupta, M. (2014). The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One 9 (9), e107016. doi:10.1371/journal.pone.0107016
PubMed Abstract | CrossRef Full Text | Google Scholar
Iwai, M., Kajino, T., Nakatochi, M., Yanagisawa, K., Hosono, Y., Isomura, H., et al. (2023). Long non-coding RNA TILR constitutively represses TP53 and apoptosis in lung cancer. Oncogene 42 (5), 364–373. doi:10.1038/s41388-022-02546-w
PubMed Abstract | CrossRef Full Text | Google Scholar
Jiang, L., Wu, Z., Meng, X., Chu, X., Huang, H., and Xu, C. (2019). LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 Axis. Hum. Gene Ther. 30 (5), 618–631. doi:10.1089/hum.2018.109
PubMed Abstract | CrossRef Full Text | Google Scholar
Jiang, W., Kai, J., Li, D., Wei, Z., Wang, Y., and Wang, W. (2020). lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J. Cell. Physiol. 235 (10), 7194–7203. doi:10.1002/jcp.29618
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, J., Meng, Q., Zhou, X., Zhao, H., Wang, K., Niu, H., et al. (2022). Gospel of malignant Glioma: oncolytic virus therapy. Gene 818, 146217. doi:10.1016/j.gene.2022.146217
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, L., Wang, Y., Song, G., Zhang, X., Gao, S., and Liu, H. (2019a). HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 450, 14–21. doi:10.1016/j.canlet.2019.02.036
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, S., Xiong, Q., Chen, M., Wang, B., Yang, X., Yang, M., et al. (2021). Long noncoding RNA HOTAIR interacts with Y-Box Protein-1 (YBX1) to regulate cell proliferation. Life Sci. Alliance 4 (9), e202101139. doi:10.26508/lsa.202101139
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, W., Zhu, Q., Zhang, S., Liu, L., Zhang, H., and Zhu, D. (2020). HOXC13-AS accelerates cell proliferation and migration in oral squamous cell carcinoma via miR-378g/HOXC13 axis. Oral Oncol. 111, 104946. doi:10.1016/j.oraloncology.2020.104946
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, X., Wang, Q., Rui, Y., Zhang, C., Wang, W., Gu, J., et al. (2019b). HOXC13-AS promotes breast cancer cell growth through regulating miR-497-5p/PTEN axis. J. Cell. Physiol. 234 (12), 22343–22351. doi:10.1002/jcp.28800
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, N., Wang, Z., Liu, D., and Xie, P. (2019). HOXC13-AS-miR-122-5p-SATB1-C-Myc feedback loop promotes migration, invasion and EMT process in glioma. Onco Targets Ther. 12, 7165–7173. doi:10.2147/ott.S220027
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, R., Wang, X., Shen, Y., and He, A. (2021). Long non-coding RNA-based glycolysis-targeted cancer therapy: feasibility, progression and limitations. Mol. Biol. Rep. 48 (3), 2713–2727. doi:10.1007/s11033-021-06247-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, X., Li, Y., Jiang, X., Deng, Y., Ma, C., Yu, Q., et al. (2022). Long non-coding RNA: multiple effects on the differentiation, maturity and cell function of dendritic cells. Clin. Immunol. 245, 109167. doi:10.1016/j.clim.2022.109167
PubMed Abstract | CrossRef Full Text | Google Scholar
Luo, H., Zhu, G., Eshelman, M. A., Fung, T. K., Lai, Q., Wang, F., et al. (2022). HOTTIP-dependent R-loop formation regulates CTCF boundary activity and TAD integrity in leukemia. Mol. Cell. 82 (4), 833–851.e11. doi:10.1016/j.molcel.2022.01.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Ma, B., Wang, S., Wu, W., Shan, P., Chen, Y., Meng, J., et al. (2023). Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 162, 114672. doi:10.1016/j.biopha.2023.114672
PubMed Abstract | CrossRef Full Text | Google Scholar
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
留言 (0)