Epidermal naevus is a relatively common cutaneous mosaic disorder, typically seen at birth or developing in early childhood and evolving during puberty. Keratinocytic epidermal naevus is usually characterised clinically by gray-brown papules or plaques distributed in a linear pattern, following the lines of Blaschko and histologically by hyperplasia of epidermal keratinocytes. Clinically, keratinocytic epidermal naevus can be classified into localised or generalised variants, and inflammatory linear verrucous epidermal naevus.1 Inflammatory linear verrucous epidermal naevus could be localised or generalised. Histopathologically, keratinocytic epidermal naevus can be categorised into many distinct types, such as epidermolytic epidermal naevus, non-epidermolytic epidermal naevus, and inflammatory epidermal naevus.2 Inflammatory epidermal naevus could overlap with the other two subtypes. Herein, we describe the clinical, histopathological, and genetic features of 22 unrelated patients with keratinocytic epidermal naevi in the Chinese population.
MethodsWe retrospectively reviewed the clinical, pathological, and genetic features of patients referred to our department for keratinocytic epidermal naevus between 2021 and 2024. Clinical information and medical history was collected. All patients underwent a complete physical examination at their first visit and every 12 months thereafter, in order to detect any transition towards epidermal naevus syndrome. Skin biopsies were performed for histopathological examination. Peripheral blood and lesional skin samples were obtained for the whole exome sequencing. Subsequently, targeted deep sequencing was carried out with an average coverage depth reaching × 20,000.
Results Clinical dataThe series consisted of 22 cases, 10 males and 12 females, from 22 unrelated families [Table 1]. Seventeen patients (17/22, 77.27%) were aged < 18 years while the remaining five (5/22, 22.73%) were adults. Lesions were observed in a majority of the patients since birth (21/22, 95.45%) with only one (1/22, 4.55%) developing lesions after the age of two. Unilateral distribution of the lesions was observed in most patients (17/22, 77.27%). Right-sided involvement (15/17, 88.24%) was much more common than the left-sided one (2/17, 11.76%). Fourteen patients had generalised epidermal naevus (14/22, 63.64%) while eight had the localised variant (8/22, 36.36%) [Figures 1a–1c]. All three patients with inflammatory linear verrucous epidermal naevus were female, with the lesions in all of them presenting on the right side of the body. Extracutaneous abnormalities were not documented in our patients.
Table 1: Clinical, histopathological and molecular features of 22 cases with keratinocytic epidermal naevus in our study
Pt N* Sex Age of visit Age of onset Family history Clinical type Side of the involvement Distribution Histopathologic type Mutation Reference Mutation allele load in skin lesions (reads) Mutation allele load in peripheral blood (reads) Germline mutation disease 1 M† 8y° 5mo§ Negative Localised Right Neck Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 13.12% (25,436/193,871) 0 Costello syndrome 2 F‡ 1y Birth Negative Generalised Right Scalp, face, trunk, groin, limbs Non-epidermolytic HRAS c.38G>T; p.Gly13Val 6 28.13% (202,230/719,040) 7.22% (66,506/921,430) / 3 M 2y Birth Negative Localised Left Hand, chest Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 15.16% (30,047/198,200) 0 Costello syndrome 4 F 28y 1y Negative Generalised Right Trunk, leg Non-epidermolytic FGFR3 c.742C>T; p.Arg248Cys 7 25.71% (13,770/53,550) 0.86% (527/61,026) Thanatophoric dysplasia 5 F 4y 1y Negative Generalised, ILVEN# Right Neck, trunk, limbs Epidermolytic, inflammatory KRT10 c.467G>A; p.Arg156His 3 8.74% (4653/53,234) 1.91% (1065/55,646) EI 6 F 10y birth Negative Generalised, ILVEN Right Trunk, arm Epidermolytic, inflammatory KRT10 c.466C>T; p.Arg156Cys 8 18.42% (32,005/173,715) 2.12% (3185/150,292) EI 7 M 4y 2mo Negative Generalised Left and right Scalp, face, neck, trunk, limbs Non-epidermolytic HRAS c.34G>A; p.Gly12Ser 9 37.11% (88,706/239,059) 2.12% (5598/263,832) Costello syndrome 8 F 5y 7mo Negative Generalised Left and right Neck, trunk, limbs Non-epidermolytic HRAS c.34G>A; p.Gly12Ser 9 32.45% (95,228/293,464) 4.56% (10,934/239,863) Costello syndrome 9 F 10y Birth Negative Localised Left and right Back, hands Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 17.46% (11,765/67,364) 0 Costello syndrome 10 M 4y Birth Negative Localised Right Chest Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 22.01% (36,256/164,697) 0 Costello syndrome 11 F 4y Birth Negative localised Right Neck Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 34.25% (29,269/85,461) 0 Costello syndrome 12 M 7y Birth Negative Generalised Right Scalp, trunk, limbs Non-epidermolytic GJB2 c.148G>A; p.Asp50Asn 5 38.46% (108,640/282,464) 1.40% (5315/380,411) Keratitis-ichthyosis-deafness syndrome 13 F 4y Birth Negative Generalised Left and right Right region of trunk, right arm, lower legs Non-epidermolytic Not identified / 0 0 / 14 M 19y 12y Negative Generalised Right Trunk, leg Non-epidermolytic Not identified / 0 0 / 15 M 13y Birth Negative Generalised Right Trunk, groin, leg Non-epidermolytic Not identified / 0 0 / 16 M 3y Birth Negative Generalised Right Trunk, limbs Non-epidermolytic Not identified / 0 0 / 17 F 5y Birth Negative Localised Right Leg Non-epidermolytic Not identified / 0 0 / 18 F 47y Birth Negative Localised, ILVEN Right Arm Non-epidermolytic, inflammatory PIK3CA c.1633G>A; p.Glu545Lys 10 20.43% (18,657/91,309) 0 / 19 M 33y Birth Yes (daughter suffers from EI◊) Generalised Left and right Trunk, limbs Epidermolytic KRT10 c.467G>A; p.Arg156His 3 25.08% (163,778/652,937) 0 EI 20 M 13y Birth Negative Localised Left Neck, face Non-epidermolytic HRAS c.37G>C; p.Gly13Arg 4 27.27% (47,256/173,272) 0 Costello syndrome 21 F 27y Birth Negative Generalised Right Trunk, arm, buttock Non-epidermolytic FGFR3 c.742C>T; p.Arg248Cys 7 28.66% (18,485/64,495) 0 Thanatophoric dysplasia 22 F 6y Birth Negative Generalised Right Neck, trunk, limbs Non-epidermolytic FGFR3 c.742C>T; p.Arg248Cys 7 24.47% (16,468/67,304) 0 Thanatophoric dysplasia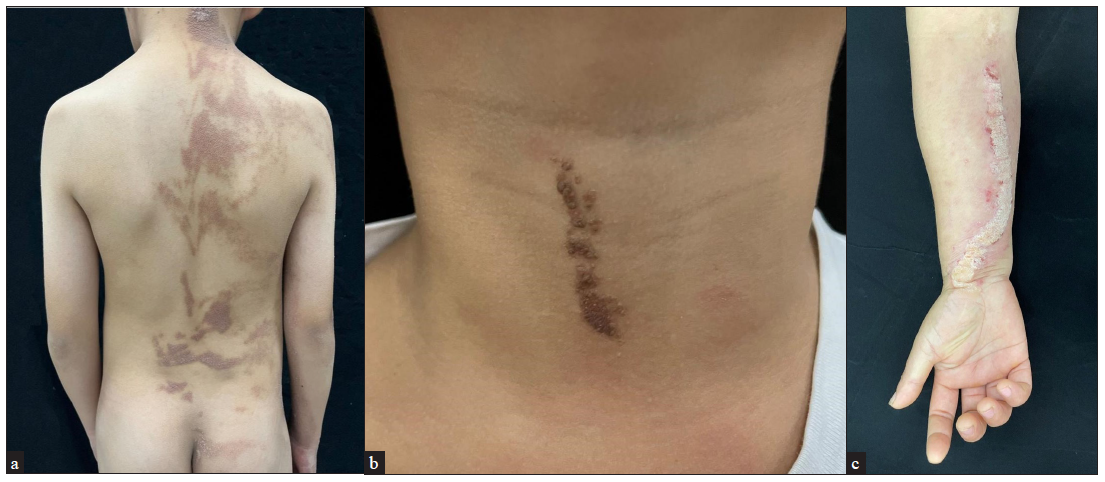
Export to PPT
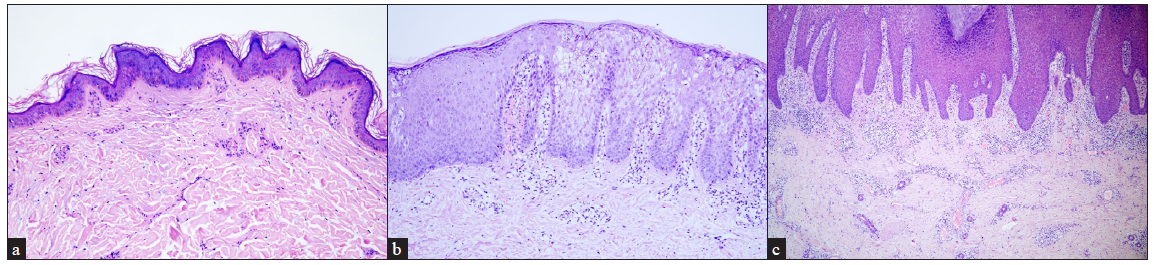
Histopathological findingsHaematoxylin-eosin staining was performed in all the 22 patients [Table 1]. Nineteen patients were affected with non-epidermolytic epidermal naevus (19/22, 86.36%) and the remaining three had epidermolytic epidermis naevus (3/22, 13.64%). Among them, two of epidermolytic epidermal naevus (2/3, 66.67%) and one of non-epidermolytic epidermal naevus (1/3, 33.33%) overlapped with inflammatory epidermal naevus. Compared to the non-inflammatory epidermal naevus (19/22, 86.36%), inflammatory epidermal naevus (3/22, 13.64%) was rarer [Figures 2a–2c].
Export to PPT
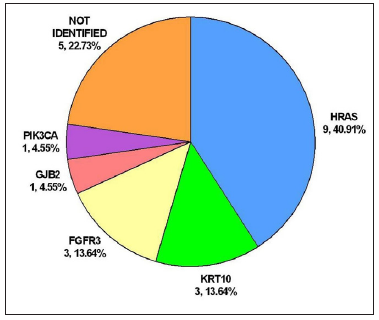
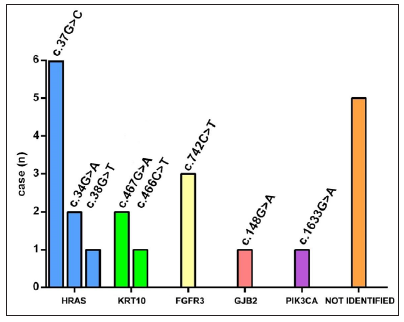
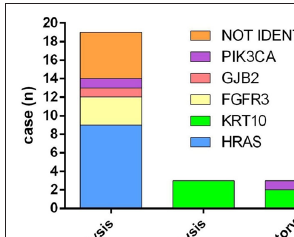
Genotype-phenotype correlationWhole exome sequencing followed by targeted deep sequencing were performed using lesional skin and peripheral blood samples from all 22 patients [Table 1]. Seventeen patients (17/22, 77.27%) were found to carry mutations in five genes related to keratinocytic epidermal naevus, including HRAS, KRT10, FGFR3, GJB2, and PIK3CA. Mutations of HRAS were mostly prevalent with an incidence of 40.91% (9/22). Mutations of KRT10, FGFR3, GJB2, and PIK3CA were identified in 3/22 (13.64%), 3/22 (13.64%), 1/22 (4.55%) and 1/22 (4.55%), respectively [Figure 3a]. A total of eight missense mutations were identified in our study, which were classified as pathogenic/likely pathogenic variants in the ClinVar database and were previously reported in epidermis naevus or epidermis naevus-related syndrome.3–10 The c.37G>C substitution in HRAS represented a possible hotspot mutation (6/22, 27.27%) [Figure 3b].
Export to PPT
Export to PPT
Export to PPT
In lesional skin samples of the 17 patients with somatic mutations, the mutation allele load ranged from 8.74% to 38.46%. Only seven patients (7/22, 31.82%) were found carrying mutations in the peripheral blood, with the mutation allele load ranging from 0.86% to 7.22%. Mutations were identified in half of the blood samples in generalised epidermal naevus (7/14, 50%), while none of the mutations was observed in the blood samples in localised epidermal naevus. On the whole, in terms of mutation allele load, it was much lower in the blood (0.92 ± 1.82%) than in the lesional skin (18.95 ± 12.82%). In addition, compared to the localised epidermis naevus, the mutation allele load was significantly higher in generalised epidermal naevus, either in the peripheral blood (1.44 ± 2.13% vs. 0) or lesional skin (19.09 ± 14.48% vs. 18.71 ± 10.17%) [Table 2].
Table 2: Mutation allele load analysis of patients with keratinocytic epidermal naevus included in this study
Localised EN* (PN†, MAL‡) Generalised EN (PN, MAL) Total EN (PN, MAL) p-value (Localised vs. Generalised) Total (N) 8 14 22 / Blood 0, 0 7, 1.44 ± 2.13% 7, 0.92 ± 1.82% 0.0730 Lesional skin 7, 18.71 ± 10.17% 10, 19.09 ± 14.48% 17, 18.95 ± 12.82% 0.9492 p-value (Blood vs. Lesional skin) 0.0001 0.0001 < 0.0001 /In our study, 14 patients (14/22, 63.64%) had generalised epidermal naevus, seven of whom were identified with mutations in HRAS (3/14, 21.43%), KRT10 (2/14, 14.29%), FGFR3 (1/14, 7.14%) and GJB2 (1/14, 7.14%). Eight patients (8/22, 36.36%) presented with localised epidermal naevus. Mutations in HRAS (6/8, 75.00%) and PIK3CA (1/8, 12.50%) were found in seven of them. HRAS mutations occurred most frequently, including c.34G>A, c.38G>T, and c.37G>C. The c.34G>A and c.38G>T mutations were found in generalised epidermal naevus, while all the c.37G>C mutations were identified in localised epidermal naevus. Of note, acantholysis (epidermolytic epidermal naevus) was found in three patients (3/22, 13.64%) exclusively carrying KRT10 mutations. Inflammatory epidermal naevus (3/22, 13.64%) was caused by mutations in KRT10 (2/3, 66.67%) and PIK3CA (1/3, 33.33%) [Figure 3c].
DiscussionClinically, no gender predilection was observed in the study and a majority of the patients developed keratinocytic epidermal naevus at birth. Unilateral distribution of the lesions was found in most patients, with right-sided involvement being much more common. Histopathologically, non-epidermolytic epidermal naevus was found more commonly.
Inflammatory linear verrucous epidermal naevus is a rare inflammatory epidermal naevus characterised with pruritic erythematous scaly papules and plaques occurring along Blaschko’s lines.11 In this study, three female cases of inflammatory linear verrucous epidermis naevus were included. Unlike a previous report from India12that, the left-sided inflammatory linear verrucous epidermis naevus was more common, in our study, all the the patients with this variant exhibited right-sided involvement, suggesting clinical heterogeneity in different populations. Inflammatory linear verrucous epidermal naevus was reported to be caused by mosaic mutations in GJA1, CARD14, HRAS, and KRT10.13 In our study, intriguingly, except for two patients with generalised inflammatory linear verrucous epidermal naevus caused by KRT10 mutations, a localised inflammatory linear verrucous epidermal naevus patient was found to carry a PIK3CA mutation (c.1633G>A), which has not been reported in this type previously. We speculate that compared to the PIK3CA mutation, KRT10 mutations seemingly lead to more extensive skin lesions in inflammatory linear verrucous epidermal naevus patients.
Keratinocytic epidermal naevus is a genotypically diverse mosaic disorder. Non-epidermolytic keratinocytic epidermal naevus is known to arise as a result of prenatal postzygotic mutations in either rat sarcoma/mitogen-activated protein kinase pathway (HRAS, KRAS, NRAS, BRAF) or the PI3K/AKT pathway (PIK3CA).4,14,15 Among the growth factor receptor genes found upstream of these pathways, mutations in FGFR2, FGFR3, or EGFR have also been reported to cause non-epidermolytic keratinocytic epidermal naevus.14 However, epidermolytic keratinocytic epidermal naevus is often associated with mutations in keratin genes, such as KRT1, KRT2, and KRT10.16,17 In addition, mutations in GJB2 have been found in both non-epidermolytic and epidermolytic keratinocytic epidermal naevus.5,18 Similar findings were observed in our study. Mutations of HRAS, GJB2, FGFR3 and PIK3CA were identified in non-epidermolytic keratinocytic epidermal naevus, while KRT10 was associated with epidermolytic keratinocytic epidermal naevus [Figure 3c].
Clinical manifestations of keratinocytic epidermal naevi vary according to the underlying pathogenic genes. Postzygotic mutations in HRAS and FGFR3 have been reported to cause systematized epidermal naevus, inducing skeletal abnormalities, thymoma, breast hypotrophy, acanthosis nigricans, and more.19,20 More than half (12/22, 54.55%) of the keratinocytic epidermal naevus patients carried the HRAS or FGFR3 mutations in our study. Although none of the patients in our study was found to be associated with extracutaneous symptoms, further periodical follow-up every 12 months is necessary to assess the risk of systematic involvement, especially in those who carry mutations in HRAS and FGFR3.21
HRAS was the most commonly affected gene, with c.37G>C substitution representing a hotspot mutation.4 In our study, compared to c.38G>T and c.34G>A found in the generalised epidermis naevus, c.37G>C was only identified in the localised epidermis naevus, suggesting c.37G>C mutation may lead to a milder epidermis naevus phenotype.
As a mosaic disorder, our study showed that the mutation allele load was significantly higher in generalised epidermal naevus either in the peripheral blood or lesional skin when compared to the localised variant. Meanwhile, the mutation detection rate was much lower in the blood than lesional skin in either generalised or localised epidermal naevus. Therefore, deep next-generation sequencing of the affected lesions is necessary to detect mutations in keratinocytic epidermal naevus. Genetic analysis performed solely in the peripheral blood could easily lead to false conclusions, particularly in localised epidermal naevus.22,23
It is worth noting that most mosaic mutations detected in our keratinocytic epidermal naevus patients were the same as the germline mutations identified in systemic diseases caused by these genes.11 This means that in keratinocytic epidermal naevus, an individual carrying a somatic heterozygous mutation could have co-existing gonadal mosaicism and would be at risk of transmission of the mutation to the offspring, who usually presents much more severe phenotypes.
Treatment for epidermis naevus is challenging. Recently, sirolimus which blocks mammalian target of rapamycin signalling was found effective in the patient with epidermis naevus carrying a FGFR3 mutation.24 Based on the widespread application of gene detection, targeted therapy could be a trend in the treatment of keratinocytic epidermal naevus in the future.
LimitationsThis study is limited by its retrospective nature, the non-standardised documentation and small sample size.
ConclusionTo the best of our knowledge, this is the largest case series of keratinocytic epidermal naevus in the Chinese population so far. Our results reveal the genotype-phenotype spectrum and its correlation in this disease. The study underscores the importance of genetic testing in characterisation and categorisation of keratinocytic epidermal naevus, having a bearing on genetic counselling as well as prenatal diagnosis. Future therapeutic strategies could also be based on these findings.
留言 (0)