Preeclampsia (PE) is a complex condition that affects various bodily systems and occurs in 2%–8% of pregnancies worldwide. It continues to be a significant cause of maternal and fetal morbidity and mortality (1). It is believed that PE is responsible for approximately 76,000 maternal and 500,000 fetal fatalities annually (2). It is a pregnancy-related disease that usually occurs after 20 weeks of gestation, characterized by high blood pressure and proteinuria. According to the definition of the World Health Organization, PE refers to a pregnant woman with a blood pressure ≥140/90 mmHg in the later stages of pregnancy and urine containing ≥300 milligrams of protein. It poses significant health hazards, including the potential onset of eclampsia, hemolysis, elevated liver enzymes, and low platelet count (HELLP) syndrome, and enduring cardiovascular issues (3). The pathophysiological mechanism of PE is intricate, encompassing various factors such as placental insufficiency, vascular endothelial dysfunction, and immune dysregulation. Research has demonstrated that in a normal pregnancy, the placenta releases specific signaling molecules to facilitate maternal blood vessel dilation and increased blood flow to accommodate the fetal growth requirements. However, in individuals with PE, there is often inhibition of placental development and function, resulting in damage to endothelial cells and a systemic inflammatory response, ultimately leading to elevated blood pressure and other complications (4). Despite advancements in prenatal care, the primary method for diagnosing PE is to monitor blood pressure and protein levels in the urine. The clinical physician also takes into consideration other potential symptoms, such as cephalalgia, visual impairments, epigastric discomfort, and renal function irregularities. The presence of these symptoms is typically associated with the severity of the condition and holds prognostic significance. Treatment includes using antihypertensive drugs to control the mother’s blood pressure and early low-dose aspirin and calcium supplements to reduce the risk of developing PE. In severe cases, termination of pregnancy is frequently required; however, this generally results in premature birth. Although these interventions are necessary, treatment options for PE are significantly limited, focusing primarily on symptom management rather than addressing the underlying cause of the condition (5). Current treatments have potential limitations, including side effects from antihypertensive drugs and early delivery-associated risks, emphasizing the importance of improving our understanding of PE management strategies.
The disruption of metabolism and metabolites in PE pathogenesis is becoming an essential component of the disease pathophysiology. Studies have reported that carbohydrate and lipid metabolism abnormalities are essential in the etiology and clinical progression of PE (6, 7). Previous studies have indicated a connection between the diverse expression of energy metabolism-related genes (EMRGs) and the emergence of several pregnancy complications, including gestational diabetes mellitus and fetal obesity. This suggests that these conditions can have a common pathophysiological foundation and can be potential targets for treatment (8). Furthermore, differential expression of EMRGs has been associated with altered mitochondrial function and oxidative stress (9), which are characteristic features of the placental pathology in PE (10). Despite the preceding insights, there are significant gaps in understanding the complex energy metabolic pathways and their interactions in PE pathogenesis.
A comprehensive comprehension of the pathogenesis, biomarkers, and associated complications of PE is imperative for enhancing early diagnosis and treatment efficacy. Our study aimed to identify and analyze EMR differentially expressed genes (DEGs) in PE and determine their functional significance. Using bioinformatics methods, including data collection, differential gene expression analysis, functional pathway enrichment, protein-protein interaction (PPI) network creation, regulatory network visualization, and immune infiltration assessment, we provide a new perspective on molecular alterations in PE. This integrated genomic and bioinformatics approach aims to develop novel diagnostic markers and therapeutic targets, increasing our understanding of the molecular foundation underlying PE and aiding in personalized medical strategies to mitigate its impact on mothers and offspring.
2 Materials and methods2.1 Data downloadThe gene expression omnibus (GEO) database (11) (https://www.ncbi.nlm.nih.gov/geo/) provided the PE datasets GSE60438 (12) and GSE75010 (13–18), which were retrieved using the R package “GEOquery”. Dataset GSE60438 was derived from Homo sapiens, originating from decidua basalis tissue, with chip platforms GPL10558 and GPL6884. Dataset GSE75010 was derived from Homo sapiens placental tissues using the chip platform GPL6244. Detailed information is provided in Table 1. Additionally, dataset GSE60438 contained 42 control and 35 PE samples on the GPL10558 platform and 23 control and 25 PE samples on the GPL6884 platform. Dataset GSE75010 comprised 77 control and 80 PE samples. Following batch correction, the combined data from both GPL platforms in dataset GSE60438 were included in the study, whereas dataset GSE75010 was used as a validation set.
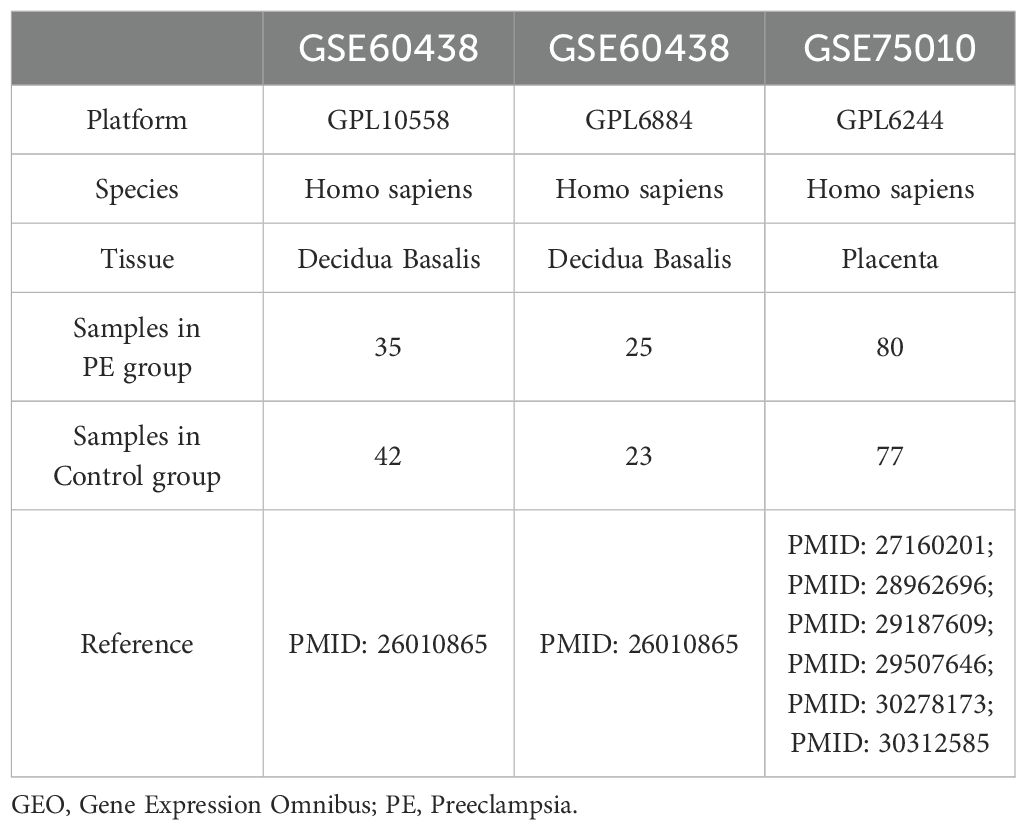
Table 1. GEO Microarray Chip Information.
We obtained EMRGs from the GeneCards database (19). The GeneCards database provides extensive provides on genes in the human body. After conducting a search using the term “Energy Metabolism” and filtering for “Protein Coding” and “Relevance Score > 2” EMRGs, 571 EMRGs were obtained. Furthermore, using “Energy Metabolism” as the keyword in PubMed, 8 EMRGs were found in the published literature (20). Following the combination and elimination of duplicates, 573 EMRGs were identified, with detailed information presented in Supplementary Table S1.
The “sva” (21) package in R was used to correct batch effects in data from two GPL platforms (GPL10558 and GPL6884) to obtain the merged GEO dataset (combined datasets). The combined datasets included 65 control and 60 PE samples. Finally, the annotation and standardization of the merged datasets were performed using the R software package “limma” (22). To determine the impact of the batch effect, we performed a principal component analysis (PCA) (23) on the expression matrix before and following its removal.
2.2 Energy metabolism-related differentially expressed genes in PEThe “limma” R package was used to analyze the differences in gene expression between PE and control groups. To identify the DEGs, criteria of |log fold change(logFC)| > 0.5 and a p < 0.05 were set. Additionally, genes with a logFC > 0.5 and a p < 0.05 were classified as upregulated DEGs. Conversely, genes with a logFC < -0.5 and a p < 0.05 were identified as downregulated DEGs. The differential analysis results were depicted using the volcano plot feature provided by the “ggplot2” package in R.
We combined datasets to identify EMRGs that were differentially expressed in association with PE. We determined variance to identify genes exhibiting significant differences (|logFC| > 0.5 and p < 0.05). Venn diagrams were used to map the intersection of the DEGs and EMRGs, enabling the identification of EMRDEGs. We generated a heatmap with the R package “pheatmap”. Furthermore, we constructed a chromosome localization map using the R package “RCircos” (24).
2.3 Enrichment analysis using gene ontology and the Kyoto encyclopedia of genes and genomesGO (25) analysis is a widely used methodology for in-depth investigations aimed to improve functionality across multiple dimensions, including biological process (BP), cellular component (CC), and molecular function (MF). The KEGG (26) database is an extensive resource for deciphering the intricate functions and uses of biological systems by connecting genetic information with biochemical pathways and cellular activities. We employed the R package “clusterProfiler” (27) to perform GO and KEGG enrichment analyses on the EMRDEGs. The parameters established for including genes were an adjusted p-value (adj. p) < 0.05 and a false discovery rate (FDR) < 0.05, both of which were considered statistically significant. The Benjamini–Hochberg (BH) procedure was used as a p-value adjustment method.
2.4 Gene set enrichment analysisGSEA (28) is a statistical technique to determine if predefined gene groups exhibit significant enrichment across various biological conditions. In this study, the genes of combined datasets were first sorted according to logFC values. Then, GSEA was performed on the entire set of genes from the merged datasets, using the “clusterProfiler” package in R. GSEA settings were accessing the “c2.cp.all.v2022.1.Hs.symbols.gmt [All Canonical Pathways] (3050)” gene set from the Molecular Signatures database (29), using 2022 seeds, performing 1000 calculations, with each gene set containing between 10 and 500 genes. The evaluation standards were established as adj. p < 0.05 and FDR (q-value) < 0.05 via the BH method for p-value adjustment.
2.5 Analysis of PPI and identification of key genesThe PPI network includes essential proteins involved in numerous biological functions, including signaling pathways, gene expression regulation, metabolism of energy and substances, and cell cycle management. This network is crucial for comprehending protein functionalities, signaling mechanisms, and physiological and pathological functional associations. The search tool for the retrieval of interacting genes/proteins (STRING) database (30) (https://cn.string-db.org/) investigates the connections among identified and anticipated proteins. This study used the STRING database to build a PPI network associated with EMRDEGs, adhering to the criteria of a minimum interaction coefficient exceeding 0.400, which corresponded to a medium confidence level. The associated regions within the PPI network could indicate molecular assemblies with distinct biological roles. Certain genes were identified as key genes within the PPI network as a result of their interactions with other genes.
The GeneMANIA database (31) (https://genemania.org/) was used to predict potential gene functions, evaluate gene lists, and pinpoint genes for further functional analyses. When provided with a list of query genes, GeneMANIA identifies functionally similar genes by analyzing a comprehensive genomics and proteomics dataset. It assigns weights to each functional genomic dataset based on the anticipated value of the query in this process. Besides, GeneMANIA can predict gene functions by identifying genes likely to share tasks with a given query gene based on their interactions. Using the GeneMANIA online website, the PPI network was created to predict genes with functions similar to those of key genes.
2.6 Construction of regulatory networkGene expression is regulated by transcription factors (TFs) through their interaction with crucial genes during the post-transcriptional phase. We used the ChIPBase database (http://rna.sysu.edu.cn/chipbase/) (32) to obtain data on TFs and determine their control over essential genes. The screening criterion for mRNA-TF interaction pairs was based on the total number of upstream and downstream samples, which was required to be > 5. Finally, the mRNA-TF regulatory network was developed using Cytoscape software.
The role of miRNA in regulation is vital for developmental and evolutionary mechanisms in organisms. Different target genes could be regulated, and several miRNAs could influence a single target gene. To investigate the association between pivotal genes and miRNA, we retrieved the miRNA that interacted with key genes from the encyclopedia of RNA interactomes (ENCORI) database (https://rnasysu.com/encori/) (33). We used a screening threshold of pancancerNum > 5 to select mRNA-miRNA interaction pairs. The interaction network between mRNA and miRNA was illustrated using Cytoscape software.
Furthermore, RNA-binding proteins (RBPs) control gene expression by engaging with crucial mRNAs after transcription. We used the ENCORI database to extract RBP information and analyze their regulation of key mRNAs.The criterion for screening mRNA-RBP interaction pairs was clusterNum >1. Ultimately, Cytoscape software was used to visualize the constructed mRNA-RBP regulatory network.
2.7 Validation of differential gene expression and analysis of key genes using receiver operating characteristic curvesTo analyze the variation in key gene expression between the PE and control groups within the combined datasets, we used the Mann–Whitney U test. Comparative maps were constructed based on the expression levels of these essential genes. Subsequently, the R package “pROC” (34) was used to generate the ROC curve for the significant genes. The area under the curve (AUC) evaluated the effectiveness of gene expression in diagnosing PE. The validation process was performed using the GSE75010 dataset.
2.8 Immune infiltration analysisWe measured the proportion of immune cell infiltration using single-sample (ss) GSEA (35). The recognized categories of immune cells comprised activated CD8 + T cells, activated dendritic cells, gamma-delta T cells, natural killer (NK) cells, regulatory T cells (Tregs), and several other human immune cell subtypes. The proportion calculated through ssGSEA was used to illustrate the relative levels of immune cell infiltration in each sample, creating an immune cell infiltration matrix. Then, immune cells indicating significant variations between the two groups were selected for additional analysis, and their relationships were evaluated using the Spearman method. Correlation heatmaps were created with the R package “pheatmap” to demonstrate the correlation between immune cells. The Spearman method determined the relationship between crucial genes and immune cells, with a significance threshold set at p < 0.05. Using the R package “ggplot2”, a bubble map was drawn to illustrate the connection between essential genes and immune cells. We selected immune cells with top1 positive and top1 negative correlation with key genes and plotted correlation scatter plots using ggplot2.
2.9 Patient and tissue samplesPlacenta samples were obtained from 52 pregnant women who underwent cesarean sections at the Third Affiliated Hospital of Wenzhou Medical University. 26 had PE, and 26 were healthy controls matched for gestational age. Each group included 14 term pregnancy and 12 preterm pregnancies. Ethical approval was obtained from the Research Ethics Committee at the Ruian People’s Hospital, under approval number YJ2024130. All participants provided written consent. The inclusion criteria for the PE group included blood pressure ≥ 140/90 mmHg and 24-h urinary protein ≥ 0.3 g/24 h after 20 weeks of gestation, age between 20 and 40 years old, and no significant abnormalities during pregnancy. Exclusion criteria included other pregnancy complications such as gestational diabetes mellitus; Prepregnancy comorbidities such as prepregnancy hypertension, prepregnancy diabetes, serious medical and surgical diseases, infectious diseases such as COVID-19, obstetric complications, congenital diseases of the fetus, or the use of drugs that may affect the results of the experiment. After delivery, a tissue sample was extracted from the central region of the placenta and preserved at –80°C for long-term storage.
2.10 Isolation of RNA and analysis using quantitative real-time-polymerase chain reactionTotal RNA was extracted from placental tissue samples using the tissue total RNA isolation kit V2 (Vazyme) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA samples with an A260/A280 ratio between 1.8 and 2.0 were considered suitable for further analysis. Subsequently, 1 µg of total RNA was reverse transcribed into complementary DNA (cDNA) using the HiScript III All-in-one RT SuperMix (Vazyme) in a 20 µL reaction volume. The reverse transcription reaction was performed at 25°C for 5 minutes, followed by 50°C for 15 minutes, and terminated by heating at 85°C for 5 minutes. Quantitative real-time PCR (qRT-PCR) was conducted using the CFX Connect real-time PCR system (BioRad, Hercules, CA, USA) with Taq Pro Universal SYBR qPCR Master Mix (Vazyme). Each qRT-PCR reaction was carried out in a 10 µL volume containing 5 µL of SYBR Green Master Mix, 0.5 µL of each forward and reverse primer (10 µM), 1 µL of cDNA template, and 3 µL of nuclease-free water. The thermal cycling conditions were as follows: initial denaturation at 95°C for 30 seconds, followed by 40 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. A melt curve analysis was performed to verify the specificity of the amplification products. The relative expression levels of the key genes were normalized to the expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCt method. All reactions were performed in triplicate, and the average Ct values were used for analysis. The results were expressed as fold changes in gene expression relative to the control group.
2.11 Isolation of protein and analysis using Western BlottingTissues were minced and homogenized in RIPA lysis buffer (P0013B, Beyotime) with PMSF (100 mM, ST506, Beyotime), using 150-250 µL of lysis buffer per 20 mg of tissue. The homogenates were centrifuged similarly to obtain the supernatant. Protein concentration was determined using the BCA protein assay kit. Samples were diluted to equal concentrations with RIPA lysis buffer containing PMSF and mixed with 5× protein loading buffer. The samples were denatured by heating at 100°C for 5-10 minutes and then cooled on ice. SDS-PAGE was performed using self-prepared gels by first casting the separating and stacking gels between clean glass plates. The samples were then loaded into the wells formed by the comb in the stacking gel, and electrophoresis was conducted to separate proteins based on their molecular weight. Proteins were transferred to PVDF membranes (ISEQ00010, Millipore) using a wet transfer system (Mini Trans-Blot, BIO-RAD). The membranes were blocked with non-fat milk blocking solution for 1-2 hours at room temperature and incubated overnight at 4°C with primary antibodies diluted in antibody dilution buffer (P0256-500ml, Beyotime). After washing, the membranes were incubated with HRP-conjugated secondary antibodies for 1 hour at room temperature. The protein bands were visualized by using BeyoECL Plus working solution (P0018S, Beyotime) and detected with a chemiluminescence imaging system. The relative expression levels of proteins were analyzed by Image J.
2.12 Statistical analysisThe study used R software (version 4.3.1) for statistical analysis and data handling. The independent student t-test was implemented to assess the statistical significance of normally distributed data and compare continuous variables between two groups unless otherwise specified. For non-normally distributed variables, the Mann–Whitney U or Wilcoxon rank sum test was used to determine differences. Furthermore, the Kruskal–Wallis test was applied to compare outcomes among three or more groups. Spearman’s rank correlation was used to determine the correlation coefficients for several molecules without particular specifications. All statistical analyses used two-tailed p-values, with a significance level set at p < 0.05.
3 Results3.1 Analytical flow diagramFigure 1 displays the technical approach of the study, providing a concise overview of the analytical processes used in this study.It begins with the combination of datasets GSE60438 (GPL10558 and GPL6884) and proceeds with the identification of differentially expressed genes (DEGs). Energy metabolism-related genes (EMRGs) are intersected with DEGs to identify energy metabolism-related differentially expressed genes (EMRDEGs). The subsequent enrichment analyses include Gene Set Enrichment Analysis (GSEA), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment. A protein-protein interaction (PPI) network is constructed to identify key genes, which are further analyzed for immune infiltration. Key genes are validated using the GSE75010 dataset through the Wilcoxon Rank Sum Test and Receiver Operating Characteristic (ROC) analysis. Finally, regulatory networks involving mRNA-TF, mRNA-miRNA, and mRNA-RBP interactions are constructed to understand the regulatory mechanisms.
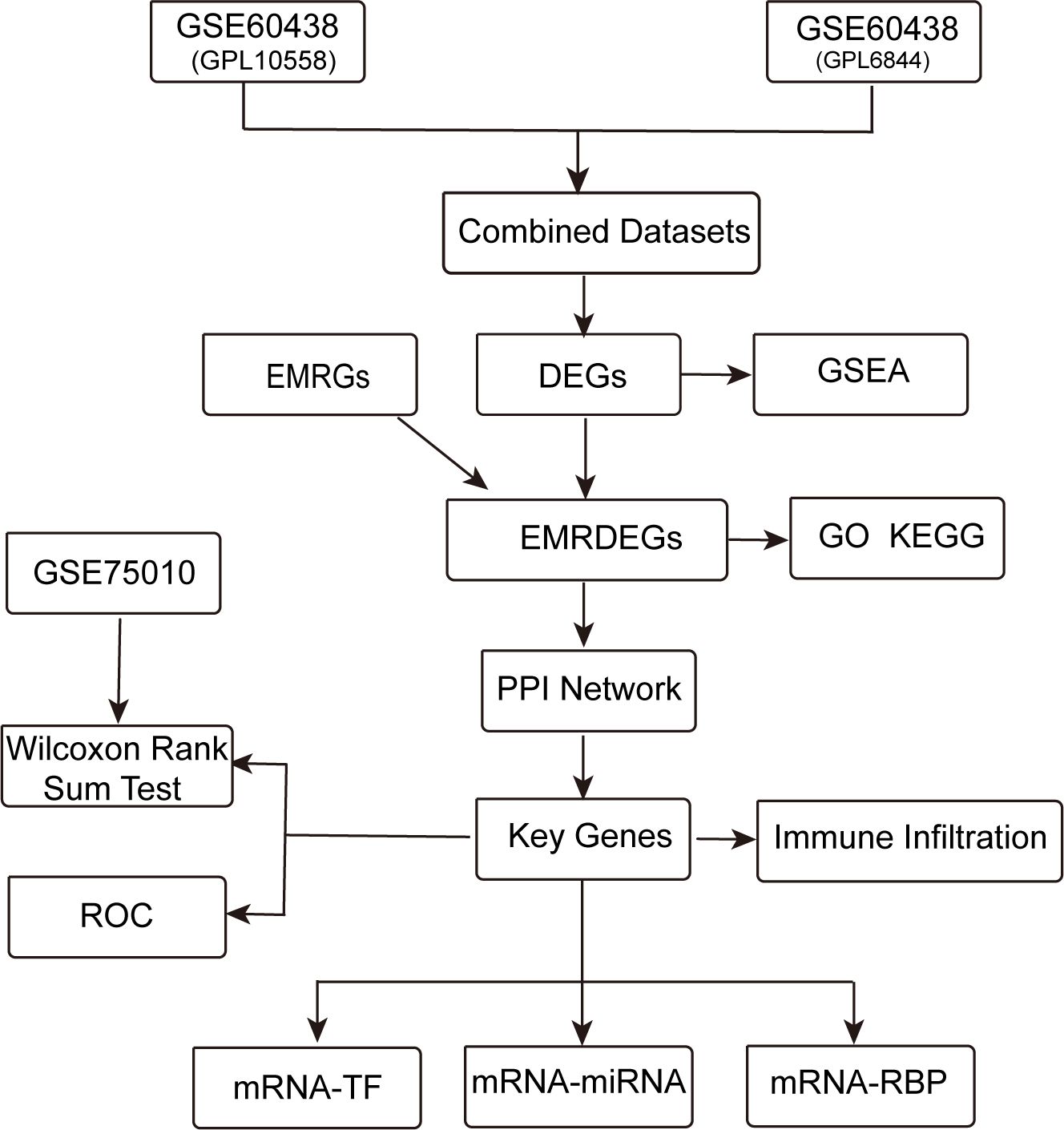
Figure 1. Technology roadmap. DEGs, Differentially Expressed Genes; EMRGs, Energy Metabolism-Related Genes. EMRDEGs, Energy Metabolism-Related Differentially Expressed Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analysis; ROC, Receiver Operating Characteristic Curve; PPI Network, Protein-protein Interaction Network; TF, Transcription Factor; RBP, RNA-Binding Protein.
3.2 Merging of PE datasetsTo eliminate batch effects from the PE datasets GSE60438 (using GPL10558 and GPL6884 platforms), the R package “sva” was used, resulting in combined datasets. Boxplots (Figures 2A, B) were used to compare the expression values of the datasets pre- and post-batch effect removal. Furthermore, a PCA plot (Figures 2C, D) compared the distribution of low-dimensional features in the dataset before and after addressing batch effects. The outcomes from the distribution box plot and PCA plot indicated that the batch effect in the PE dataset samples was significantly reduced after batch correction.
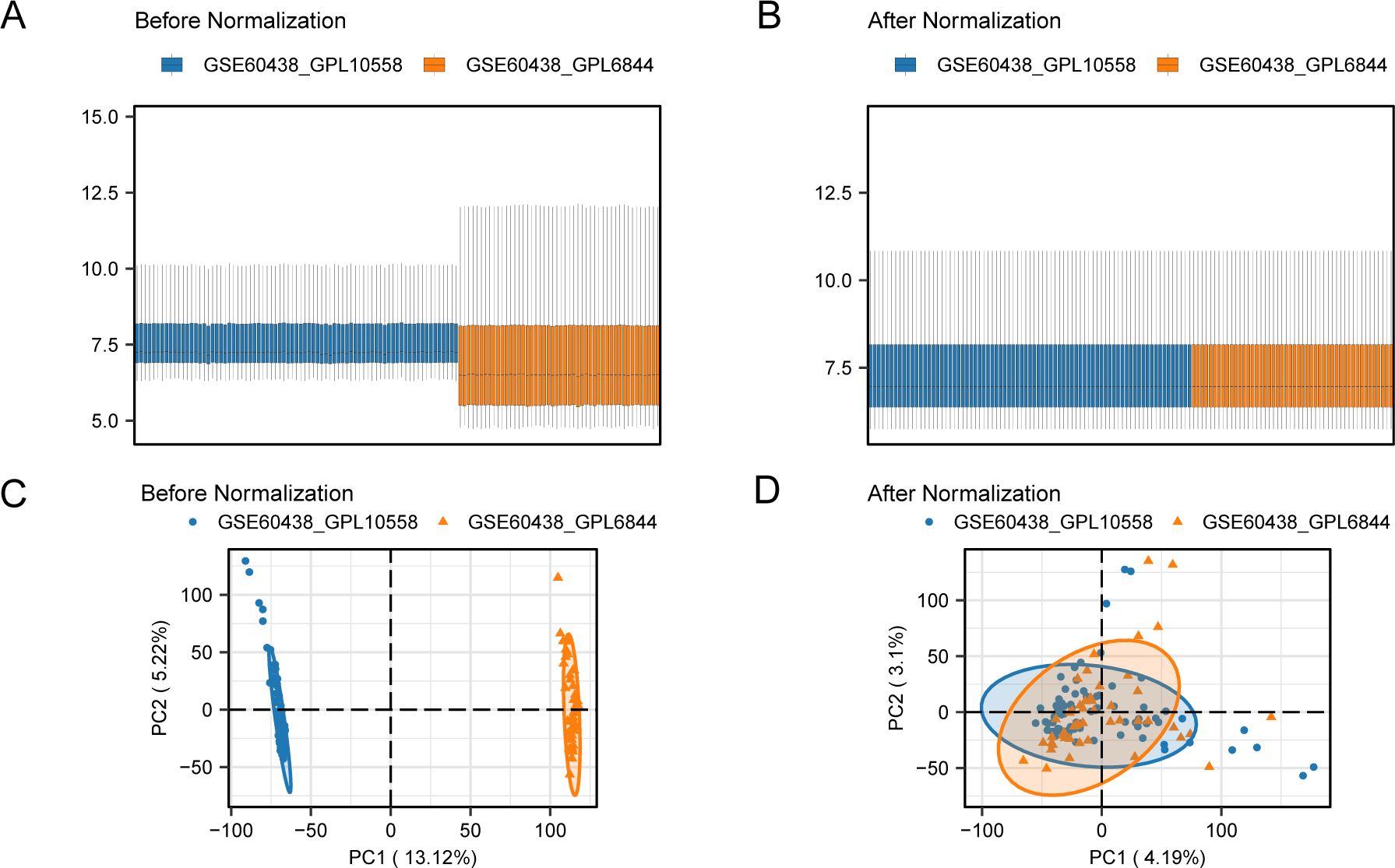
Figure 2. Batch effects removal of GSE60438 (GPL10558, GPL6884). (A) Boxplots of combined datasets distribution before batch removal. (B) Post-batch integrated combined datasets distribution boxplots. (C) PCA plot of the datasets before debatching. (D) Go to the PCA map of the combined datasets after batch processing. PCA, Principal Component Analysis; PE, Preeclampsia. The PE dataset GSE60438 (GPL10558 platform) is blue, and the PE dataset GSE60438 (GPL6884 platform) is orange.
3.3 Genes with altered expression associated with energy metabolism in PEThe data from the combined datasets were separated into PE and control groups. We performed a comparative analysis of gene expression levels between PE and control groups across the combined datasets using the R package “limma”. The findings identified 55 genes with differential expression, satisfying the criteria of |logFC| > 0.5 and a p < 0.05 in the combined datasets. Out of these DEGs, 15 indicated increased expression (logFC > 0.5, p < 0.05), whereas 40 exhibited decreased expression (logFC < –0.5, p < 0.05), which was illustrated in the volcano plot analysis of the dataset (Figure 3A).
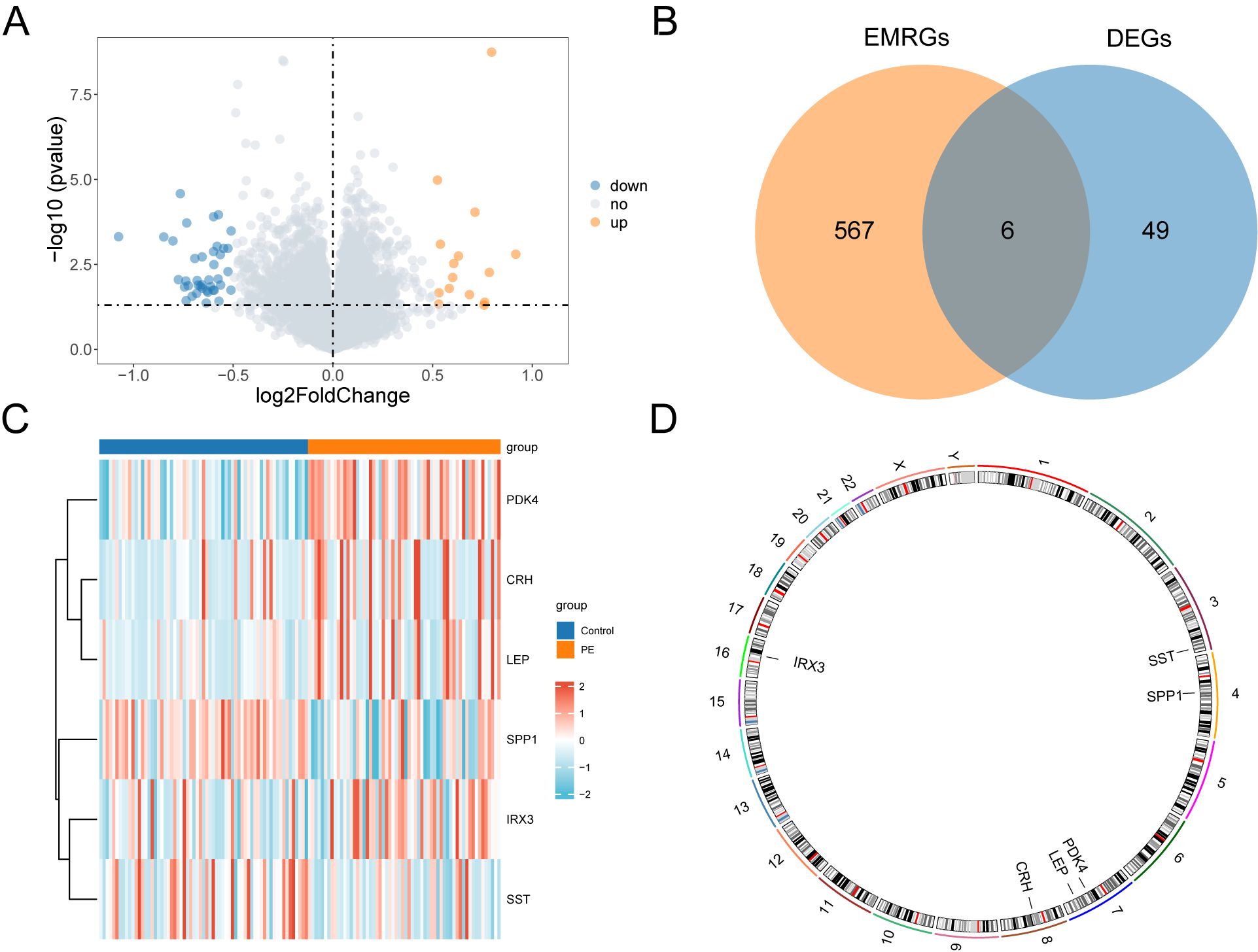
Figure 3. Differential gene expression analysis. (A) Volcano plot of differentially expressed genes analysis between PE and Control groups in combined datasets. (B) DEGs and EMRGs Venn diagram in the combined datasets. (C) Heat map of EMRDEGs in the combined datasets. (D) Chromosomal mapping of EMRDEGs; DEGs, Differentially Expressed Genes; EMRGs, Energy Metabolism Related Genes; EMRDEGs, Energy Metabolism Related Differentially Expressed Genes. The orange is the PE group, and the blue is the Control group. The red in the heat map represents high expression, and the blue represents low expression.
To identify genes that exhibited differential expression and were associated with energy metabolism, we selected genes with |logFC| > 0.5 and a p < 0.05 from the overlap of DEGs and EMRGs (Figure 3B). Six EMRDEGs, including CRH, IRX3(Iroquois Homeobox 3), LEP, PDK4, SPP1, and SST, were identified (Table 2). The variations in the expression of identified EMRDEGs among different sample groups in the combined datasets were investigated through the intersection results. The analysis results were visualized in a heatmap created with the “pheatmap” package in R (Figure 3C). Besides, the R package “RCircos” was used to plot the positions of these six EMRDEGs on human chromosomes and constructed a chromosome localization map (Figure 3D). The mapping revealed that most of these EMRDEGs were located on chromosome 7, particularly LEP and PDK4.
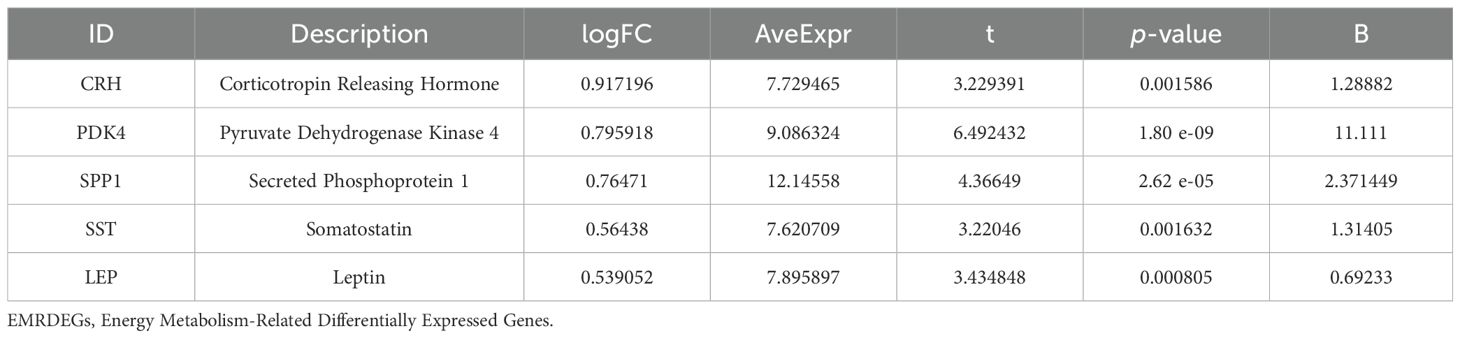
Table 2. Description of EMRDEGs.
3.4 Enrichment analysis using GO and KEGGWe employed GO and KEGG enrichment analyses to investigate the association between BP, CC, MF, and biological pathways (KEGG) of six EMRDEGs and PE. The six EMRDEGs underwent GO and KEGG enrichment analyses (Table 3). The results indicated that the six EMRDEGs were primarily involved in several BP, including cell lipid export, regulation of bone remodeling, tissue and bone restructuring, and glucagon release. Furthermore, they were associated with CC, neuronal cell bodies, and MFs related to hormone activity, receptor signaling activation, peptide hormone receptor binding, neuropeptide hormone activity, and other hormone receptor interactions. Additionally, the biological pathway associated with neuroactive ligand-receptor interaction (KEGG) exhibited an increase. The findings from GO and KEGG enrichment analyses were presented using bar graphs (Figure 4A).
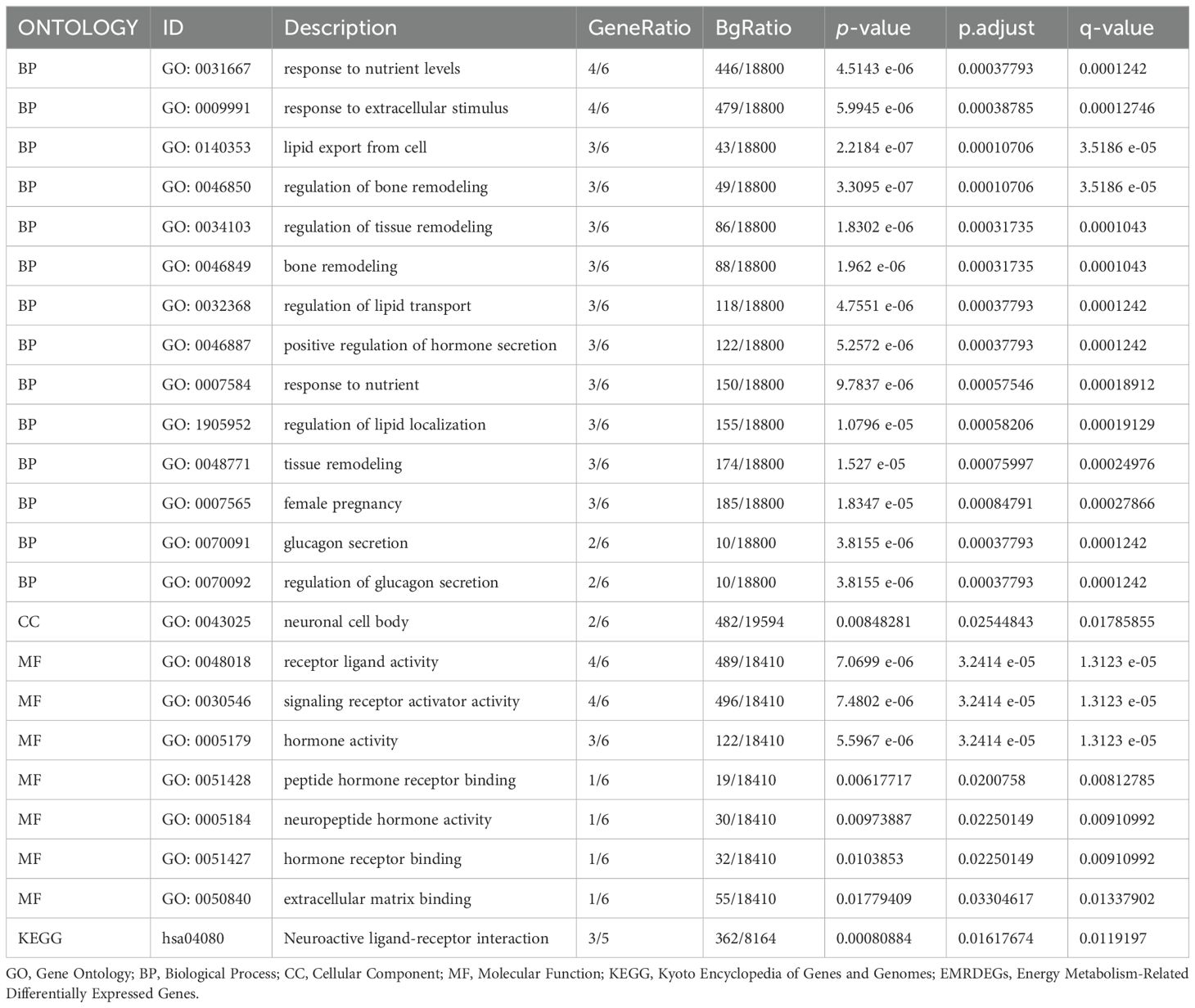
Table 3. Results of GO and KEGG enrichment analysis for EMRDEGs.
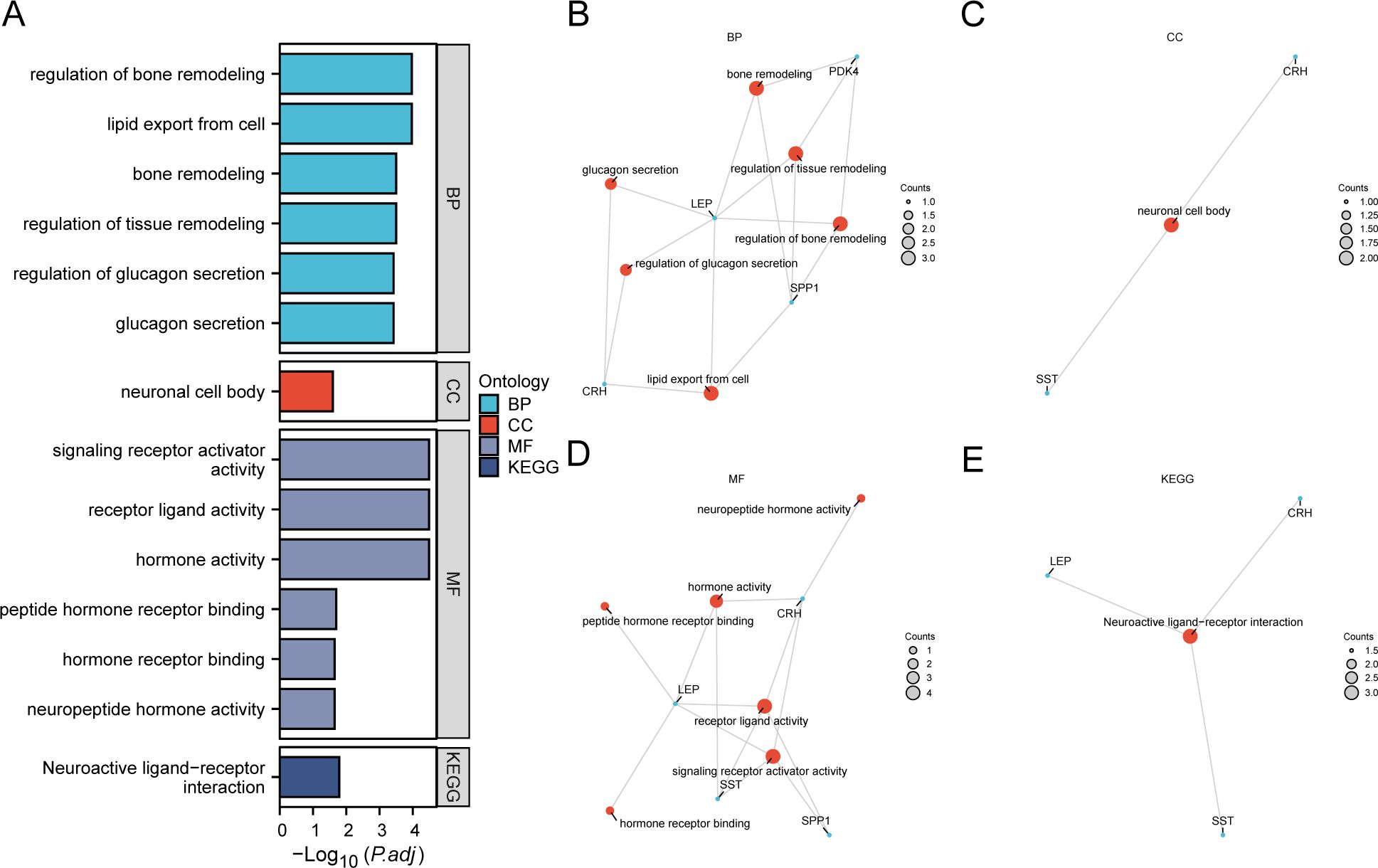
Figure 4. GO and KEGG enrichment analysis for EMRDEGs. (A) Bar graph of GO and KEGG enrichment analysis results of EMRDEGs: BP, CC, MF, and KEGG. GO terms and KEGG terms are indicated on the ordinate. B-E. GO and KEGG enrichment analysis results of EMRDEGs network diagram exhibiting BP (B), CC (C), MF (D), and KEGG (E). The orange nodes represent items, the blue nodes represent molecules, and the lines represent the relationship between items and molecules. EMRDEGs, Energy Metabolism-Related Differentially Expressed Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, Biological Process; CC, Cellular Component; MF, Molecular Function. The screening criteria for GO and KEGG enrichment analysis were adj. p < 0.05, and FDR value (q-value) < 0.05, and the p-value correction method was Benjamini-Hochberg (BH).
Following GO and KEGG enrichment analyses, BP, CC, MF, and biological pathways (KEGG) were schematically presented (Figures 4B–E). The connections display the molecules corresponding to the entries, with annotations for each. The magnitude of the nodes indicates the quantity of molecules present in each record.
3.5 GSEAWe conducted GSEA to determine how gene expression levels across the combined datasets influenced PE and to identify the associated BPs. The relationship between affected CCs and performed MFs is depicted in Figure 5A, with specific results provided in Table 4. The findings indicated that all genes in the combined datasets were significantly enriched in glycolysis and gluconeogenesis (Figure 5B), faerie-mediated Ca2+ mobilization (Figure 5C), NK cell-mediated cytotoxicity (Figure 5D), interleukin (IL) 10 signaling (Figure 5E), IL12 pathway (Figure 5F), an overview of proinflammatory and profibrotic mediators (Figure 5G), neutrophil degranulation (Figure 5H), and other biologically related functions and signaling pathways.
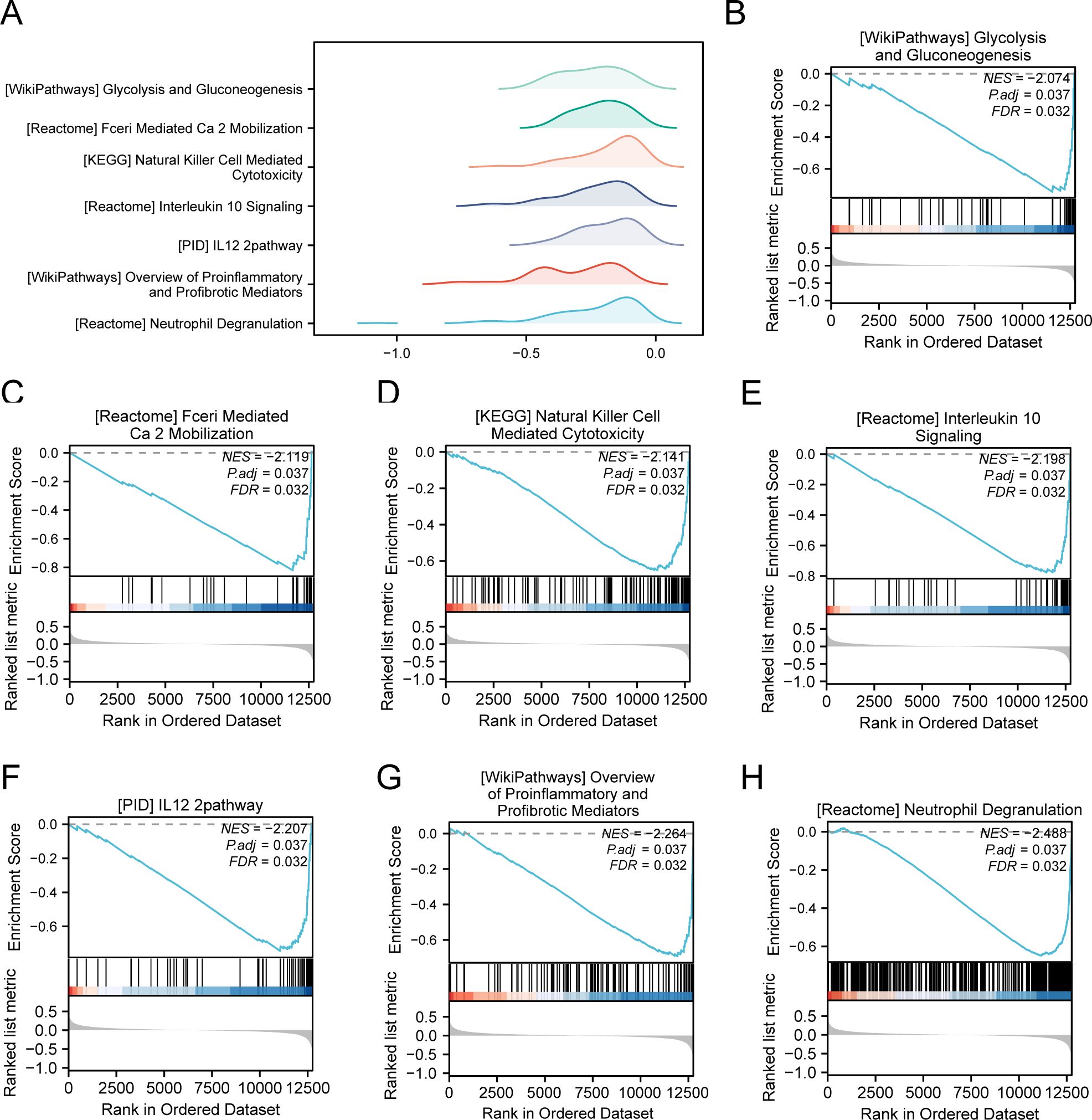
Figure 5. GSEA for combined datasets. (A) GSEA mountain map presentation of 7 biological functions of the combined datasets. B-h. GSEA revealed that EMRDEGs were significantly enriched in WP_GLYCOLYSIS_AND_GLUCONEOGENESIS (B), REACTOME_FCERI_MEDIATED_CA_2_MOBILIZATION (C), KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY (D), Reactome_interleukin_10 signaling (E), PID_IL12_2PATHWAY (F), WP_OVERVIEW_OF_PROINFLAMMATORY_AND_PROFIBROTIC_MEDIATORS (G), REACTOME_NEUTROPHIL_DEGRANULATION (H). GSEA, Gene Set Enrichment Analysis; EMRDEGs, Energy Metabolism-Related Differentially Expressed Genes; The screening criteria of GSEA were adj. p < 0.05 and FDR value (q-value) < 0.05, and the p-value correction method was Benjamini-Hochberg (BH).
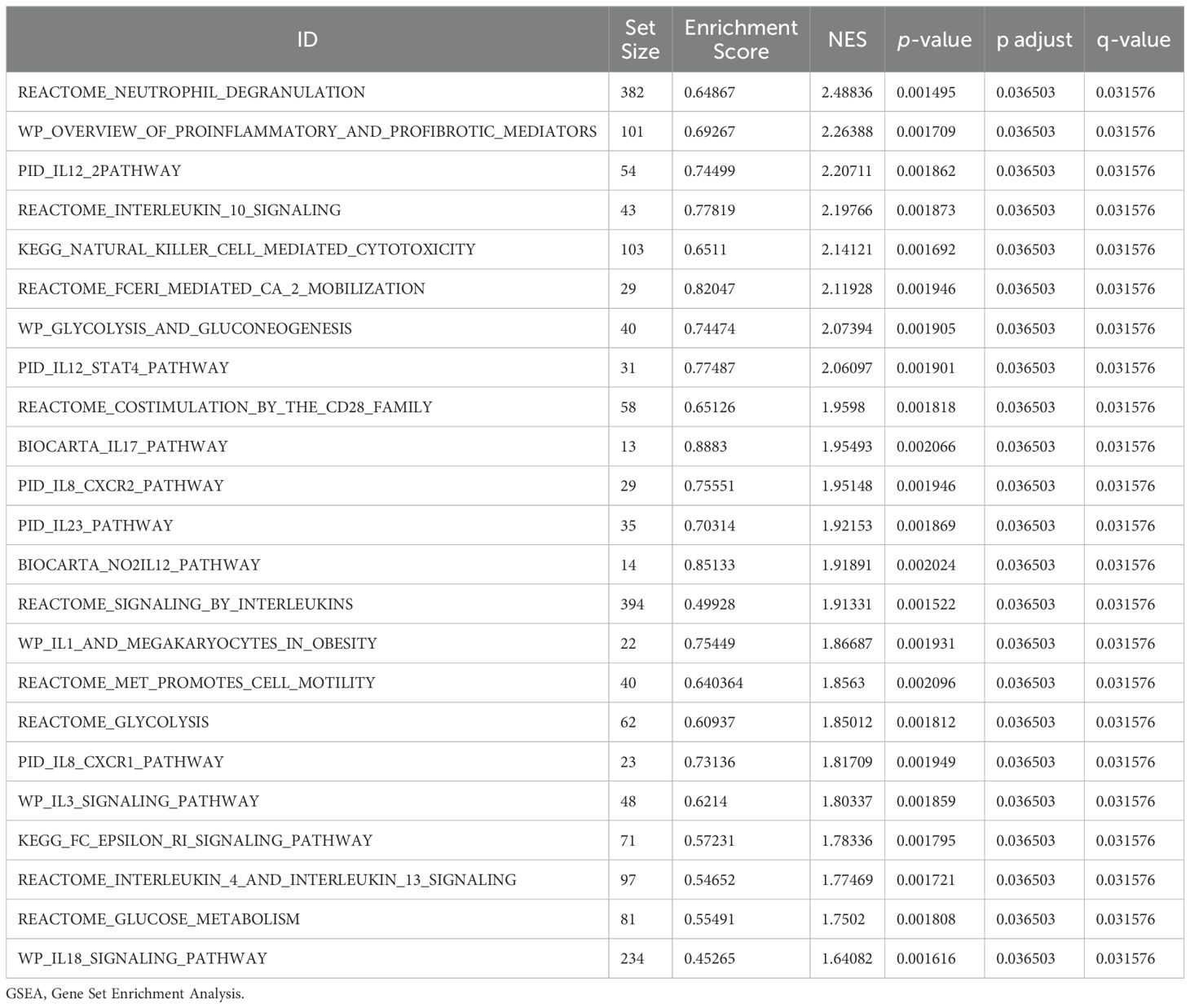
Table 4. Results of GSEA for combined datasets.
3.6 PPI networkPPI interaction analysis was performed, and the PPI network of six EMRDEGs was constructed using the STRING database (Figure 6A). The PPI network findings indicated a connection among five EMRDEGs: CRH, LEP, PDK4, SPP1, and SST. Furthermore, the interaction network of five EMRDEGs and their functionally similar genes (Figure 6B) was predicted and constructed using the GeneMANIA website. The colored lines represent their co-expression and share protein domains and other information. Among them, there were 5 EMRDEGs and 20 functionally similar proteins.
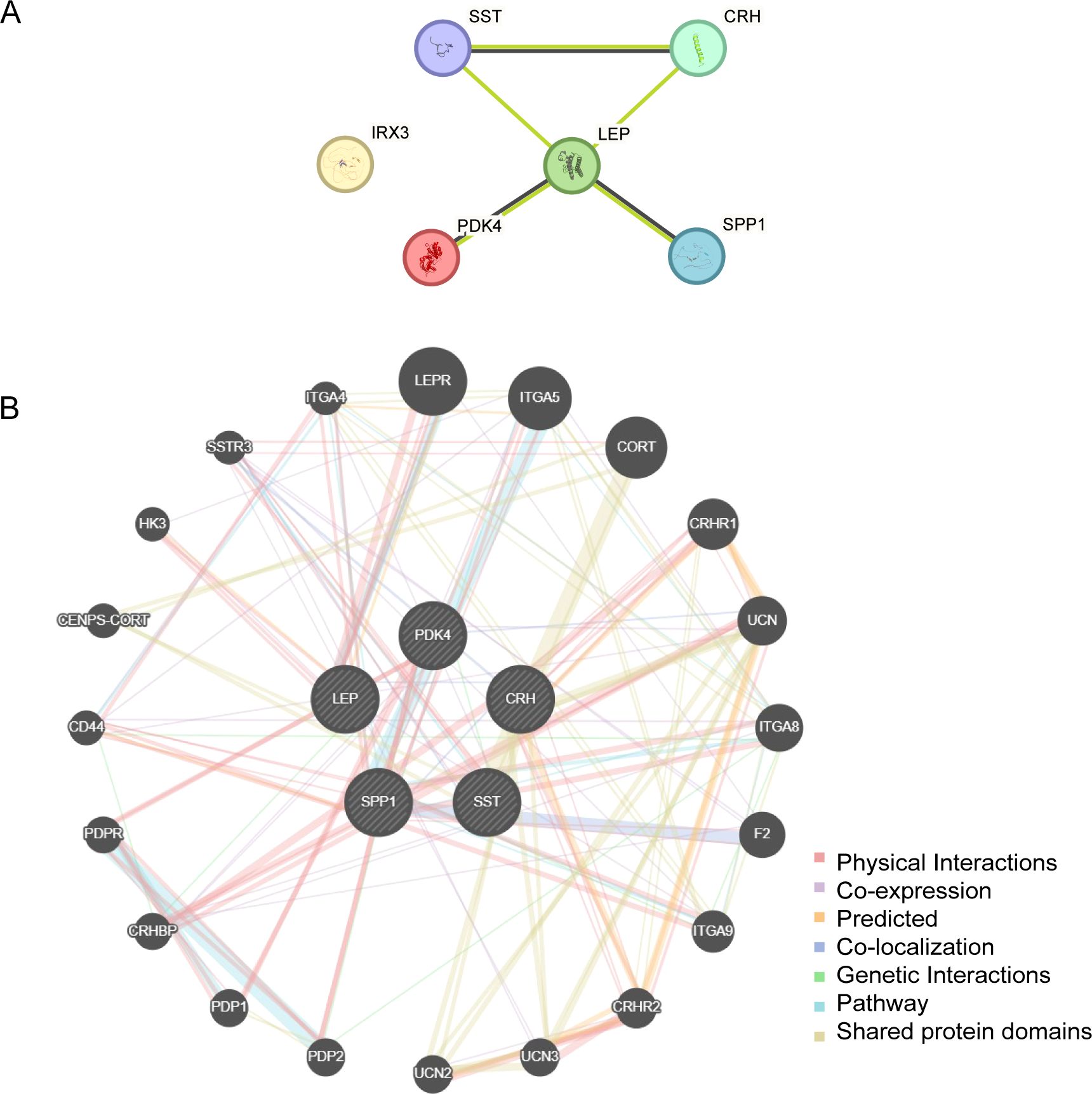
Figure 6. PPI network analysis. (A) PPI Network of EMRDEGs calculated from the STRING database. (B) The GeneMANIA website predicts the interaction network of functionally similar genes of EMRDEGs. The circles in the figure indicate the EMRDEGs and their functionally identical genes, and the corresponding colors of the lines represent the interconnected functions. PPI, Protein-protein Interaction; EMRDEGs, Energy Metabolism-Related Differentially Expressed Genes.
3.7 Construction of regulatory networkWe constructed the mRNA-TF regulatory network, which included five key genes (CRH, LEP, PDK4, SPP1, and SST) and 39 TFs, resulting in 51 mRNA-TF interactions (Figure 7A). Detailed information is provided in Supplementary Table S2. The mRNA-miRNA regulatory network consisted of two key genes (PDK4 and SPP1) and 56 miRNAs, resulting in 59 mRNA-miRNA interactions (Figure 7B). Detailed data is presented in Supplementary Table S3. Our derived mRNA-RBP network included three key genes (LEP, PDK4, and SPP1) and 30 RBP molecules, resulting in 32 mRNA-RBP interactions (Figure 7C). Detailed data is provided be found in Supplementary Table S4.
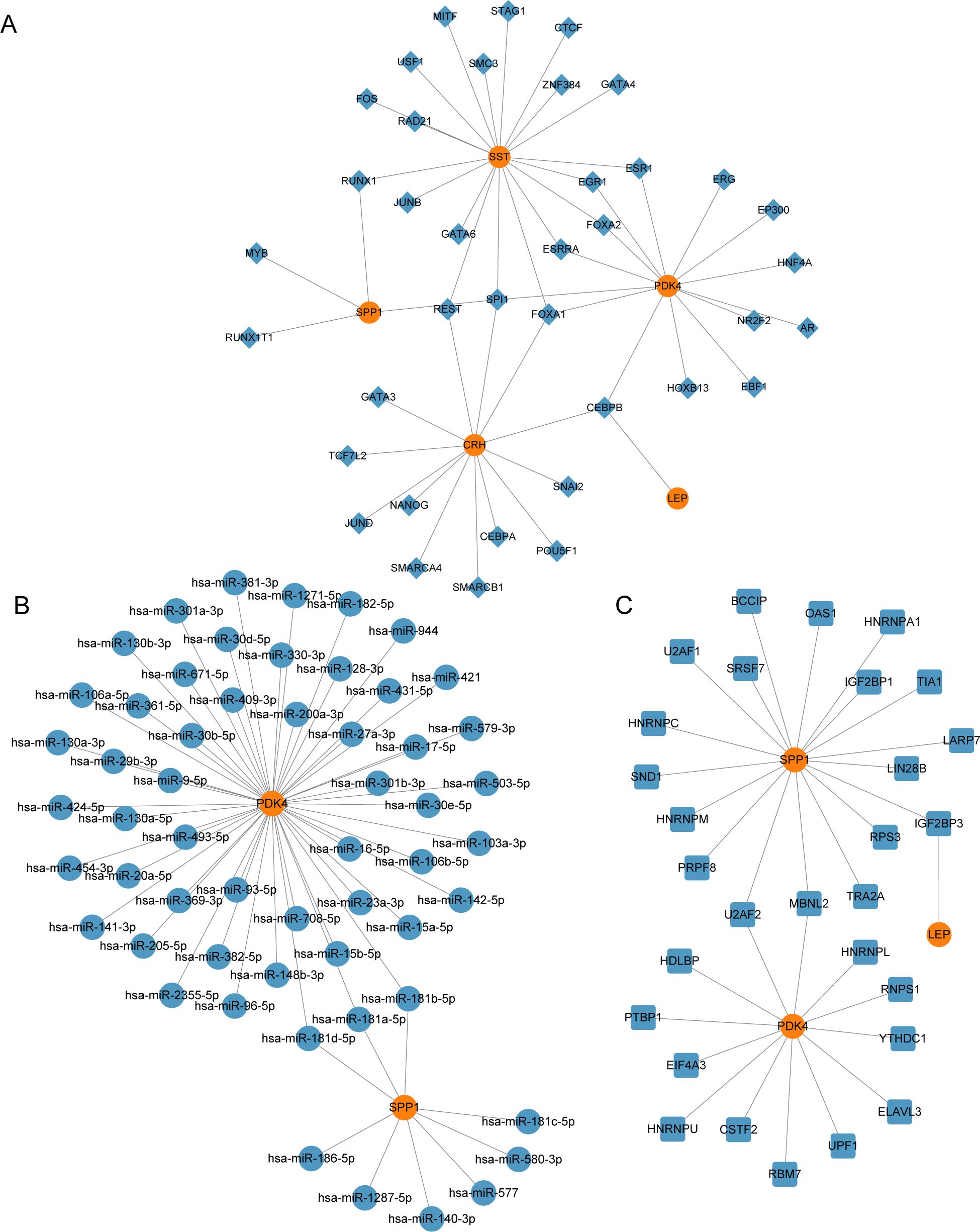
Figure 7. Regulatory network of EMRDEGs. (A) mRNA-TF Regulatory Network of Key Genes. (B) mRNA-miRNA Regulatory Network of Key Genes. (C) mRNA-RBP Regulatory Network of Key Genes. EMRDEGs, Energy Metabolism-Related Differentially Expressed Genes; TF, Transcription Factor; RBP, RNA-Binding Protein. Orange is mRNA, blue diamonds are TF, blue circles are miRNA, and blue squares are RBP.
3.8 Validation of differential gene expression and analysis of key genes using ROC curvesTo investigate the key genes (CRH, LEP, PDK4, SPP1, and SST) across the combined datasets, a comparative analysis was performed through a group comparison (Figure 8A), indicating the outcomes of the differential expression analysis for these five key genes in PE samples compared with control samples from the combined datasets. The findings from the differential analysis (Figure 8A) indicated that two crucial genes (PDK4 and SPP1) exhibited a significant statistical difference (p < 0.001) in both PE and control groups across the combined datasets. Furthermore, SST demonstrated a statistical significance (p < 0.01) across both types of samples. The other two crucial genes (CRH and LEP) exhibited significant expression in both PE and control groups, with a p < 0.05. Moreover, the expression levels of crucial genes in combined datasets were evaluated by creating ROC curves with the “pROC” package in R. The ROC curve (Figures 8B–F) revealed that the expression levels of key genes, including PDK4 in PE samples, exhibited moderate to high accuracy across different groups (AUC: 0.7–0.9). The expression levels of crucial genes (CRH, LEP, SPP1, and SST) in PE samples demonstrated low precision across various groups (AUC: 0.5–0.7). The ROC curves for key genes in dataset GSE75010 (Figures 8G–K) revealed that the expression levels of CRH and LEP in PE samples demonstrated moderate to high precision among various groups (AUC between 0.7 and 0.9). Conversely, the expression levels of crucial genes (PDK4, SPP1, and SST) in PE samples demonstrated reduced precision across different groups (AUC: 0.5–0.7).
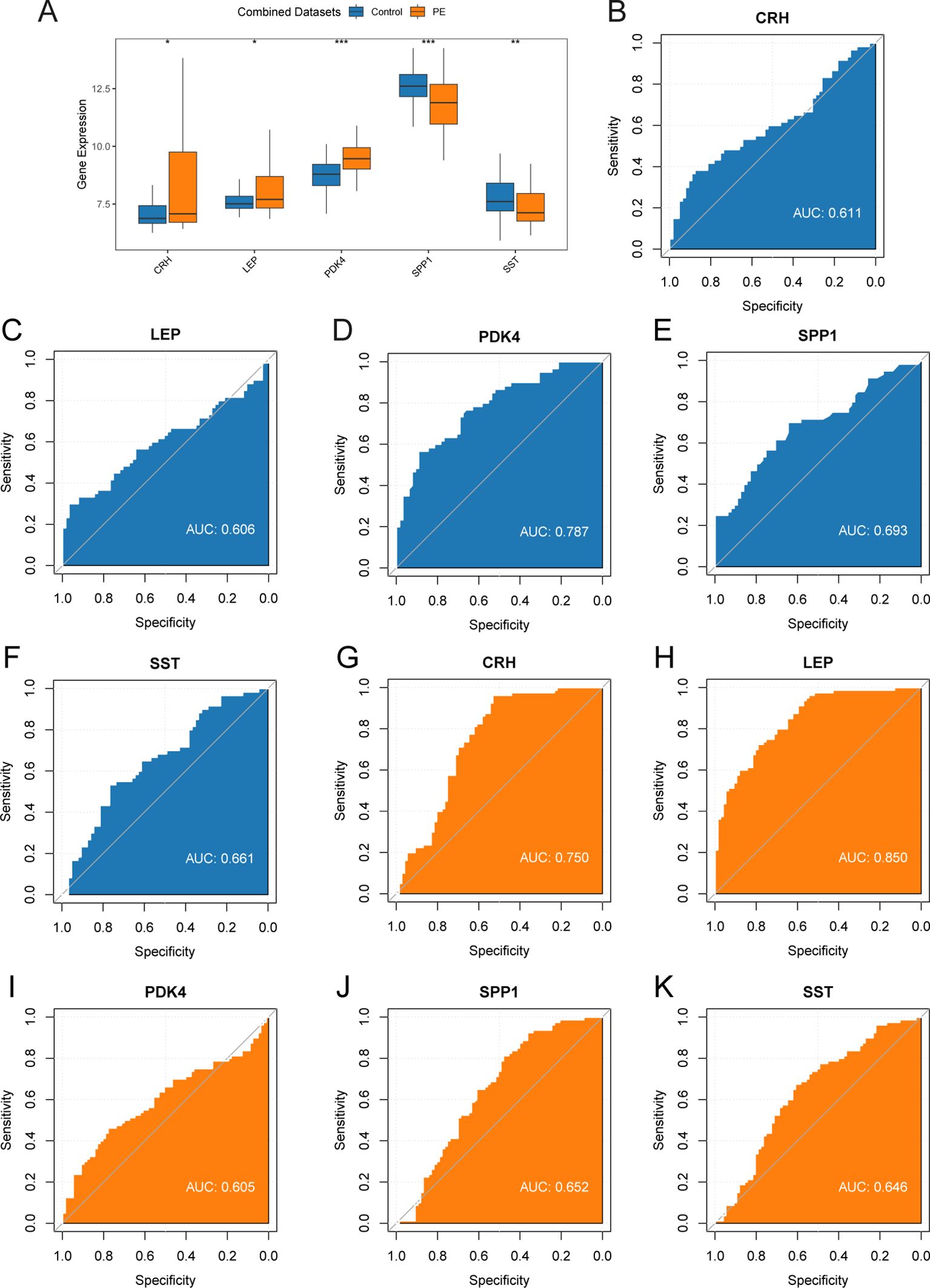
Figure 8. Differential expression validation and ROC curve analysis. (A) Group comparison plot of Key Genes in PE samples and Control samples of combined datasets. B-F. ROC curves of Key Genes CRH (B), LEP (C), PDK4 (D), SPP1 (E), and SST (F) in combined datasets. G-K. ROC curves of Key Genes CRH (G), LEP (H), PDK4 (I), SPP1 (J), and SST (K) in dataset GSE75010. ROC, Receiver Operating Characteristic; AUC, Area Under The Curve. ROC, Receiver Operating Characteristic Curve; TPR, True Positive Rate; FPR, False Positive Rate. * represents p-value < 0.05, statistically significant; ** represents p-value < 0.01, highly statistically significant; *** represents p-value < 0.001 and highly statistically significant. AUC between 0.5-0.7 had low accuracy, and AUC of 0.7-0.9 had moderate accuracy. In the group comparison figure, the PE group is orange, and the Control group is blue.
3.9 Immune infiltration analysisThe ssGSEA algorithm evaluated the presence of 28 types of immune cells using expression data from combined datasets. Immune cells were selected with a p < 0.05 using a comparative group plot. The differences in immune cell infiltration levels across various groups were observed. The comparative chart (Figure 9A) indicateds that 16 immune cell types, including activated CD4+ T cells, activated CD8+ T cells, activated dendritic cells, CD56bright NK cells, CD56dim NK cells, effector memory CD8+ T cells, eosinophils, gamma-delta T cells, immature B cells, macrophages, myeloid-derived suppressor cells (MDSCs), monocytes, plasmacytoid dendritic cells, Tregs, T follicular helper cells, and type 1 T helper cells, exhibited significant differences between PE and control samples (p < 0.05).The correlation results from the combined datasets (Figure 9B) exhibited the abundance of 16 different immune cell infiltrations in the immune infiltration study. The results revealed a significant correlation among immune cells. The correlation between 5 key genes and 16 immune cells was examined and visualized using a correlation bubble diagram (Figure 9C). The results indicated a significant positive correlation between SPP1 and Tregs, with an r-value = 0.458 and a p < 0.05. The key gene LEP exhibited a significantly negative correlation with CD56dim NK cells (r-value = –0.359, p < 0.05). Finally, a correlation scatter plot demonstrated the relationship between top1 positive and top1 negative key genes and immune cells (Figures 9D, E).
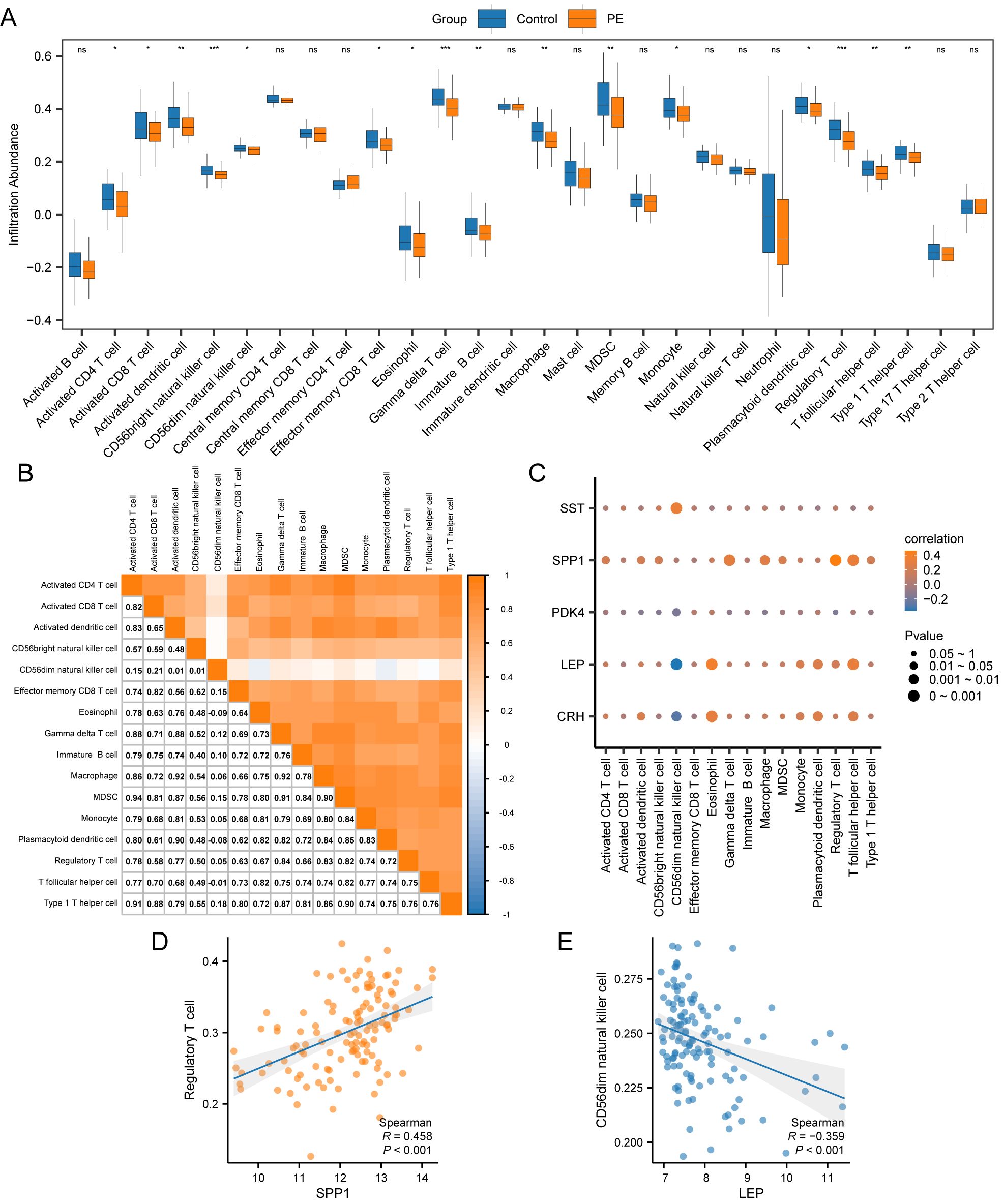
Figure 9. Immune infiltration analysis by ssGSEA algorithm. (A) Group comparison plot of immune cells in PE and Control samples from the combined datasets. (B) Correlation heatmap of immune cell infiltration abundance in the combined datasets. (C) Bubble correlation plot between Key Genes and immune cell infiltration abundance in the combined datasets. (D) Scatter plot of the correlation between Top1 positively correlated Key Genes and immune cells. (E) Scatter plot of correlation between TOP1-negatively correlated Key Genes and immune cells. ssGSEA, single-sample Gene-Set Enrichment Analysis; ns stands for p-value ≥ 0.05, not statistically significant; * represents p-value < 0.05, statistically significant; ** represents p-value < 0.01, highly statistically significant; *** represents p-value < 0.001 and highly statistically significant. The absolute value of the correlation coefficient (r-value) below 0.3 was weak or no correlation; between 0.3 and 0.5 was a weak correlation, between 0.5 and 0.8 was a moderate correlation, and above 0.8 was a strong correlation. In the group comparison plot, the PE samples are orange, and the Control samples are blue. In the heat map and correlation map, orange is a positive correlation, blue is a negative correlation, and the depth of color represents the strength of the correlation.
3.10 Validation of key genes in PETo determine the mRNA expression levels of five crucial genes in PE, qRT-PCR analysis was performed on 26 patients with PE and 26 placental samples of comparable gestational age. Table 5 presents the primer sequences. The clinical features of the patient are presented in Table 6. The two groups exhibited no significant variances in gestational age and birth weight. The PE group revealed higher systolic and diastolic blood pressure levels than the control group. The results of qRT-PCR indicated that, in contrast to the control group, the expressions of LEP and CRH in placental samples of PE patients were significantly elevated, while the expression level of SPP1 was significantly reduced (Figures 10A–E). Through the Western blotting experiment, we further examined the protein expression levels of LEP, CRH, and SPP1 (Figures 10F). The findings demonstrated that, compared with the control group, the expression of LEP and CRH proteins in placental samples of PE patients increased markedly, while the expression level of SPP1 decreased conspicuously (Figures 10G–I).
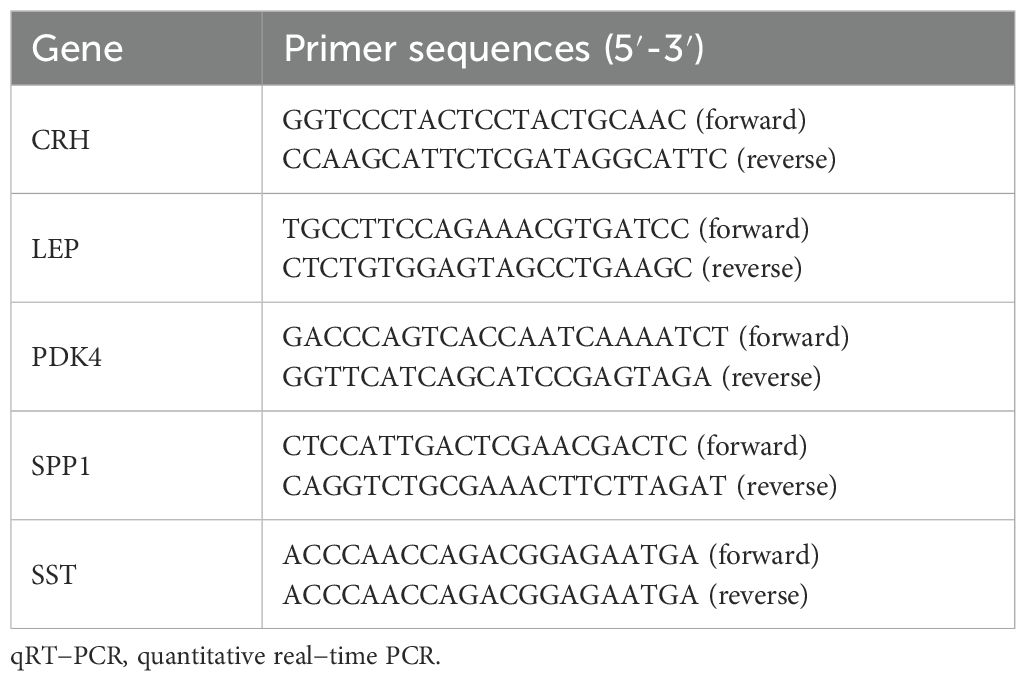
Table 5. Primer sequences for qRT-PCR.
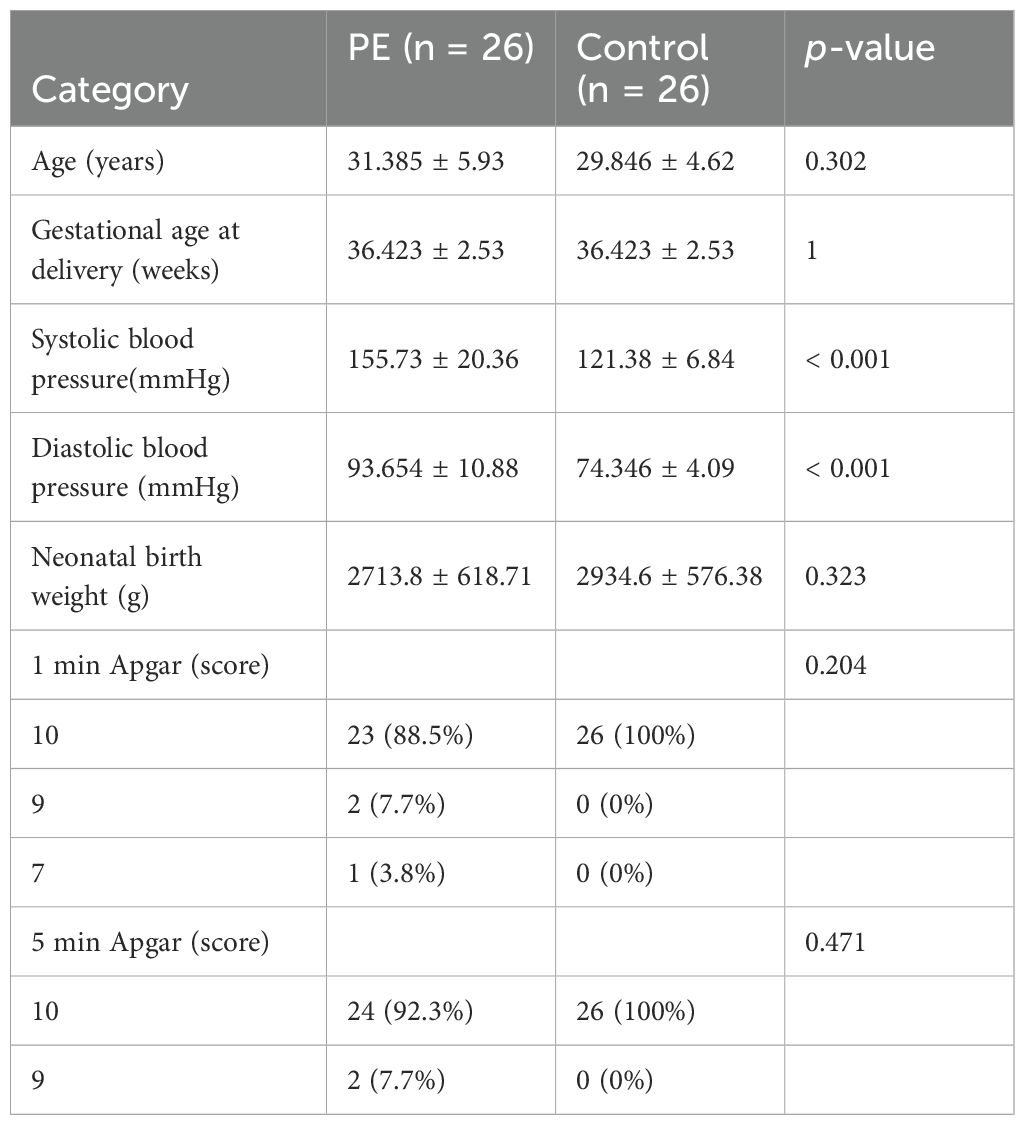
Table 6. Clinical information of the patients.
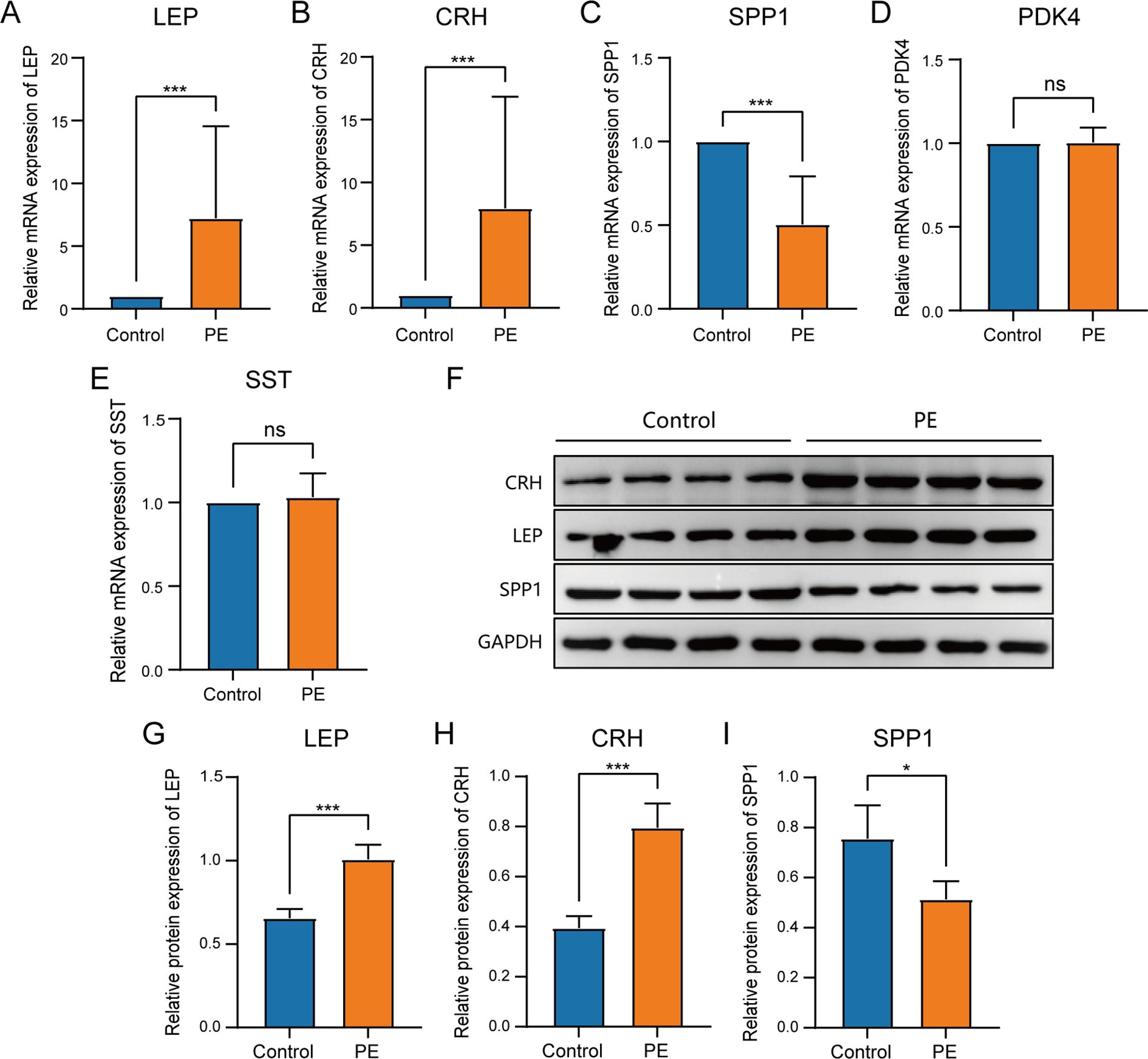
Figure 10. Comparison of key genes expression in placental samples of the control group and PE group. The expression bars of LEP
留言 (0)