Tomatoes (Solanum lycopersicum) and potatoes (Solanum tuberosum) are essential crops due to their widespread consumption and economic value. Potatoes are the third most important food crop globally, with an annual production of over 330 million tons (FAO, 2016). They play a crucial role in food security, particularly in developing regions, where they serve as a key source of income (Lutaladio and Castaldi, 2009; Lutz and Kc, 2010). Similarly, tomatoes contribute 16% to global vegetable production, with a market value of $58 billion, making them a vital crop for local consumption and export (FAO, 2016). Tomatoes are rich in essential nutrients such as vitamin C and potassium and contain antioxidants such as lycopene, which are associated with reduced cancer risks and heart disease (Blanca et al., 2015; Mercado-Meza et al., 2023). Major producers of these crops include China, India, and the US, where they are cultivated across various climatic conditions (Costa and Heuvelink, 2018).
Ralstonia solanacearum (hereafter Ralstonia) and Clavibacter michiganensis (Clavibacter) are two major bacterial pathogens that infect plants, particularly members of the Solanaceae family, including potatoes and tomatoes (Fernandez-Pozo et al., 2015). Ralstonia, the causal agent of bacterial wilt, infects over 200 plant species, including tomatoes and potatoes. The pathogen enters the plant through the roots, colonizing the xylem vessels and causing wilting, yellowing of the leaves, and necrosis (Salanoubat et al., 2002). It thrives in warm, humid climates, particularly in tropical and subtropical regions such as Sub-Saharan Africa, where it persists in soil, plant debris, and water, making it difficult to control (Hayward and Lederberg, 2000).
Clavibacter, which causes bacterial canker in tomatoes, infects plants through natural openings and wounds. Nevertheless, there are currently no highly effective control strategies against this pathogen, primarily due to its ability to enter plants through natural openings, causing cankers on leaves and stems, infecting the xylem, and leading to wilting and necrosis (Chalupowicz et al., 2017). First identified in North America in 1910, the pathogen has since spread to all major tomato-growing regions globally. Its spread had been particularly severe in Europe, significantly affecting tomato crop yields (Baysal et al., 2011). Both pathogens pose serious threats to crop production, resulting in significant economic losses globally, as they affect the quality and yield of tomatoes and potatoes.
Climate change poses significant challenges to global agriculture, particularly for crops such as tomatoes and potatoes. Rising global temperatures and altered precipitation patterns are major threats to agricultural productivity. Higher temperatures accelerate plant stress, reduce crop fertility, and increase pests and disease prevalence (Abbass et al., 2022; Farooq et al., 2022; Nyamukondiwa et al., 2022). Potatoes, for example, thrive within an optimal temperature range of 5°C to 30°C, and temperatures above this threshold reduce tuber formation and overall yield (Haverkort and Verhagen, 2008; Levy and Veilleux, 2007). Reduced water availability, a consequence of climate change, further exacerbates these crops’ challenges as they are particularly sensitive to soil moisture deficits (Daccache et al., 2012). Similarly, tomato plants are sensitive to high temperatures, leading to reduced fertility, flower drop, and diminished fruit set when daytime temperatures exceed 29°C and nighttime temperatures rise above 21°C (Berry and Uddin, 1988).
Increased rainfall and humidity in some regions may also favor the spread of bacterial pathogens such as Ralstonia and Clavibacter, increasing the risk to tomato and potato production (Bhanwar, 2022; Singh et al., 2023). Moreover, climate change can extend the geographic range of these pathogens, exposing new areas to infection, particularly in temperate regions where these diseases were previously rare (Singh et al., 2023). In the face of such a changing scenario, it is necessary to predict the geographic patterns of host and bacteria’s range shifts caused by climate change.
Species distribution models (SDMs), also known as ecological niche models (ENMs), are widely used tools for predicting the suitable range of species under both current and future environmental conditions (Franklin, 2010; Guisan et al., 2013; Zurell et al., 2020). These models explore factors influencing species distributions by identifying statistical relationships between species occurrence and both biotic and abiotic predictors (Guisan et al., 2013). Despite their assumptions and uncertainties (Elith et al., 2010; Shabani et al., 2016a; Zurell et al., 2020), SDMs are widely used to predict the distribution of diverse taxa, including insects (Ward, 2007), plants (Araújo and Luoto, 2007; Williams et al., 2009; Shabani et al., 2016b), bacterial species such as Bacillus thuringiensis (Ejaz, 2025), and animals (Ahmadi et al., 2024; Lorestani et al., 2022; Luo et al., 2015; Peterson et al., 2001; Shabani et al., 2019). While some studies have predicted the future distributions of host species such as potatoes and tomatoes, along with their associated bacteria, in specific countries (Simon et al., 2010), global-scale predictions for both hosts and pathogens simultaneously remain rare in the scientific literature.
This study aims to model the current and future global distributions of both comprehensively Solanum lycopersicum and Solanum tuberosum alongside their bacterial pathogens, Clavibacter michiganensis and Ralstonia solanacearum. By utilizing state-of-the-art SDMs and incorporating climate projections under two shared socioeconomic pathways (SSP4.5 and SSP8.5), this research provides critical insights into how climate change will alter the spatial overlap between these crops and pathogens, identifying regions at increased risk of infection. We utilized four species distribution models (SDMs), including the generalized linear model (GLM), generalized boosting model (GBM), random forest (RF), and maximum entropy (MaxEnt), combined in an ensemble approach. For future projections, we used four global circulation models (GCMs): ACCESS-CM2, CMCC-ESM2, HadGEM3-GC31-LL, and MIROC6, focusing on two shared socioeconomic pathway scenarios developed for the Coupled Model Intercomparison Project Phase 6 (CMIP6): SSP4.5 and SSP8.5. The findings of this study are crucial for informing targeted agricultural interventions, guiding disease management strategies, and ensuring food security in the face of a rapidly changing climate. Given the global importance of tomatoes and potatoes, the results have far-reaching implications for policymakers, farmers, and researchers seeking to mitigate the impacts of climate change on crop production. The publication of this study is essential as it not only fills a critical knowledge gap in the field of agricultural and environmental science but also provides actionable insights that can help safeguard global food systems.
2 Materials and methods 2.1 Species dataSpecies data were collected from the Global Biodiversity Information Facility (GBIF) using the “rgbif” package within the R environment. GBIF is one of the largest open-source archives of biodiversity data in the world. However, before directly incorporating the GBIF data into SDMs, their precision, quality, and uncertainty should be assessed, as recommended by Zizka et al. (2019). To achieve this, we applied several filters to the GBIF data before downloading it. These filters removed duplicates, records without coordinates, potentially corrupt coordinates, data pre-dating 1990, coordinates with <4 decimals, and points with identical latitude and longitude (Zizka et al., 2019). In addition, for each species, we excluded occurrence points that were <5 km apart from each other, to reduce the adverse effects of spatial autocorrelation (SAC) (Dormann et al., 2007). Furthermore, data from greenhouse cultivation, protected environments, and controlled-temperature systems were excluded from the modeling process to ensure that only open-field occurrence points were retained. This resulted in 5,915; 7,946; 97; and 36 occurrence points for potatoes (Solanum tuberosum), tomatoes (Solanum lycopersicum), Ralstonia solanacearum, and Clavibacter michiganensis, respectively, which were used in the following analysis (Figure 1).
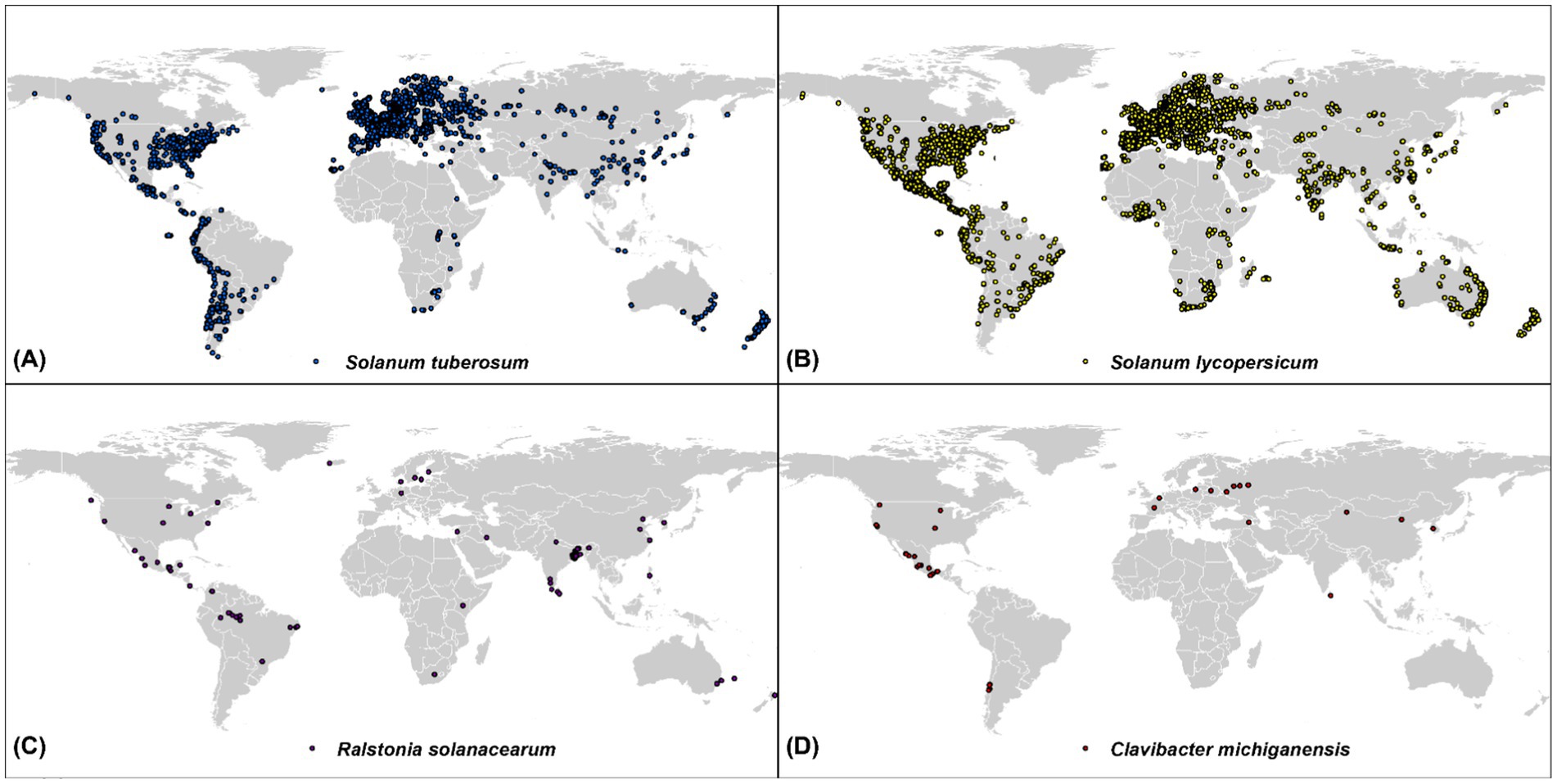
Figure 1. Global distribution of occurrence points for four species: Solanum tuberosum (potatoes, blue), Solanum lycopersicum (tomatoes, yellow), Ralstonia solanacearum (purple), and Clavibacter michiganensis (red). The occurrence data were compiled from the Global Biodiversity Information Facility (GBIF) and filtered for quality and accuracy.
2.2 Environmental dataWe obtained climate data for both current conditions and a future projection (2050) at a spatial resolution of 2.5 min (~4 Km2) from the WorldClim database (version 2.1). Given the potential multicollinearity among climatic variables, we employed the variance inflation factor (VIF) using the “usdm” package (Naimi, 2015). This analysis helped us identify the highly correlated climate variables (correlation exceeding 0.7). Following the VIF analysis, we selected eight factors as distribution predictors including bio1 (annual mean temperature), bio4 (temperature seasonality), bio5 (maximum temperature of the warmest month), bio6 (minimum temperature of the coldest month), bio12 (annual precipitation), bio13 = precipitation of the wettest month, bio14 (precipitation of the driest month), and bio15 (precipitation seasonality).
2.3 Modeling procedureThe potential current distributions of two hosts, namely, potatoes and tomatoes, and their associated bacteria were forecasted using an ensemble modeling technique (Araújo and New, 2007). We selected four SDMs including the generalized linear model (GLM), generalized boosting model (GBM), random forest (RF), and maximum entropy (MaxEnt) to develop the ensemble models, due to their widespread usage and high predictive performance compared to other methods (Phillips et al., 2006). All analyses were carried out using the BIOMOD2 package (Thuiller et al., 2009) in R Environment, version 4.3.2. The SDMs were performed separately for each host and bacteria, by using the same random sample of 10,000 background points. We employed a 10-fold cross-validation method for modeling. In this approach, presence points are randomly divided into 10 subsets. Then, models are constructed using 1 subsets for calibration and the remaining subsets for training, in each iteration. This process is repeated until all subsets have been used once for training. To measure the performance of individual models, we utilized the area under the curve (AUC) of the receiver operating characteristics (ROC) (Fielding and Bell, 1997), and the True Skill Statistic (TSS) (Allouche et al., 2006).
To assess the impact of climate change on the future distribution of four target species and compare their responses to climate change, climatic projections were conducted using four global circulation models (GCMs): ACCESS-CM2, CMCC-ESM2, HadGEM3-GC31-LL, and MIROC6. We focused on two shared socioeconomic pathway scenarios developed for the Coupled Model Intercomparison Project Phase 6 (CMIP6): SSP4.5 and SSP8.5. By employing multiple GCMs, we aimed to address the inherent uncertainties and limitations in climate modeling, providing a more comprehensive, and robust assessment of potential future climatic conditions suitable for the species. Similar to the current SDM, we integrated the model outputs into an ensemble model (Araújo and New, 2007). Through raster calculator analysis in ArcMap (v 10.80.2) for each SSP scenario, we averaged the obtained climatic suitability maps from the four GCMs. All these maps were converted to binary maps based on the threshold the maximum sum of sensitivity (Liu et al., 2016). The binary maps were overlaid to identify areas where hosts and bacteria could potentially coexist. By doing so, we generated the following scenarios:
i. Tomatoes and Potatoes at risk: the distribution area of tomatoes and potatoes, where their ranges overlap with Clavibacter michiganensis and Ralstonia solanacearum bacteria.
ii. Tomatoes and Potatoes without risk: the distribution area of tomatoes and potatoes where Clavibacter michiganensis or Ralstonia solanacearum bacteria were absent.
iii. Potatoes at risk: the distribution area of potatoes, where their range overlaps with Ralstonia solanacearum.
iv. Potatoes without risk: the distribution area of potatoes, where Ralstonia solanacearum bacteria were absent.
v. Tomatoes at risk: the distribution area of the tomatoes, where their range overlaps with Clavibacter michiganensis.
vi. Tomatoes without risk: the distribution area of tomatoes where Clavibacter michiganensis bacteria were absent.
In addition, to compare the differences in distributional changes of hosts and their bacteria, we calculated the three metrics for each: the number of grid cells that are suitable in both current and future time (stable), suitable in the current but will become unsuitable in the future (loss), and unsuitable in the current but will become suitable in the future (gain).
3 ResultsFigure 1 shows the global distribution of occurrence for four species. Tomatoes (Solanum lycopersicum) are widely distributed across North and South America, Europe, parts of Asia, parts of Africa, Australia, and New Zealand, with dense concentrations in North America and central Europe. However, the distribution of Clavibacter michiganensis, which causes bacterial canker, is more geographically restricted, occurring mainly in temperate regions such as North and South America, Europe, and parts of Asia.
Potatoes (Solanum tuberosum) are distributed in North and South America, Europe, and across Asia, particularly in China. Meanwhile, Ralstonia solanacearum (the bacterial wilt pathogen) is concentrated in North and South America, Southeast Asia, and northern Europe. These regions where crop and pathogen distributions overlap are critical for understanding potential risk areas for infection.
3.1 Model performanceThe SDMs for hosts and their associated bacteria showed a range of moderate-to-excellent predictive performances. Potatoes had the highest AUC and TSS scores of 0.974 and 0.84, respectively. In contrast, the lowest AUC and TSS values were observed for Clavibacter michiganensis, with AUC = 0.778 and TSS = 0.018. Comparing the predictive performance of four SDM methods across all species indicated that the RF model exhibited the highest performance (AUC = 0.974, TSS = 0.84), followed by GBM (AUC = 0.963, TSS = 0.808) and MaxEnt (AUC = 0.952, TSS = 0.760) (see Figure 2).
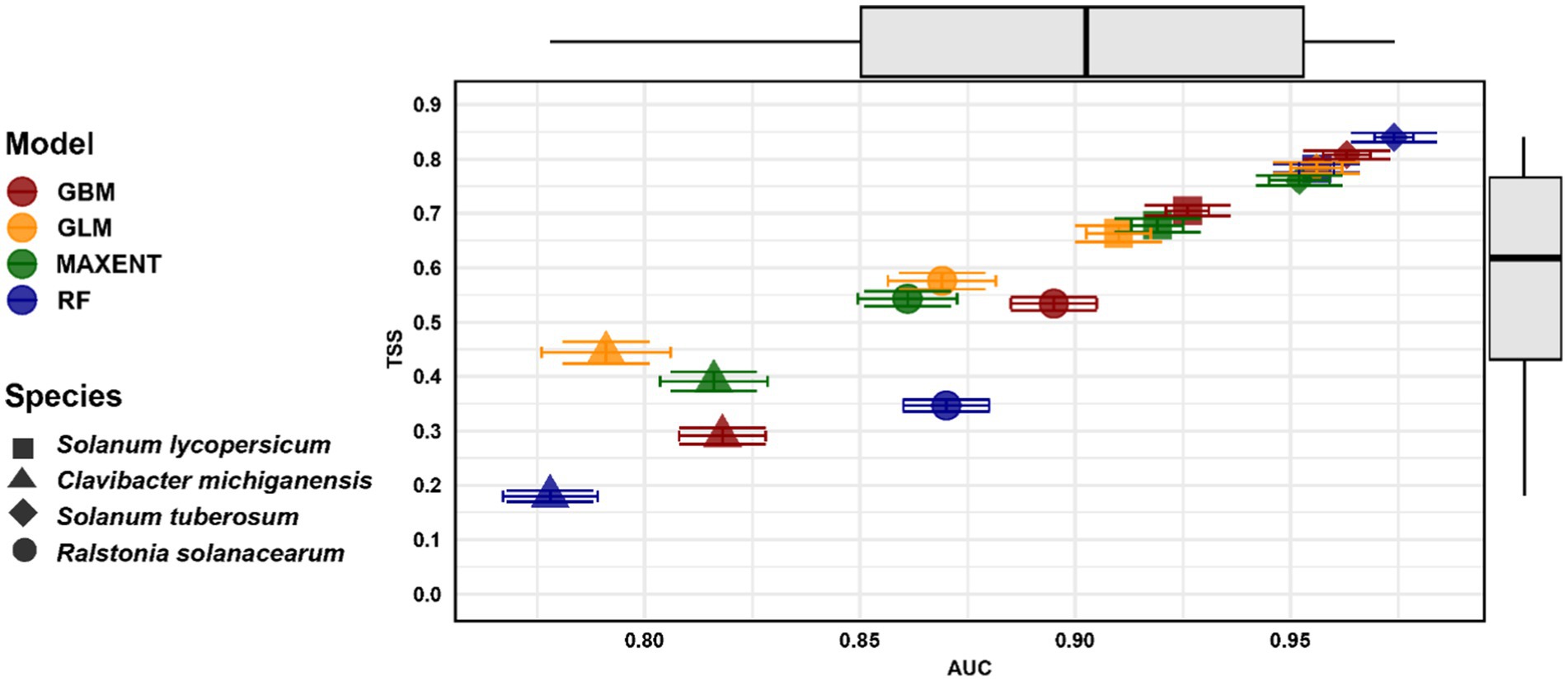
Figure 2. Mean AUC and TSS values for four species distribution models (GBM, GLM, MaxEnt, and RF) across four species: Solanum lycopersicum (tomatoes = Square), Clavibacter michiganensis (Triangle), Solanum tuberosum (potatoes = Diamond), and Ralstonia solanacearum (Circle). The error bars indicate the standard deviation of both AUC and TSS values (Refer to Supplementary Table S1 for detailed model performance data).
3.2 The comparative importance of climatic variablesFor each species, we calculated the relative importance of climatic variables in their suitability model (see Table 1). For tomatoes and their pathogen Clavibacter michiganensis, the minimum temperature of the coldest month (bio6) had the highest contribution to climatic suitability, followed by the annual mean temperature (bio1). Similarly, for potatoes and their associated pathogen Ralstonia solanacearum, annual mean temperature (bio1) had the highest relative importance. The second most important variable for potatoes was the minimum temperature of the coldest month (bio6), while for Ralstonia solanacearum, the second most important variable was temperature seasonality (bio4).
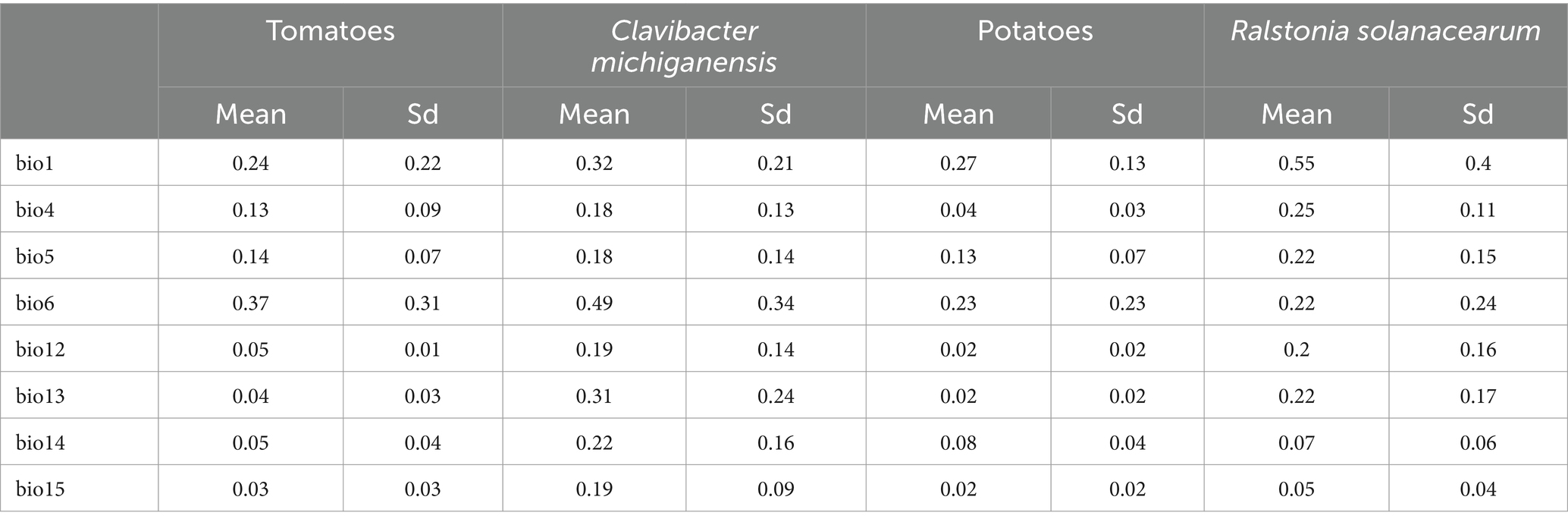
Table 1. Mean and standard deviation (Sd) of the relative importance of climatic variables in the habitat suitability.
3.3 Current spatial distribution and habitat suitability for host crops and pathogensFigure 3 illustrates the predicted suitable habitats for four species under current climatic conditions. The habitat suitability for tomatoes and their pathogen Clavibacter michiganensis shows significant overlap, particularly in most of Europe, South China, Eastern Australia, Southern Africa, and parts of North and South America. In contrast, the predicted suitable areas for potatoes and Ralstonia solanacearum differ somewhat. However, there is overlap in critical regions, including Southeastern America, Eastern Australia, Southeastern China, and parts of Africa.
3.4 Percentage gain and loss in the distribution of hosts and bacteriaProjecting the current SDMs to future climate change scenarios (2050 SSP4.5 and SSP8.5) reveals significant shifts in the suitable habitats for both host crops and their associated bacterial pathogens (refer to Supplementary Figures S1–S4 for continuous suitability maps of the four species separately).
For potatoes, suitable habitats are expected to shift from western Europe to eastern Russia and from northwestern America toward Canada. However, potatoes will lose suitability in regions such as the US, Brazil, most countries in Africa, Spain, Portugal, Ukraine, Poland, China, Japan, and South Korea. In contrast, Ralstonia solanacearum bacteria will retain most of its current suitable areas in southern China, and parts of northern and northwestern Brazil, according to future scenarios. Although its future projections show expansion across all of Brazil and central Africa, there will be a loss of suitable areas in northern Australia and some parts of the US. Suitable areas for tomatoes are expected to expand from western Europe to western Russia and from northwestern America to Canada. However, significant habitat loss will occur in the US, Spain, most African countries, and parts of China and Australia. Clavibacter michiganensis follows a similar pattern, with wide coverage in Russia and Mongolia. While future projections indicate reductions in northern Australia, southeastern China, and parts of the Americas and Asia, there will still be substantial overlap between the areas suitable for both tomatoes and Clavibacter michiganensis (Figure 4). The highest habitat gain was observed in Ralstonia solanacearum, with an increase of 65.34% under SSP4.5 and 71.32% under SSP8.5 by 2050. It also experienced the lowest habitat loss, at 12.29% (SSP4.5) and 15.5% (SSP8.5). In contrast, tomatoes showed the least habitat gain, with 24.46% under SSP4.5 and 28.85% under SSP8.5. Potatoes had the highest habitat loss, with projected losses of 23.95% under SSP4.5 and 29.2% under SSP8.5 (Table 2).
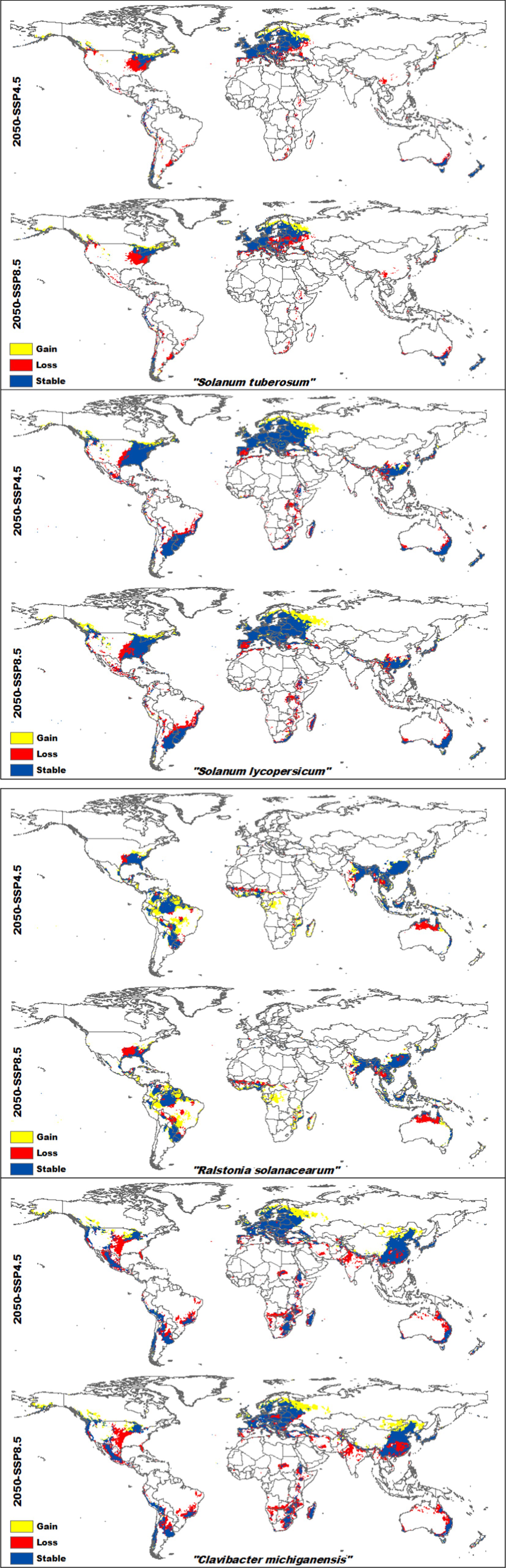
Figure 4. Projected future habitat changes (gain, loss, and stable areas) for Solanum tuberosum (potatoes), Solanum lycopersicum (Tomatoes), Ralstonia solanacearum, and Clavibacter michiganensis under climate change scenarios SSP4.5 and SSP8.5 by 2050. Yellow regions represent areas of habitat gain, red regions indicate habitat loss, and dark blue regions mark areas that remain suitable over time.
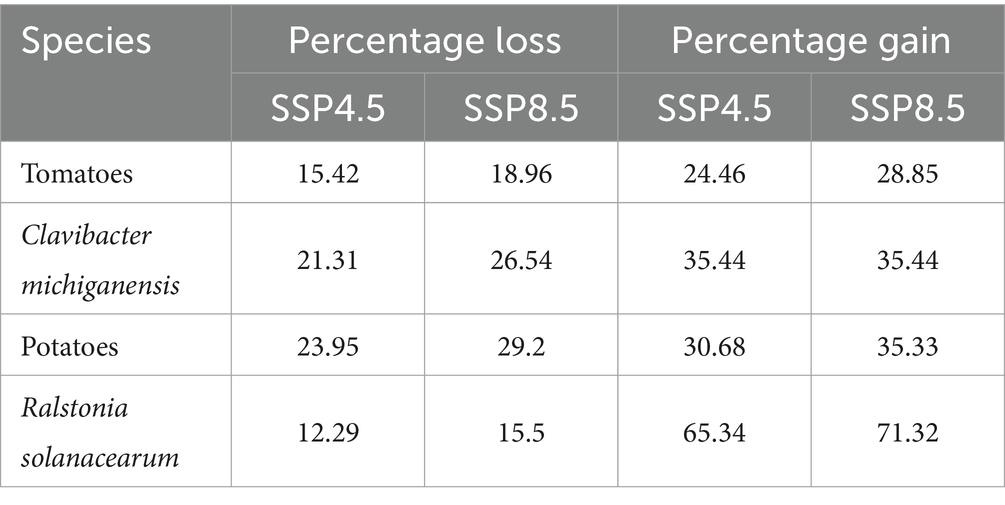
Table 2. Projected percentage of habitat gain and loss for each species, under future climate scenarios SSP4.5 and SSP8.5 by 2050.
3.5 OverlayThe red areas in Figure 5 represent regions where both tomatoes and potatoes are at risk due to overlapping suitable habitats with both Clavibacter michiganensis and Ralstonia solanacearum. Currently, these regions are concentrated in a few specific areas globally, including parts of eastern Australia, China, Japan, France, Spain, and Portugal. Collectively, these regions represent a relatively small fraction (~1%) of the total suitable habitat for both crops. However, future climate projections indicated a decline in the extent of these risk areas (SSP4.5 = 0.05%; SSP8.5 = 0.04%), particularly in China and Japan.
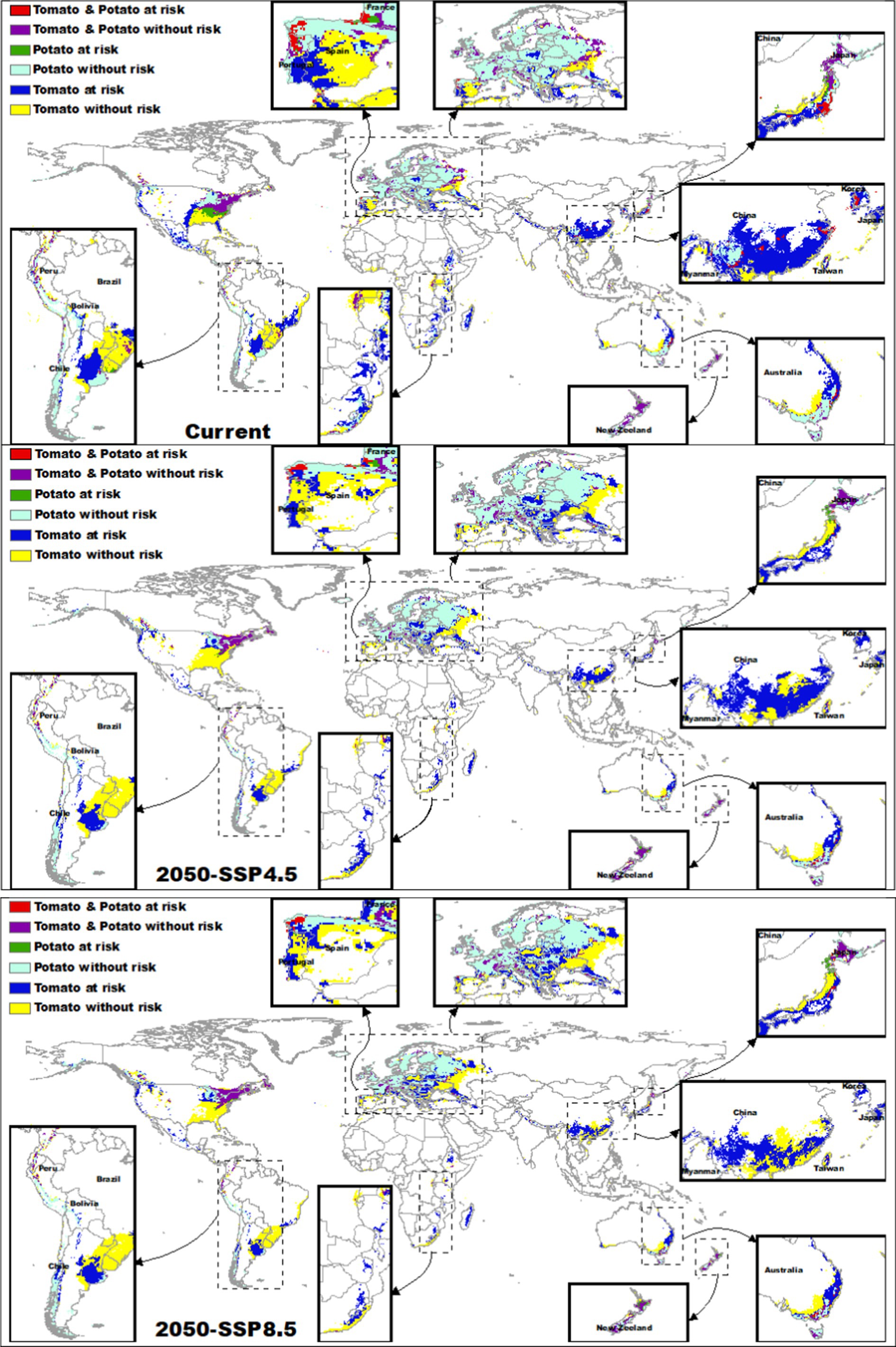
Figure 5. Projected global distribution of tomatoes and potatoes at risk from Clavibacter michiganensis and Ralstonia solanacearum under current and future climate scenarios (SSP4.5 and SSP8.5 for 2050). Red areas show the overlap of both pathogens with tomatoes and potatoes. Dark blue and green areas represent regions where only tomatoes or potatoes, respectively, are at risk. Yellow and light blue areas indicate regions where tomatoes and potatoes remain suitable without pathogen risk, while purple areas show regions without risk for either crop.
For tomato, areas at risk from Clavibacter michiganensis (represented in dark blue in Figure 5) cover a significantly larger extent than other risk areas. Currently, the overlap between tomatoes and Clavibacter michiganensis suitable habitats is approximately 61.11%. By 2050, this overlap is projected to decrease to 58.2% under SSP4.5 and 56.35% under SSP8.5, particularly in regions such as the US, Brazil, and Spain. Despite this projected reduction, significant risk areas will remain in southeastern Australia, Japan, China, eastern Africa, and Argentina, especially under the SSP8.5 scenario. Conversely, parts of Europe, the northeastern US, and southern Brazil are expected to remain without risk (depicted in yellow in Figure 5).
In contrast, potatoes show the lowest overlap with Ralstonia solanacearum (represented in green in Figure 5). Currently, the overlap between potatoes and this pathogen is only 5.36%, concentrated in regions such as the eastern US and parts of France. This overlap is projected to decrease further to 1.87% under SSP4.5, with residual risk areas remaining in France, and to 1.97% under SSP8.5, although some increase is projected in parts of Japan. Areas not at risk for potatoes (depicted in light blue in Figure 5) are projected to remain stable across most of Europe and the northeastern US, although decreases in suitability are expected in Australia and China.
Regions without risk from either Clavibacter michiganensis or Ralstonia solanacearum (represented in purple in Figure 5) are currently concentrated in eastern North America, New Zealand, central Japan, Russia, and parts of Europe, particularly France. These regions are expected to remain relatively stable in the future, particularly in the US, France, New Zealand, and Japan.
4 DiscussionThis study assessed climate change impacts on the habitats of two key crops, tomatoes and potatoes, and their associated pathogens, Clavibacter michiganensis and Ralstonia solanacearum. The key focus of this analysis was to identify the most critical areas at risk for both crops using spatially explicit species distribution models (SDMs) under future climate change scenarios (SSP4.5 and SSP8.5). All SDMs demonstrated strong predictive performance across species. Random forest (RF) models demonstrated the highest accuracy for predicting potato distribution, providing robust support for our climate-based projections.
Temperature emerged as the dominant factor affecting potato distribution, corroborating previously mentioned findings on the crop’s sensitivity to temperature fluctuations (Champooiseau et al., 2008; Kim and Lee, 2019; Obiero et al., 2022; Rana et al., 2020). Increased temperatures negatively affect the growth, yield, and quality of potatoes. Key physiological processes such as net photosynthesis, tuber emergence, and leaf appearance rate are all impacted by temperature fluctuations. Studies suggest that while temperatures below 20°C delay sprout growth, temperatures above the optimal range also hinder sprout development in potatoes (refer to Singh et al., 2013 for more detail). However, these projections are based solely on climatic factors and do not account for critical agricultural variables such as soil type and land management practices, which may significantly affect real-world outcomes.
For tomatoes, the minimum temperature of the coldest month was the most important factor, consistent with previous research showing that extreme temperatures reduce tomato yield and fruit quality (Silva et al., 2017b), which has shown that high temperatures can lead to considerable yield losses due to reduced fruit set, smaller fruit size, and diminished fruit quality in tomatoes (Singh et al., 2013). Overall, climate change impacts, such as flooding and drought conditions, significantly affect tomato growth as this crop is highly sensitive to climate stress (Shabani et al., 2012; Silva et al., 2017a, 2017b). For example, water deficits can reduce fruit number and weight, induce flower abscission, and decrease reproductive activity (Heuvelink, 2005). Previous studies also highlighted the importance of other climatic variables; for example, annual precipitation (bio12) and precipitation of the driest quarter (bio17) have been identified as key environmental variables influencing the suitability of tomatoes in China (Liu et al., 2024). Suitable habitats for tomatoes are projected to shift northward (Silva et al., 2017a), with expansions in northern Europe and Russia, and contractions in traditional growing regions such as the US and Spain (Carrasco et al., 2021). The projected losses in Spain may be attributed to a decrease in the length of the crop season (LCS) for tomatoes, which is expected to shrink to 48 days by 2050, compared to a range of 76 to 180 days in 2000, as predicted by Saadi et al. (2014). Given these projected shifts, it is crucial for countries facing declining suitability for tomato cultivation, such as Mexico—considered the center of tomato domestication in America (Ramírez-Ojeda et al., 2022)—to incorporate these findings into agricultural planning.
Similarly, Clavibacter michiganensis, the pathogen that leads to bacterial canker, is projected to follow a similar spatial shift to regions such as western Russia, northeastern China, and central Mongolia. Some areas in Europe, southern Africa, and most of China are expected to remain suitable for bacterial canker under future climate scenarios. Based on our results and previous studies on tomato cultivation in these stable regions, such as the study by Liu et al. (2024) in China, a significant overlap between the future suitable habitats of tomatoes and their pathogen is likely to persist over time.
Our results indicated an ongoing shift in the suitable range for potatoes toward higher latitudes, from western Europe and North America to eastern Russia and Canada. We found that potato suitability decreases in regions such as the US, Brazil, several European countries, Africa, China, Japan, and South Korea, reflecting a global reduction in suitable habitats for this crop due to rising temperatures. However, Kim and Lee (2019) predicted that potato yields in South Korea, particularly for spring cultivation, are expected to increase under climate change. Nonetheless, they indicated that yield decreases in hotter regions and during high-temperature episodes remain a concern. Our findings also showed that southeastern Australia will remain suitable for potato cultivation over time. At the same time, Ralstonia solanacearum is expected to lose the majority of its suitable areas in Australia. However, potatoes in Australia remain at risk from other pests and pathogens, such as the tomato potato psyllid (TPP) and its associated bacterial plant pathogen Candidatus Liberibacter solanacearum (CLso), as reported in previous studies (Wan et al., 2020). In addition, Ralstonia solanacearum was projected to maintain its current range in southern China and parts of northern and southern Brazil, while expanding significantly across Brazil, Central Africa, and India.
Finally, after overlapping the suitable habitats of both host crops and their bacterial pathogens, we identified critical areas that may represent significant risks under both current and future climate scenarios. Although the total overlap regions constitute a relatively small fraction of the global habitat for both crops, agriculturally important regions such as eastern Australia, parts of China, Japan, Spain, Portugal, and France are projected to remain at risk for both pathogens, Clavibacter michiganensis and Ralstonia solanacearum, in both current and future scenarios. The greater overlap between tomatoes and Clavibacter michiganensis, compared to potatoes and Ralstonia solanacearum, suggests that tomato cultivation may face more substantial pathogen risks in the future, particularly in regions such as southeastern Australia, eastern Africa, Argentina, and much of China. Conversely, while potatoes show less overlap with Ralstonia solanacearum, notably in the US and Japan, other factors may still pose significant threats to potato cultivation. These results underscore the urgency of developing targeted management strategies in regions where crop–pathogen overlap is projected to increase, particularly for maintaining food security in vulnerable agricultural zones.
This study offers critical insights into the future distribution of tomatoes and potatoes under climate change, providing a foundation for informed agricultural planning. However, our models are based solely on climatic variables, without considering essential factors such as soil properties, land use, or pathogen–host competition. As a result, while our projections highlight potential broad-scale geographic shifts, real-world outcomes could vary significantly based on local environmental conditions and management practices. In addition, other pests and pathogens, such as the potato tuber moth (Phthorimaea operculella) (Jung et al., 2020), Phytophthora infestans (González-Jiménez et al., 2023), and Neoleucinodes elegantalis (Silva et al., 2017b), could further influence the suitability of these crops but were not included in this analysis. Future research should integrate non-climatic variables, biotic interactions, and other pests and pathogens to comprehensively assess crops’ suitability under changing climate conditions.
While our findings provide a valuable forecast of climate-driven habitat shifts for tomatoes and potatoes, it is important to note that our models are based solely on climatic factors. Other crucial variables, such as soil type, land use, and biotic interactions, which also impact crop and pathogen distributions, were not included. Therefore, the results should be interpreted as indicative of broad-scale geographic shifts, rather than precise forecasts at a local or regional level. Furthermore, the inherent uncertainties in future greenhouse gas emission levels, as represented by the two shared socioeconomic pathways (SSP4.5 and SSP8.5), introduce an additional layer of uncertainty to the predictions. The maps and results should thus be viewed with caution, recognizing that they are scenarios based on assumptions about future climate trajectories, and real-world outcomes could differ depending on how these variables evolve.
5 ConclusionThis study highlights the potential impacts of climate change on the distributions of Solanum lycopersicum (tomatoes), Solanum tuberosum (potatoes), and their bacterial pathogens, Clavibacter michiganensis and Ralstonia solanacearum. By 2050, we identified critical areas where both crops and pathogens may experience significant shifts in habitat suitability, with expansions projected toward higher latitudes in regions such as northern Europe, Canada, and Russia and significant declines anticipated in traditional production zones such as parts of the US, Brazil, and China. These findings underscore the urgent need for targeted agricultural planning and disease management strategies to mitigate the risks posed by these shifts.
While our models provide a critical baseline for understanding broad-scale climatic suitability, they are limited by their reliance on climatic variables alone. Factors such as land use, soil characteristics, biotic interactions, and agricultural management practices, which significantly influence crop and pathogen distributions, were not included. These omissions highlight the need for future studies to integrate additional environmental, biological, and socioeconomic data to enhance model accuracy and applicability.
Despite these limitations, this research offers valuable insights for policymakers and agricultural stakeholders, serving as a foundation for proactive strategies to safeguard food security under changing climatic conditions.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributionsMH: Conceptualization, Writing – original draft. NL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft. RA-A: Conceptualization, Writing – original draft. FS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by an internal grant from Qatar University (CG24/25-383), which also covered the publication fees associated with this work.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementAs outlined in the Artificial Intelligence guidelines provided by the journal (https://www.frontiersin.org/journals/artificial-intelligence/for-authors/author-guidelines) (accessed November 5, 2024), the use of AI technologies, such as ChatGPT, in the writing or editing of manuscripts submitted to Frontiers is permitted. The authors affirm that no generative AI was employed in generating the methods, codes, modelling results, Figures, or Tables in this work. Instead, AI and Grammarly was used to improve the text’s readability and style and to ensure accuracy in grammar, spelling, punctuation, and tone.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1520104/full#supplementary-material
ReferencesAbbass, K., Qasim, M. Z., Song, H., Murshed, M., Mahmood, H., and Younis, I. (2022). A review of the global climate change impacts, adaptation, and sustainable mitigation measures. Environ. Sci. Pollut. Res. 29, 42539–42559. doi: 10.1007/s11356-022-19718-6
PubMed Abstract | Crossref Full Text | Google Scholar
Ahmadi, M., Nawaz, M. A., Asadi, H., Hemami, M. R., Naderi, M., Shafapourtehrany, M., et al. (2024). Protecting alpine biodiversity in the Middle East from climate change: implications for high-elevation birds. Divers. Distrib. 30:e13826. doi: 10.1111/ddi.13826
Crossref Full Text | Google Scholar
Allouche, O., Tsoar, A., and Kadmon, R. (2006). Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232. doi: 10.1111/j.1365-2664.2006.01214.x
Crossref Full Text | Google Scholar
Araújo, M. B., and Luoto, M. (2007). The importance of biotic interactions for modelling species distributions under climate change. Glob. Ecol. Biogeogr. 16, 743–753. doi: 10.1111/j.1466-8238.2007.00359.x
Crossref Full Text | Google Scholar
Baysal, Ö., Mercati, F., İkten, H., Yıldız, R. Ç., Carimi, F., Aysan, Y., et al. (2011). Clavibacter michiganensis subsp. michiganesis: tracking strains using their genetic differentiations by ISSR markers in southern Turkey. Physiol. Mol. Plant Pathol. 75, 113–119. doi: 10.1016/j.pmpp.2010.10.002
Crossref Full Text | Google Scholar
Berry, S. Z., and Uddin, M. R. (1988). Effect of high temperature on fruit set in tomato cultivars and selected germplasm. Hort Sci. 23, 606–608. doi: 10.21273/hortsci.23.3.606
Crossref Full Text | Google Scholar
Bhanwar, R. (2022). Effect of temperature and pH level on growth of bacterial wilt causing R. solanacearum under in vitro condition. Pharma Innov. 1, 1656–1659.
Blanca, J., Montero-Pau, J., Sauvage, C., Bauchet, G., Illa, E., Díez, M. J., et al. (2015). Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics 16, 1–19. doi: 10.1186/s12864-015-1444-1
PubMed Abstract | Crossref Full Text | Google Scholar
Carrasco, L., Papeş, M., Lochner, E. N., Ruiz, B. C., Williams, A. G., and Wiggins, G. J. (2021). Potential regional declines in species richness of tomato pollinators in North America under climate change. Ecol. Appl. 31:e02259. doi: 10.1002/eap.2259
PubMed Abstract | Crossref Full Text | Google Scholar
Chalupowicz, L., Barash, I., Reuven, M., Dror, O., Sharabani, G., Gartemann, K. H., et al. (2017). Differential contribution of Clavibacter michiganensis ssp. michiganensis virulence factors to systemic and local infection in tomato. Mol. Plant Pathol. 18, 336–346. doi: 10.1111/mpp.12400
PubMed Abstract | Crossref Full Text | Google Scholar
Champooiseau, P. G., Allen, C., Jones, J. B., Harmon, C., and Momol, T. M. (2008). Ralstonia solanacearum race 3 biovar 2: Disease cycle & epidemiology. Available at: https://plantpath.ifas.ufl.edu (accessed October 4, 2024).
Costa, J. M., and Heuvelink, E. (2018). “The global tomato industry” in Tomatoes. ed. E. Heuvelink (Wallingford: CABI Wallingford UK), 1–26.
Daccache, A., Keay, C., Jones, R., Weatherhead, K., Stalham, M., and Knox, J. (2012). Climate change and land suitability for potato production in England and Wales: impacts and adaptation. J. Agric. Sci. 150, 161–177. doi: 10.1017/S0021859611000839
Crossref Full Text | Google Scholar
Dormann, C., McPherson, J., Araújo, M., Bivand, R., Bolliger, J., Carl, G., et al. (2007). Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628. doi: 10.1111/j.2007.0906-7590.05171.x
Crossref Full Text | Google Scholar
Ejaz, M. R. (2025). “Global climate change and its impact on the distribution and efficacy of Bacillus thuringiensis as a biopesticide.” Science of the Total Environment. 958:178091.
Elith, J., Kearney, M., and Phillips, S. (2010). The art of modelling range-shifting species. Methods Ecol. Evol. 1, 330–342. doi: 10.1111/j.2041-210X.2010.00036.x
Crossref Full Text | Google Scholar
FAO (2016). The state of world fisheries and aquaculture 2016. Contributing to food security and nutrition for all. Rome: FAO.
Farooq, M. S., Uzair, M., Raza, A., Habib, M., Xu, Y., Yousuf, M., et al. (2022). Uncovering the research gaps to alleviate the negative impacts of climate change on food security: a review. Front. Plant Sci. 13:927535. doi: 10.3389/fpls.2022.927535
PubMed Abstract | Crossref Full Text | Google Scholar
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The sol genomics network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
PubMed Abstract | Crossref Full Text | Google Scholar
Fielding, A. H., and Bell, J. F. (1997). A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49. doi: 10.1017/S0376892997000088
Crossref Full Text | Google Scholar
Franklin, J. (2010). Mapping species distributions: Spatial inference and prediction. Cambridge: Cambridge University Press.
留言 (0)