Helicobacter pylori was discovered as a pathogen of the human stomach by Robin Warren and Barry Marshall in 1983, followed by the establishment of the genus Helicobacter in 1989 (Goodwin et al., 1989). Helicobacter species infecting the stomach are called gastric Helicobacter that neutralizes around itself by producing urease in order to survive in acidic condition. Gastric Helicobacter species infects various animals, including dolphins, whales, pigs, cats, dogs, non-human primates, and humans. All these exhibits a corkscrew-like spiral form different from that of H. pylori. H. pylori is known to infect the human stomach and cause gastric diseases such as gastric cancer, while non-H. pylori gastric Helicobacter species such as Helicobacter suis infecting pigs and Helicobacter ailurogastricus infecting cats have been reported as possible causes of gastric diseases such as gastric mucosa associated lymphoid tissue (MALT) lymphoma and gastric ulcers in human (Rimbara et al., 2021; Sano et al., 2023).
Helicobacter species include 54 species so far, and among them, the following seven gastric Helicobacter species have been identified in dogs and cats: H. ailurogastricus (Joosten et al., 2016), Helicobacter baculiformis (Baele et al., 2008), Helicobacter bizzozeronii (Hänninen et al., 1996), Helicobacter cynogastricus (Van den Bulck et al., 2006), Helicobacter felis (Paster et al., 1991), Helicobacter heilmannii (Smet et al., 2012), and Helicobacter salomonis (Jalava et al., 1997). Previous study on gastric Helicobacter infecting dogs and cats in Japan suggested that Helicobacter species infect Japanese dogs and cats include the species, which are not identifiable with known gastric Helicobacter species. Although culture is necessary for species identification, cultured strain was not obtained in the previous study. We have recently established methods to isolate H. suis and H. ailurogastricus from human stomachs (Rimbara et al., 2021; Sano et al., 2023). In this study, we isolated and cultured gastric Helicobacter species infecting dogs and cats in Japan, and analyzed the genome of the isolates. As a result, we isolated nine gastric Helicobacter species strains, including three novel species, from dogs and cats suffering from gastrointestinal disease in 2019 and 2021 in Japan. According to the recent standards for the identification of novel Helicobacter species proposed by On et al. (2017) and Riesco and Trujillo (2024) we verify the phenotypic, phylogenetic and genotypic characterization of these strains. Based on these results, we propose the names Helicobacter gastrocanis sp. nov., Helicobacter gastrofelis sp. nov. and Helicobacter felistomachi sp. nov. for the three novel species.
Materials and methods Strains and morphology studiesNine strains isolated from pets that were visited the Veterinary Medical Center, The University of Tokyo between 2019 and 2022 were analyzed in this study. Pets suffering from various conditions such as hypoalbuminemia and chronic vomiting underwent endoscopic examination and endoscopically obtained gastric specimens were used for the isolation of Helicobacter species when the characteristic corkscrew-like bacterium was observed by Giemsa staining. The isolation of Helicobacter species was performed according to a previously reported method (Rimbara et al., 2021). In brief, these specimens were homogenized in Brucella Broth with 0.05% HCl (pH around 5) and spread on non-Helicobacter pylori Helicobacter (NHPH) agar plates containing 1.5% agar (Difco Becton Dickinson) and NHPH medium consisting of Brucella broth (Difco Becton Dickinson), 20% fetal bovine serum (Gibco), Vitox supplement (Oxoid), Campylobacter-selective supplement, Skirrow (Oxoid), 5 mg/mL amphotericin B (FUJIFILM Wako), and 0.05% HCl (FUJIFILM Wako). The plates were incubated for 6 to 14 days at 37°C under microaerobic conditions with a gas mixture of 5% O2 and 12% CO2. A single colony was sub-cultured on the NHPH agar plates, followed by inoculation into the two-layer NHPH medium. The biopsy specimen taken for clinical diagnostic purpose was used for this study. For characterization of the strain according to its morphological features, we performed scanning electron micrograph and Giemsa staining (Rimbara et al., 2021).
Genome sequencingWhole-genome sequencing of all strains was performed using the MiniSeq (NHP19-002, NHP19-003T, NHP19-009, NHP19-012T, NHP21-005T, NHP21-011), HiSeq (NHP20-010, NHP20-013), and NovaSeq (NHP22-001) platforms (Illumina, San Diego, USA). Genomic DNA from strains was extracted using DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. The library for Illumina sequencing (paired-end, insert size of 500–900 bp) was prepared using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, USA). Illumina reads were assembled using Shovill v1.1.0. and default parameters, to acquire the draft genome sequences. For NHP19-003T, NHP19-012T, and NHP21-005T, which were considered novel species, complete genomes were determined on the MinION platform (Oxford Nanopore Technologies [ONT], Oxford, UK). Genomic DNA from isolated strains was extracted using Genomic-tips 20/G and buffers (Qiagen) according to the manufacturer’s instructions. The library for MinION sequencing was prepared using Rapid Sequencing Kit (SQK-RBK004) (ONT, Oxford, UK). ONT reads were base-called using Guppy v4.2.2 (NHP19-003T and NHP19-012T) or v6.4.2 (NHP21-005T) and were assembled de novo using Canu v1.8 (NHP19-003T and NHP19-012T) or v2.1 (NHP21-005T) (Koren et al., 2017). The overlap region in the assembled contig was detected using LAST (Frith et al., 2010) and was trimmed manually. Illumina reads were mapped onto the resulting circular sequences, and sequencing errors were corrected twice with ONT reads using Racon v1.4.13 (Vaser et al., 2017), and then polished twice with Illumina reads combined using Pilon v1.20.1 (Walker et al., 2014), resulting in the complete genomes. The obtained complete and draft genomic sequences were annotated using the DFAST server.
The average nucleotide identity (ANI) values were calculated using Pyani 0.2.12 (Pritchard et al., 2016). Since it has been reported that a value of 70% DNA–DNA hybridization for species delineation corresponds to about 95% ANI, 95% ANI cut-off values were used for species identification (Goris et al., 2007). According to the standard recommended by Riesco and Trujillo (2024), the average amino acid identity (AAI) values were calculated using EzAAI (Kim et al., 2021). Digital DNA–DNA hybridization (dDDH) values were calculated using GGDC and 70% dDDH cut-off values were used for species identification (Meier-Kolthoff et al., 2013).
Phylogenetic characteristicsThe alignment of the VacA-like proteins was generated by MAFFT version 7.49 and the tree was constructed using RAxML-NG version 1.1.0 with a LG + G4 model and 1,000 bootstrap replicates. The accession numbers of the VacA-like proteins of the Helicobacter species used in the analysis are summarized in Supplementary Table S1.
A phylogenetic analysis using a core-gene and 16S rRNA sequence alignment, was generated using Roary (Page et al., 2015) and MAFFT version 7.49 (Katoh et al., 2002), respectively. Aligned sequences were used for phylogenetic reconstruction using RAxML-NG version 1.1.0 with a GTR + G + I model and 1,000 bootstrap replicates (Kubota-Aizawa et al., 2017a; Kubota-Aizawa et al., 2017b). The genomic information and accession numbers of the 16S rRNA and genomic sequences of the known Helicobacter species used in the analysis are summarized in Supplementary Tables S2, S3.
The ureAB sequences of Helicobacter species amplified and sequenced from gastric biopsies of dogs (47 samples) and cats (24 samples) in Japan, which were obtained in previous studies (Kubota-Aizawa et al., 2017a; Kubota-Aizawa et al., 2017b), were re-analyzed in this study. Sequences obtained in previous studies were assigned “LCXXXXXXXX” as accession numbers. The accession numbers of the ureAB sequences used in the analysis are summarized in Supplementary Table S4. The alignment of the partial ureA and ureB genes was generated by MAFFT version 7.49 and the tree was constructed using RAxML-NG version 1.1.0 with a GTR + G + I model and 1,000 bootstrap replicates.
Phenotypic characteristicsThe physiological characteristics of NHP19-003T, NHP19-012T and NHP21-005T were demonstrated using API Campy (bioMérieux) according to the manufacturer’s instruction. Tolerance to 1% bile, 1% glycine, and 1.5% NaCl was assessed by culturing organisms on NHPH agar plates supplemented with each compound. The plates were inoculated with 10 μL cultures prepared according to McFarland standard 6.0 (bioMérieux) and incubated for 7 days at 37°C under microaerobic conditions. Bacterial growth was visually confirmed. Catalase activity of the isolates was examined by adding a 3% H2O2 solution and observing the reaction within 5 s. Oxidase activity was determined using the Poremedia® oxidase test indicator (Eiken Chemical). To clarify the optimum temperature, strains were incubated at 25°C, 37°C, and 42°C under microaerobic conditions. Oxygen requirements were tested at 37°C under aerobic, microaerobic, and anaerobic conditions.
MALDI-TOF MS assaysProtein profiles were generated from MALDI Biotyper (BrukerBiotyper, BrukerDaltonics). Bacterial isolates were prepared according to an acetonitrile–formic acid-extraction protocol provided by the manufacturer and analyzed using flexAnalysis software (version 3.4; BrukerDaltonics). MALDI-TOF mass spectra were recorded with an LT microflex mass spectrometer (BrukerDaltonics) within the range of 2,000 Da to 20,000 Da according to the manufacture, m/z stands for mass to charge ratio.
Animal experimentsMice infections were performed using three-week-old female C57BL/6 N mice (CLEA Japan, Inc. Tokyo, Japan). Three novel Helicobacter sp. strains (NHP19-003T, NHP19-012T, and NHP21-005T), H. ailurogastricus ASB7T, and H. suis NHP19-4004 were infected intragastrically using a feeding needle with 0.2 mL of 1 × 109 colony-forming units (CFU) ml−1 of every other day, repeated three times. Five mice were infected for each strain. After 1 month, mice were sacrificed, and the stomachs were cut along the greater curvature and the gastric content were removed and rinsed with PBS. Half of the stomach was used for histopathological examination and the other half for DNA extraction. For histopathology the stomach was fixed with 10% (wt/vol) neutral-buffered formalin (FUJIFILM Wako), embedded in paraffin, and sectioned to approximately 4 μm thickness. The sections were stained with hematoxylin–eosin (H&E) and immunohistochemically stained for CD3(+) and CD19(+) using specific antibodies. Paraffin sections (4 μm) were deparaffined, rehydrated, antigen-retrieved in the Immunosaver (Nishin EM, Tokyo, Japan), antigen-retrieved, blocked, and incubated with the primary antibodies (anti-CD3, Cell Signaling #99940, 1/600dilution; CD19, Cell Signaling #90176, 1/3200 dilution) overnight at 4°C. The antigen–antibody complexes on the slides were visualized using VECTASTAIN Elite ABC HRP Kit and DAB Substrate Solution (#SK-4105, VECTOR). Finally, cell nuclei were stained with hematoxylin (MUTO Pure Chemicals Co., Ltd., Japan). For the fundic and pyloric regions, 30 glands were randomly selected for each sample, and quantification was performed by counting the number of positive cells within these glands to determine the ratio. For the forestomach-glandular border, the number of positive cells in the border was counted and the result was categorized as follows: <20 positive cells was 1, 20–100 positive cells was 2, and >100 positive cells was 3.
The relative Helicobacter sp. count in the mouse stomachs was evaluated by the comparative ΔCt method using probe-based quantitative PCR targeting the region of 16S rRNA specific to NHPH species as shown in a previous report (Matsui et al., 2023). β-actin was used as an internal control using the following primers: β-actin_forward (5’-TGAAGTGTGACGTTGACATCC-3′), β-actin_reverse (5′-TCCTTCTGCATCCTGTCAGC-3′), and β-actin_probe (/56-FAM/ATTCCATAC/ZEN/CCAAGAAGGAAGGCTGG/3IABkFQ/). DNA was extracted from the mouse stomach using the DNeasy Blood & Tissue Kit (Qiagen) and PCRs were performed in an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). Probe-PCR was performed in duplicate for each sample, and the 2-ΔCt (16S-β-actin) value of each sample was used for comparison. The relative value was calculated by dividing the 2-ΔCt (16S-β-actin) of each sample by the average of the 2-ΔCt (16S-β-actin) values of the H. ailurogastricus ASB7-infected group. All animal experiments and the study protocol were approved by the Committee for Animal Experimentation of the National Institute of Infectious Diseases (approval number: 123036).
Statistical analysisFor data on the number of CD3(+) and CD19(+) cells that violated parametric assumptions such as normality and homogeneity of variances, the Kruskal–Wallis test was employed, with post hoc analysis conducted using Dunn’s multiple comparison test. Means were adjusted by ± one standard deviation (SD). In an RNA assay, each Helicobacter sp.-infected group was compared with the ASB7T-infected group using one-way ANOVA with Tukey’s test. Prism 10 (GraphPad Software, La Jolla, CA, USA) was used for statistical analysis. Data represent means ± SD. p < 0.05 was considered statistically significant.
Results Helicobacter species identified by genomic analysisThe list of the nine strains isolated in this study are shown in Table 1. The nine strains of Helicobacter species obtained were isolated from different individual animals. Among the nine strains, two strains [NHP19-002 (Accession No. GCA_030270085.1) and NHP19-009 (Accession No. GCF_030270105.1)] were identified as Helicobacter ailurogastricus, two strains [NHP20-010 (Accession No. GCF_030270125.1) and NHP20-013(Accession No. GCA_036248205.1)] were Helicobacter bizzozeronii, and NHP21-011 (Accession No. GCF_030270145.1) was Helicobacter heilmannii by the average nucleotide identity (ANI) value calculation using whole genomic sequences. The four remaining strains (NHP19-003T, NHP19–012T, NHP21-005T, and NHP22-001) had no more than ANI threshold value (95–96%) with any Helicobacter species, suggesting that these strains are novel Helicobacter species (Figure 1 and Table 1). The ANI value between NHP19-012T and NHP22-001 was 98.5%, suggesting that these two strains were of the same species (Table 1). Therefore, subsequent experiments were conducted using only the three representative strains: NHP19-003T, NHP19-012T, and NHP 21-005T. It was also clear that NHP19-003T, NHP19-012T, and NHP21-005T are distinct from any Helicobacter species, since the ANI value between these strains were 85.7% (NHP19-003T and NHP19-012T), 86.5% (NHP19-003T and NHP21-005T), and 86.5% (NHP19-012T and NHP21-005T), respectively. The average amino acid identity (AAI) values among NHP19-003T, NHP19-012T, NHP21-005T, H. heilmannii ASB1T, and H. ailurogastricus ASB7T ranged between 87.0 and 89.8%. These values are lower than the AAI values of 90.2% between H. pylori ATCC43504T and H. acynonychis NCTC12686T, and 92.6% between H. bizzozeronii CCUG35545T and H. mehei L15T. This supports the conclusion that NHP19-003T, NHP19-012T, and NHP21-005T are three novel Helicobacter species (Table 1). The digital DNA–DNA Hybridization (dDDH) analysis also indicated that these three strains are novel Helicobacter species as well as ANI and AAI value (Table 1).
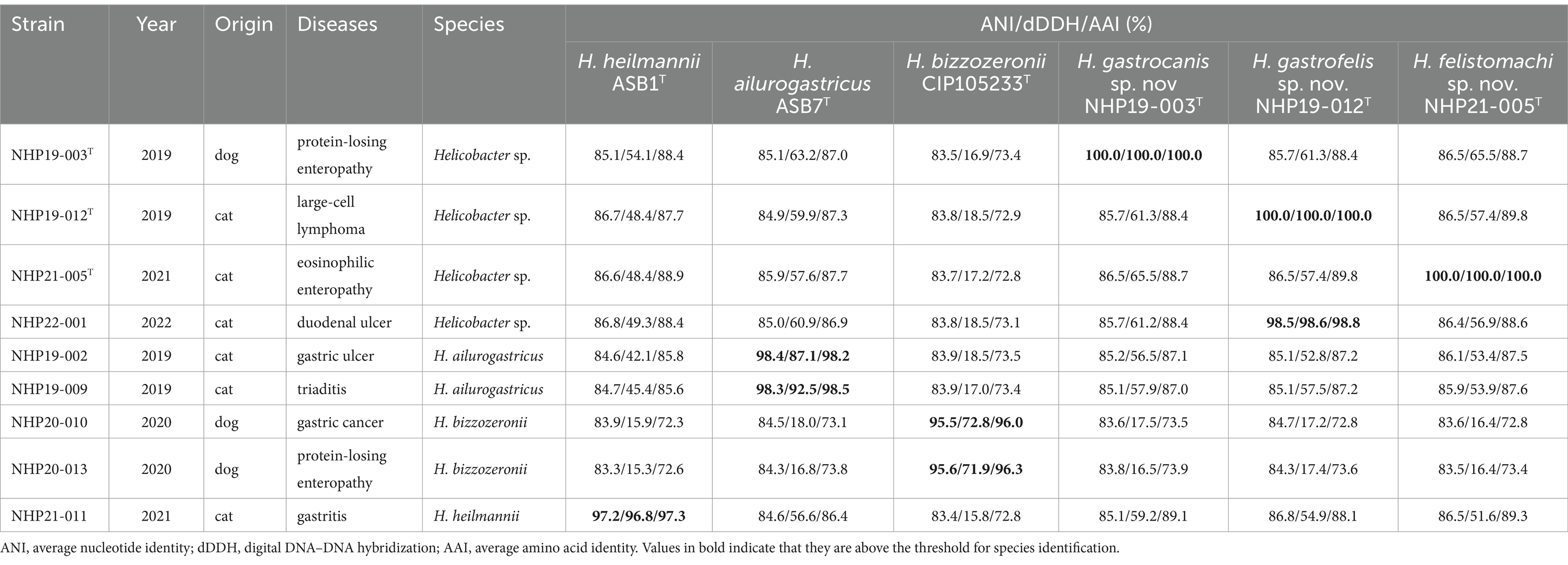
Table 1. Strains isolated in this study.
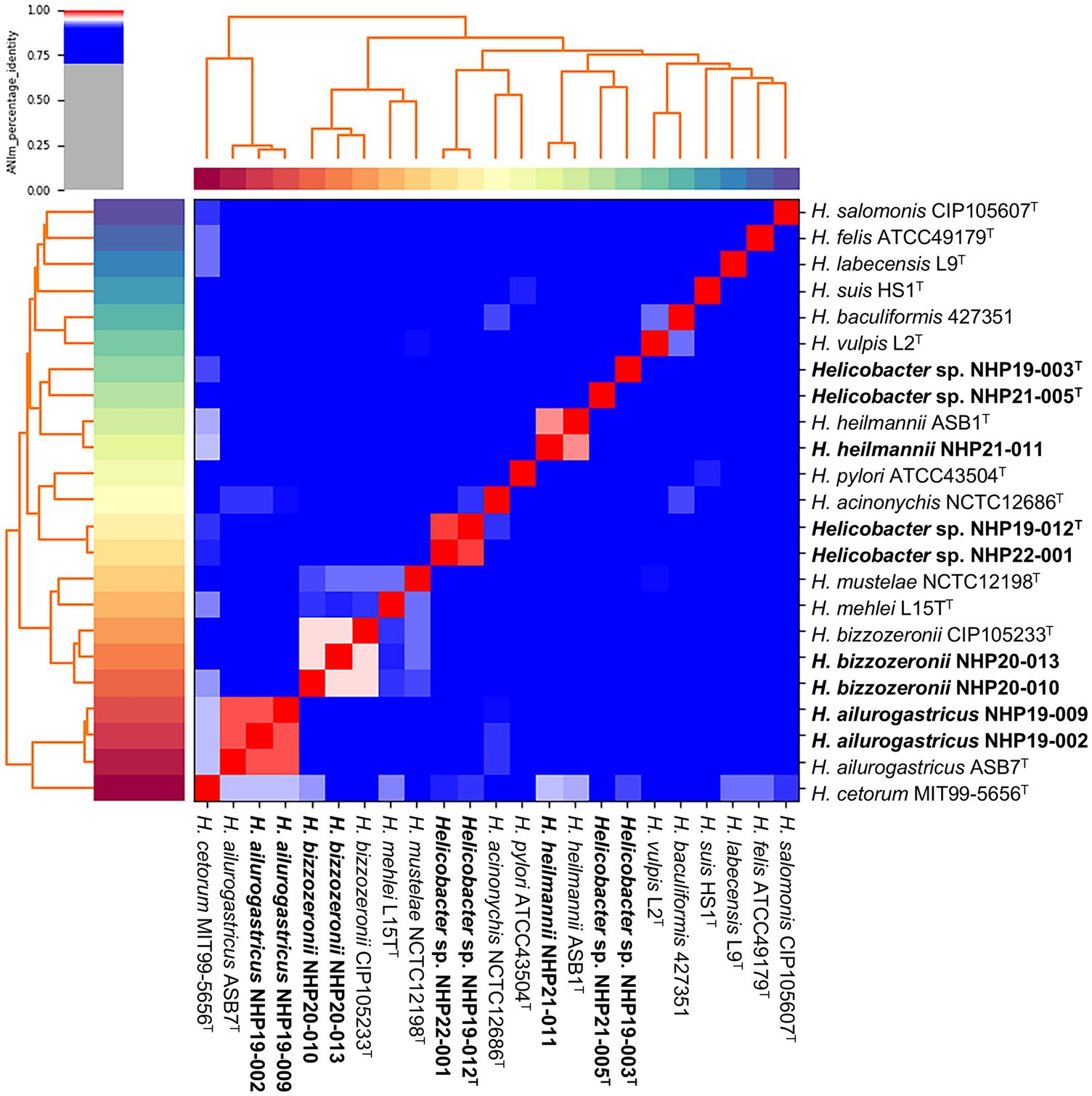
Figure 1. Average nucleotide identity (ANI) of the nine strains analyzed in this study compared to other Helicobacter species. A heatmap and hierarchical clustering are shown. Pairs of genomes with ANI > 95% can be considered as from the same species. The nine strains shown in bold are those isolated in this study.
Isolation source and electron microscopic images of novel Helicobacter speciesThree strains of novel Helicobacter species, NHP19-003T, NHP19–012T, and NHP21-005T, were isolated from a dog (a 6-year-old female, suffering from protein-losing enteropathy), a cat (a 14-year-old female, suffering from large-cell lymphoma), and another cat (a nine-year-old female, suffering from eosinophilic enteropathy), respectively. No specific endoscopic findings due to Helicobacter species infection were observed in all three cases (Figures 2A–C). The presence of spiral-shaped bacteria was confirmed by Giemsa staining of gastric biopsies, and all strains exhibited a corkscrew-like form in vitro (Figures 2D–F). Electron microscopy showed that NHP19-003T and NHP21-005T displayed spiral bacilli with a characteristic twisted structure with multiple flagella at both ends (Figures 2G,I), while NHP19-012T appeared rod-shaped without torsion after passaging (Figure 2H).
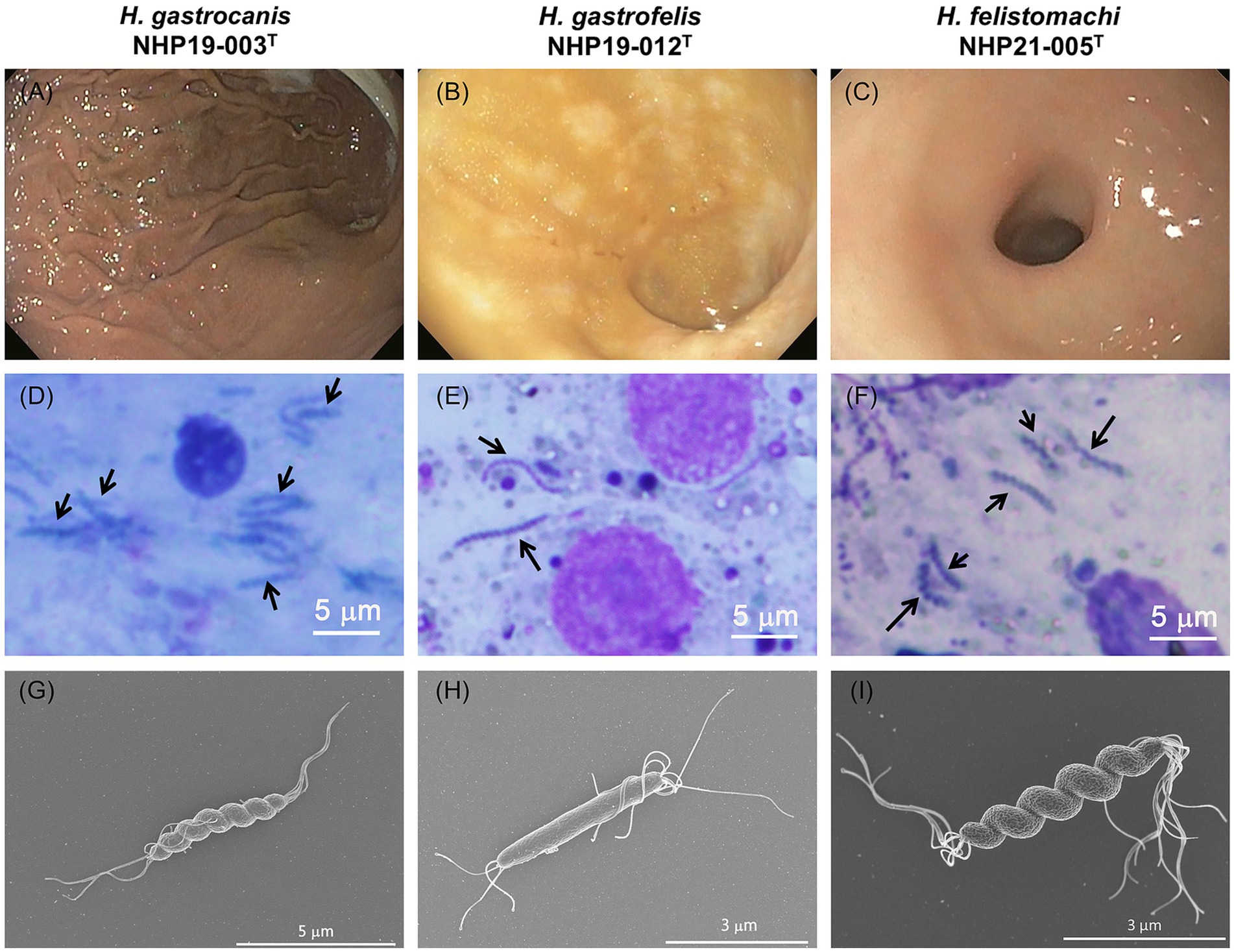
Figure 2. Endoscopic and histological images from NHP19-003T–infected dog, NHP19-012T–infected cat, and NHP21-005T–infected cat. Representative endoscopic images NHP19-003T–infected dog suffering from protein-losing enteropathy (A), NHP19-012T–infected cat suffering from large-cell lymphoma (B), and NHP21-005T–infected cat suffering from gastric lymphoma (C). Giemsa staining of gastric mucosa of infected dog (D) and infected cat (E,F). Giemsa staining showed the presence of corkscrew-like morphology of the bacteria (arrows). Scanning electron micrograph of strains NHP19-003T(G), NHP19-012T(H), and NHP21-005T(I). Electron microscopy showed the spiral bacilli with a characteristic twisted structure with multiple flagella at both ends.
Genomic characterization of novel Helicobacter speciesTable 2 displays the complete genomic information of NHP19-003T, NHP19-012T, NHP21-005T, and three relative gastric Helicobacters. The complete genome of NHP19-003T consists of one chromosome (Accession No. AP024814) and four plasmids (Accession No. AP024815, AP024816, AP024817 and AP024818), with a G + C content of 48.3%. The G + C content of NHP19-003T was found to be slightly higher than that of H. heilmannii (47.8%). The genome of NHP19–012T consists of one chromosome (Accession No. AP024819) and seven plasmids (Accession No. AP024820, AP024821, AP024822, AP024823, AP024824, AP024825 and AP024826, respectively), with a G + C content of 46.9%. The G + C content of NHP19-012T was found to be slightly lower than those of H. heilmannii, H. ailurogastricus (47.6%) and NHP19-003T. The complete genome of NHP21-005T, consists of one chromosome (1,737,144 bp, Accession No. AP028022) and eight plasmids (Accession No. AP028023, AP028024, AP028025, AP028026, AP028027, AP028028, AP028029, and AP028030), with a G + C content of 47.1%. The G + C content of NHP21-005T was found to be slightly lower than those of H. heilmannii, H. ailurogastricus and NHP19-003T.
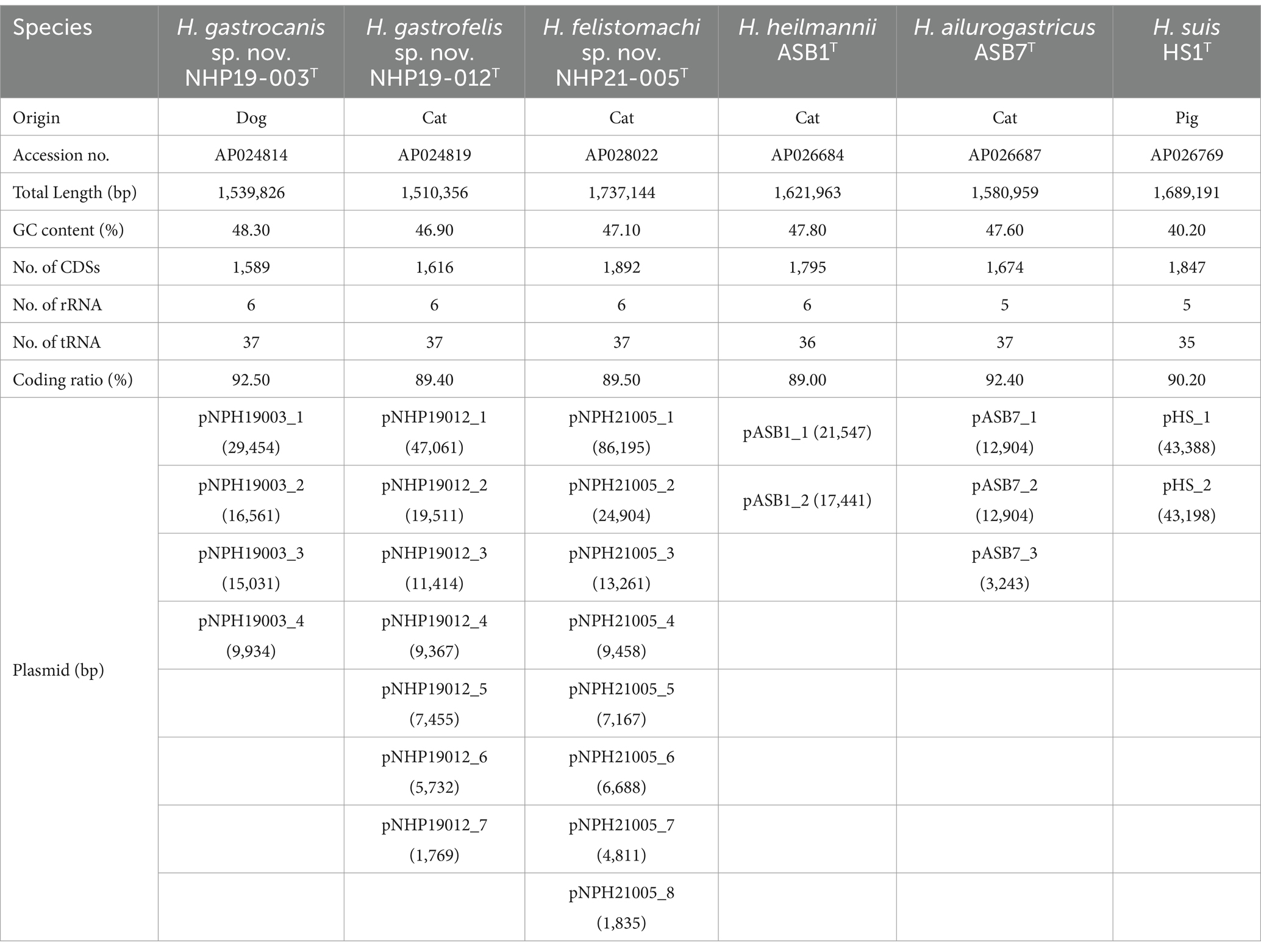
Table 2. Complete genome information of Helicobacter gastrocanis sp. nov. NHP19-003T, Helicobacter gastrofelis sp. nov. NHP19-012T, Helicobacter felistomachi sp. nov. NHP21-005T, and three related gastric Helicobacter species.
The chromosomes of NHP19-003T, NHP19-012T and NHP21-005T as circular genome maps (Figures 3A–C). NHP19-003T, NHP19-012T and NHP21-005T all exhibited urease activity in biochemical tests using API Campy, as shown later, and genomic analysis confirmed the presence of the urease gene complex containing ureA genes. In addition, genes related to motility (flagellin, FlaA), colonization in host cells (cholesterol-α-glucosyltransferase, CGT), and induction of apoptosis in host cells (gamma-glutamyltranspeptidase, GGT), which contribute to the pathogenesis in other gastric Helicobacter species (Leying et al., 1992; Shibayama et al., 2003; Hsu et al., 2021), are also present in the chromosomes of NHP19-003T, NHP19-012T and NHP21-005T. Vacuolating toxin A (VacA) and Cytotoxin-associated-gene A (CagA), which are virulence factors of H. pylori, were not found in NHP19-003T, NHP19-012T, and NHP21-005T, as in other gastric Helicobacter species. Meanwhile, the gene encoding a VacA-like protein was located in the genome of NHP19-003T, NHP19-012T and NHP21-005T. H. pylori has three VacA-like proteins other than VacA, which are ImaA, VlpA, and FaaA and H. suis also has a VacA-like protein HsvA. Phylogenetic comparison of the amino acid sequences of these VacA-like proteins in gastric Helicobacter spp. (Figure 4) revealed that the VacA-like proteins in NHP19-003T, NHP19-012T and NHP21-005T most closely resemble those in H. heilmannii ASB1T and H. ailurogastircus ASB7T. All VacA-like proteins lack the VacA domain shown in red in Figure 4, and the lengths of the VacA-like proteins differ from that of VacA, suggesting VacA-like proteins have functions different from those of VacA.
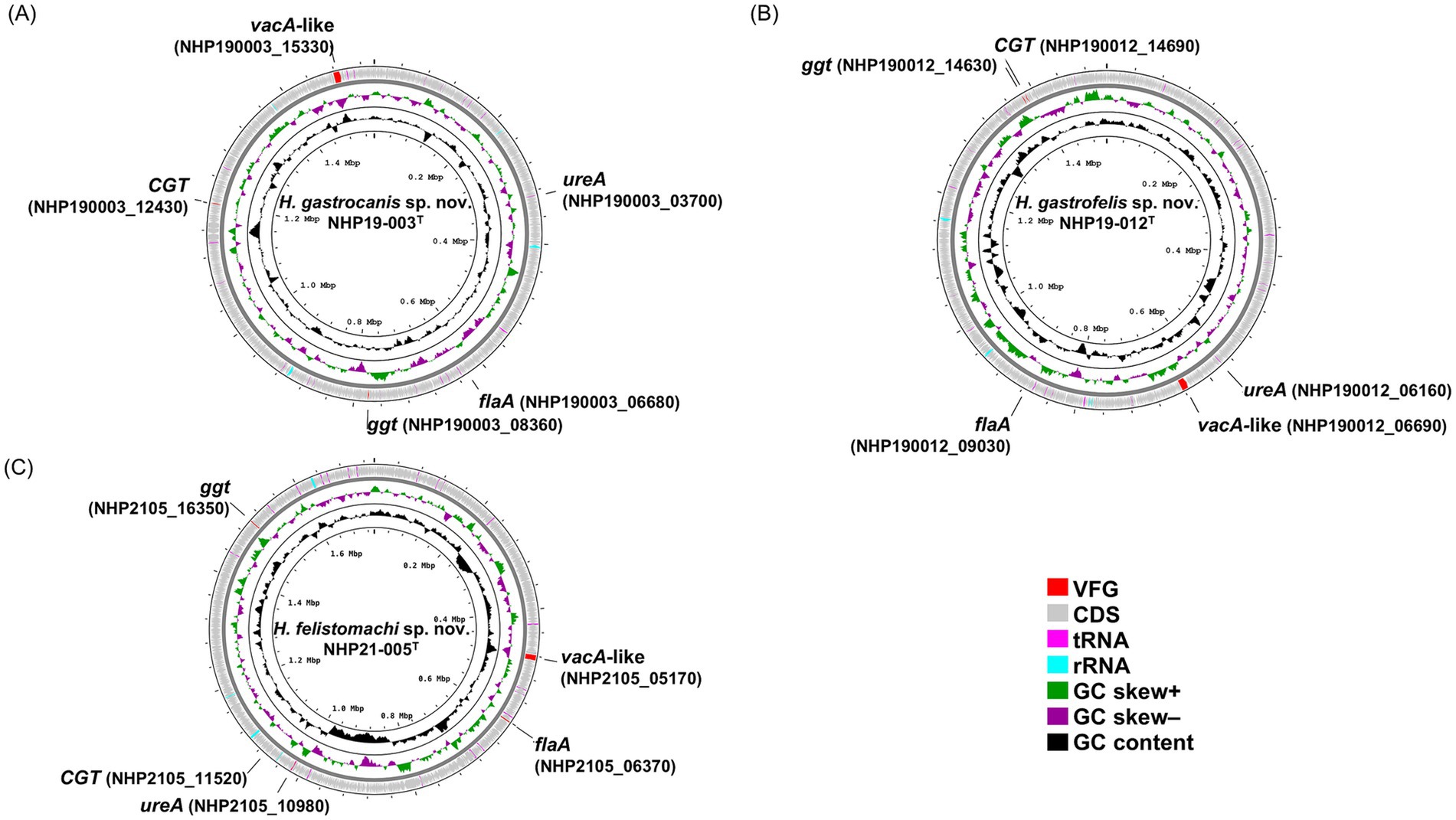
Figure 3. A circular genomic map of NHP19-003T, NHP19-012T, and NHP21-005T, comparing a circular graphical display of the distribution of genome information. The gene maps of (A) Helicobacter gastrocanis sp. nov. NHP19-003T; (B) Helicobacter gastrofelis sp. nov. NHP19-012T; and (C) Helicobacter felistomachi sp. nov. NHP21-005T. VFG, virulence factor gene; CDS, coding sequence.
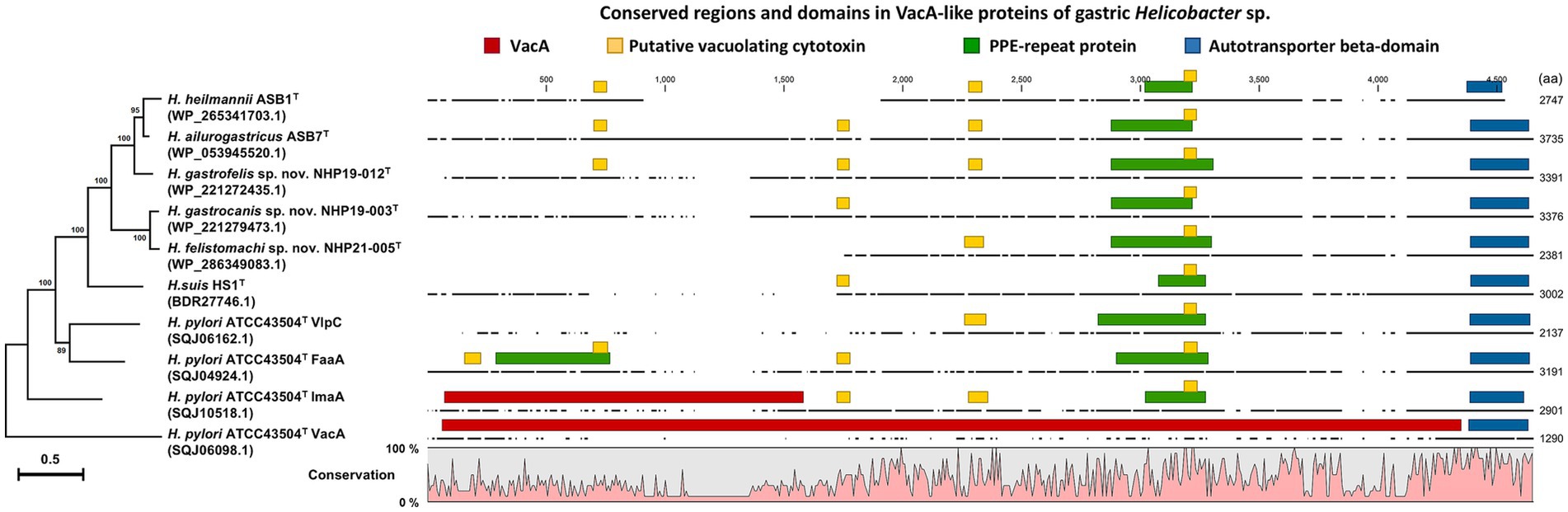
Figure 4. Phylogenetic tree, conserved regions, and domains in VacA of H. pylori ATCC43504T and VacA-like proteins of H. heilmannii ASB1T, H. ailurogastricus ASB7T, H. suis HS1T, NHP19-003T, NHP19-012T, NHP21-005T, and H. pylori ATCC43504T. The amino acid sequences of VacA and VacA-like proteins were aligned by MAFFT version 7.49, and phylogenetic tree was constructed using RAxML-NG version 1.1.0 with an LG + G4 model and 1,000 bootstrap replicates. Numbers indicate bootstrap percentages, and the scale bar indicates the number of base substitutions per site. Conserved domains were identified using the NCBI platform’s CD-search tool. The conserved regions and identified domains were visualized using CLC Genomics Workbench Version 22.0.2.
Phylogenetic characterizationThe phylogenetic tree was generated from the 16S rRNA sequences of NHP19-003T, NHP19-012T, NHP21-005T and 54 other Helicobacter sp. (Figure 5A). NHP19-003T, NHP19-012T and NHP21-005T were in the clade containing H. bizzozeronii, H. cynogastricus, H. felis, H. ailurogastricus, H. heilmannii, and H. salomonis, which are all gastric Helicobacters from cats and dogs. Among them, NHP19-003T, NHP19-012T, and NHP21-005T were most closely related to H. heilmannii ASB1T. The identity of the 16S rRNA sequences of NHP19-003T, NHP19-012T and NHP21-005T with H. heilmannii ASB1 from cats and dogs in the same clade were 98.8, 99.2 and 99.1%, respectively. This result suggested that phylogenetic analysis using 16S rRNA sequence failed to determine the taxonomic position of NHP19-003T, NHP19-012T, and NHP21-005T within the genus Helicobacter. By phylogenetic analysis using whole genomic sequences, we successfully identified 230 genes as the core genome among gastric Helicobacter species and Helicobacter cinaedi, defined as those genes present in over 99% of the species, with an identity higher than 60%. Phylogenetic analysis revealed that NHP19-003T, NHP19-012T, and NHP21-005T belonged to a clade containing H. heilmannii and H. ailurogastricus, both isolated from cat, and were clearly distinct from both species (Figure 5B).
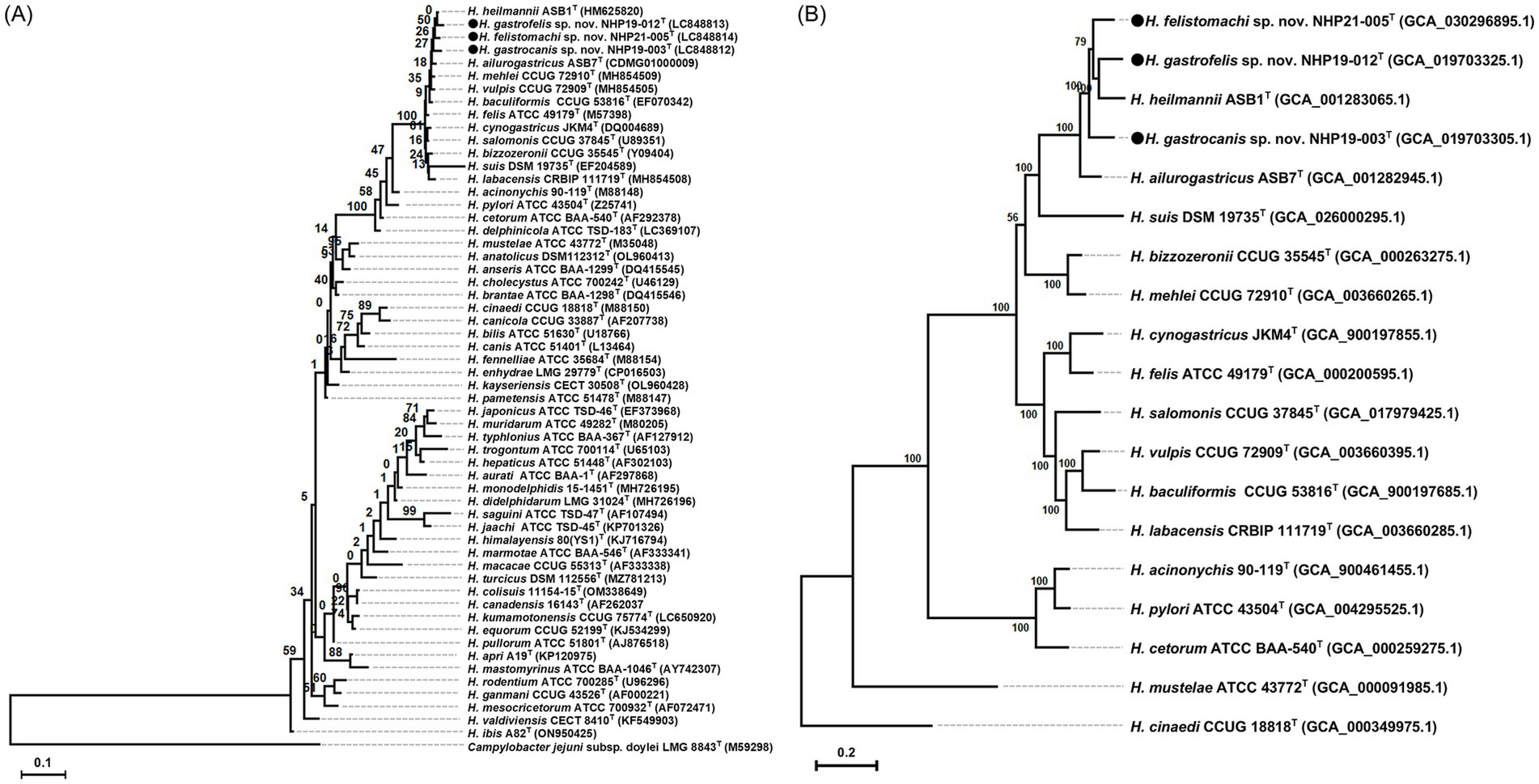
Figure 5. Phylogenetic trees showing the relationship of NHP19-003T, NHP19-012T and NHP21-005T to their closely related species. (A) Is created from 16S rRNA gene sequences of Helicobacter species including NHP19-003T, NHP19-012T, NHP21-005T and 54 Helicobacter species. Campylobacter jejuni LMG 8841T was added as an out group. The sequences were aligned by MAFFT version 7.49 and the phylogenetic tree was constructed using RAxML-NG version 1.1.0 with a GTR + G + I model and 1,000 bootstrap replicates. Numbers indicate bootstrap percentages, and the scale bar indicates the number of base substitutions per site. Circles indicated Helicobacter sp. strain NHP19-003T, NHP19-012T, and NHP21-005T, respectively. (B) Is generated from 230 core genes of gastric Helicobacter species. Core genes alignments were obtained from Roary and aligned sequences were used for the phylogenetic tree reconstruction using RAxML-NG version 1.1.0 with a GTR + G + I model and 1,000 bootstrap replicates. H. cinaedi was included as an out group. Numbers and the scale bar indicate bootstrap percentages and substitutions per nucleotide position, respectively. Circles indicate Helicobacter sp. strain NHP19-003T, NHP19-012T, and NHP21-005T, respectively.
Phenotypic characterizationNHP19-003T, NHP19-012T and NHP21-005T grew on NHPH agar plates (Rimbara et al., 2021). Tolerance to 1% bile, 1% glycine, and 1.5% NaCl was not observed in either NHP19-003T, NHP19-012T, and NHP21-005T. On the other hand, catalase- and oxidase-activities were positive. Growth of NHP19-003T, NHP19-012T and NHP21-005T occurred at 37°C, but not at 25 and 42°C under microaerobic conditions (Table 3). They also grew well in microaerophilic conditions and very weakly in anaerobic conditions, but did not grow under aerobic conditions.
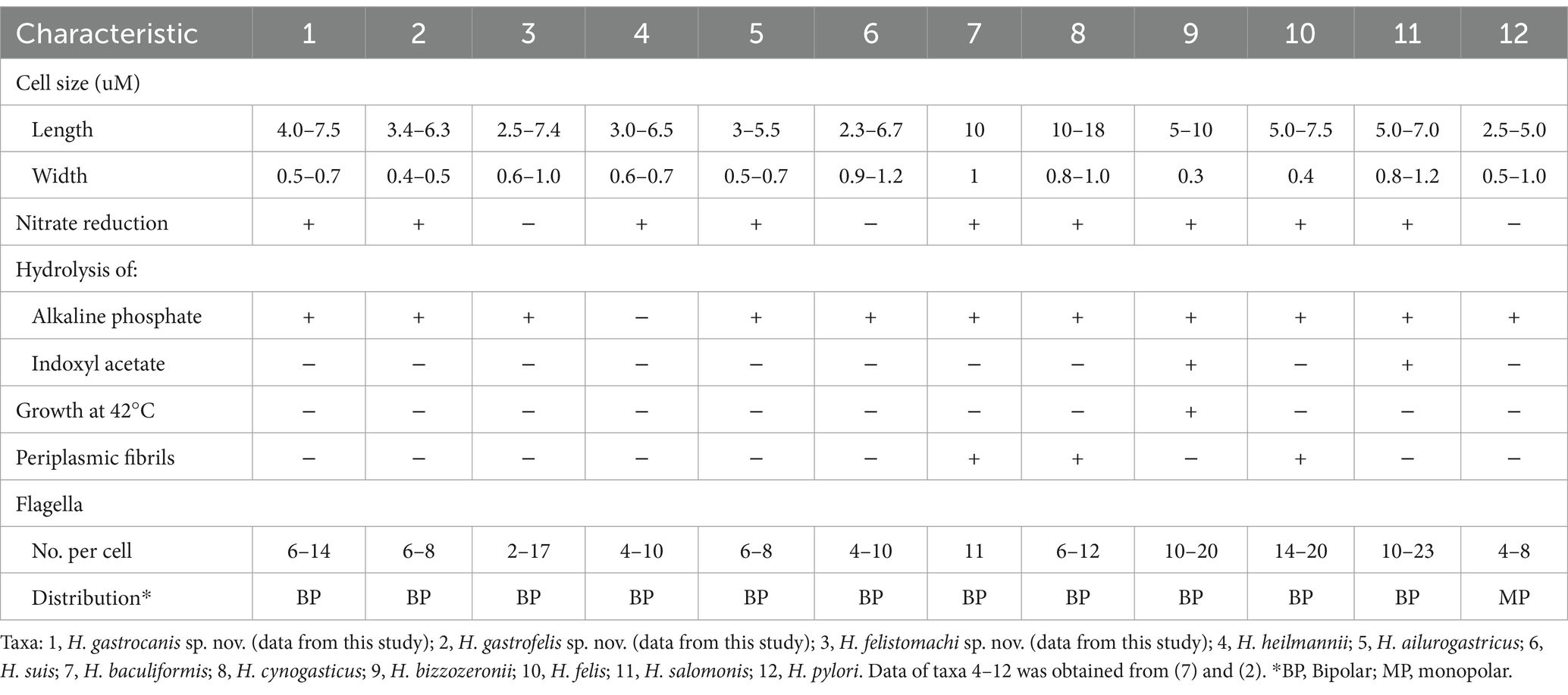
Table 3. Characteristics of Helicobacter gastrocanis sp. nov. NHP19-003T, Helicobacter gastrofelis sp. nov. NHP19-012T, Helicobacter felistomachi sp. nov. NHP21-005T, and related gastric Helicobacter species.
MALDI-TOF MS assaysProtein profiles of NHP19-003T, NHP19-012T and NHP21-005T generated from MALDI Biotyper (BrukerBiotyper, BrukerDaltonics) were compared with those of H. ailurogastricus ASB7T, H. suis HS1T and H. heilmannii ASB1T (Figure 6). The profiles of NHP19-003T, NHP19-012T and NHP21-005T differed from those of known type strains close to them. Compared with the type strain H. ailurogastricus, NHP19-003T had two specific peaks at 4,120 and 7,227 m/z. Compared with the type strain H. heilmannii, NHP19-012T had unique peaks at 3,266 and 7,816 m/z. Compared with the type strain H. heilmannii, NHP21-005T had unique peaks at 2,062 and 7,051 m/z. In addition, the peak at m/z approximately 13,000 pertaining to three other gastric Helicobacters, was not detected in NHP19-003T, NHP19-012T and NHP21-005T making them different from other related strains.
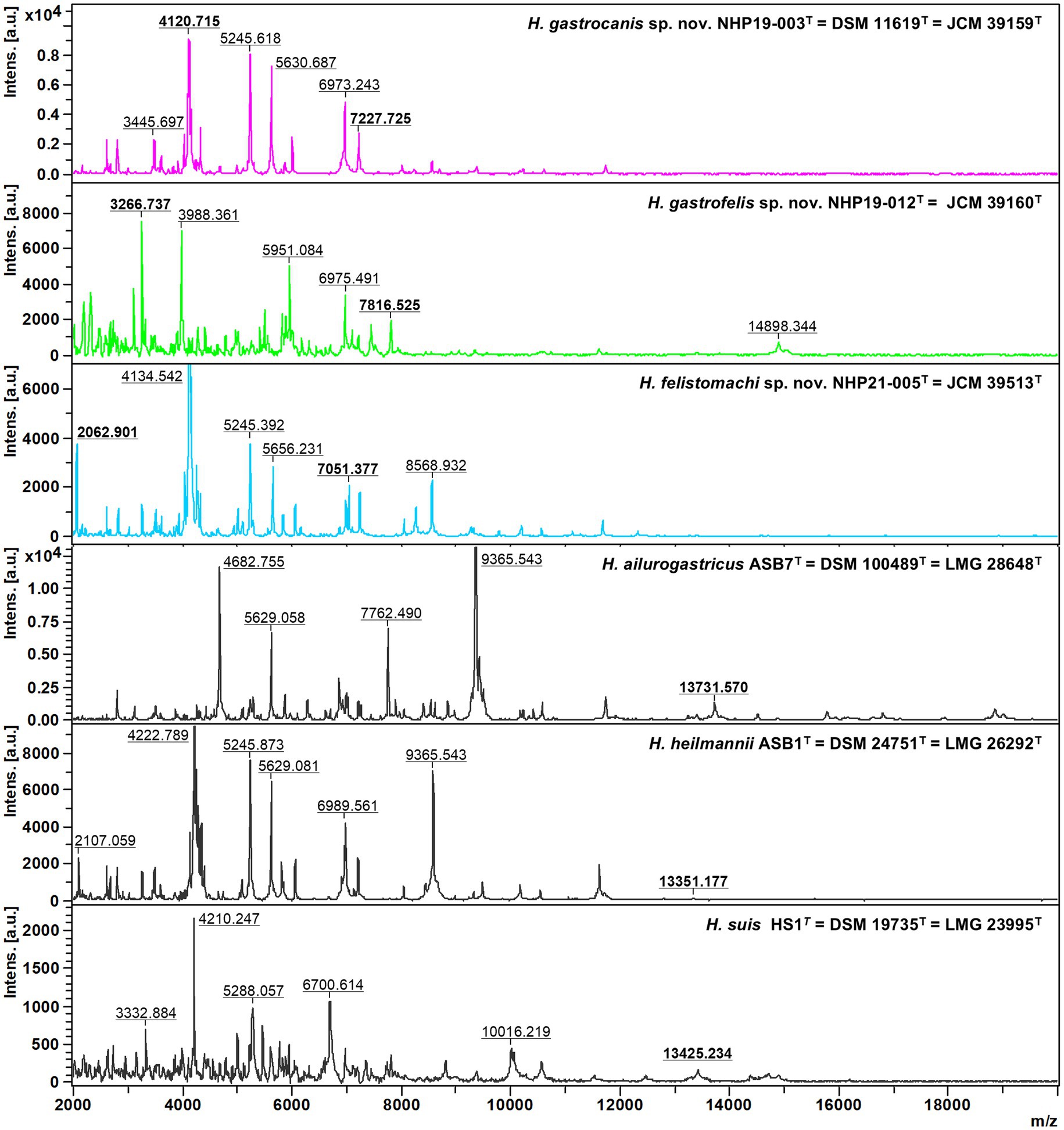
Figure 6. Comparative MALDI-TOF spectra profiles of type strains of novel species and related Helicobacter species strains obtained with flexAnalysis software. The baseline was subtracted. The y-axis shows the relative intensities of the ions, and the x-axis shows ion masses (Da). The peak at m/z approximately 13,000 pertaining to three other gastric Helicobacter species, H. ailurogastricus, H. heilmannii, and H. suis was not detected in NHP19-003T, NHP19-012T, and NHP21-005T.
Prevalence of gastric Helicobacter species in cats and dogs in JapanThe ureAB genes sequences obtained from gastric specimens of Helicobacter-species-infected dogs (n = 47) and cats (n = 24) in Japan analyzed in a previous study (Kubota-Aizawa et al., 2017a; Kubota-Aizawa et al., 2017b) were re-evaluated by phylogenetic analysis using the nine strains obtained in this study (Figure 7). The analysis included 90 sequences of gastric Helicobacter species for which genomic sequences were available from NCBI. The clades containing NHP19-003T, NHP19-012T, and NHP21-005T included 14 (19.7%), 24 (33.8%) and 10 (14.1%) of the 71 sequences from dogs and cats in Japan (Table 4). The clades of known Helicobacter sp., including H. heilmannii, H. ailurogastricus, H. bizzozeronii, H. felis, and H. pylori included one (1.4%), eight (11.3%), three (4.2%), five (7.0%), and one (1.4%) sequences from dogs and cats in Japan, respectively (Table 4).
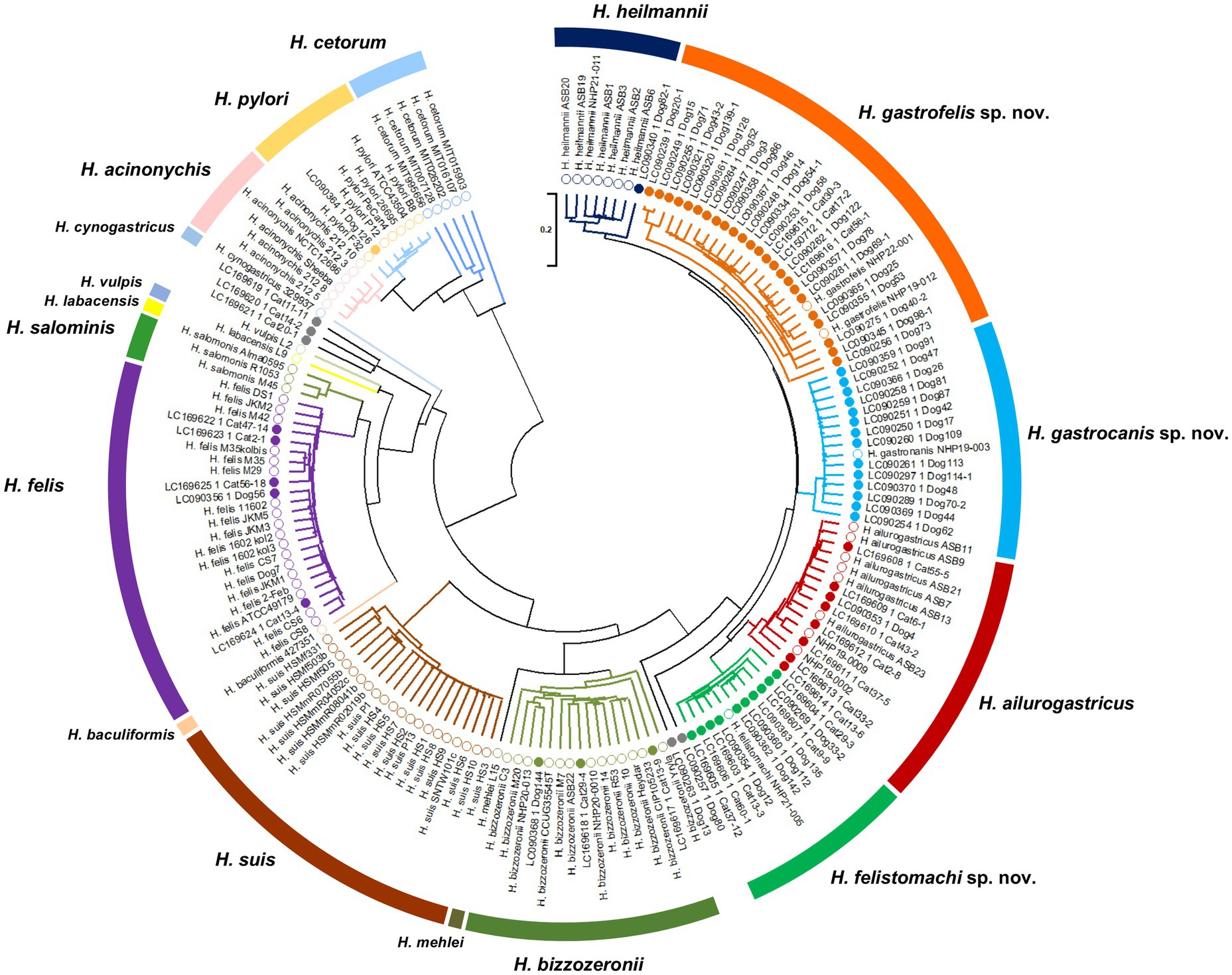
Figure 7. Phylogenetic tree generated from ureAB gene sequences of gastric Helicobacter species. The ureAB sequences shown as the GenBank no. (LCXXXXXX) from cats (n = 24) and dogs (n = 47) in Japan analyzed in previous studies (13, 14). The ureAB sequences obtained from nine strains analyzed in this study (NHP19-003T, NHP19-012T, NHP21-005T, NHP22-001, NHP19-002, NHP19-009, NHP20-010, NHP20-013, and NHP21-011), and 88 reference strains of gastric Helicobacter species strains whose genomic sequences were available from NCBI were added to the analysis. The accession numbers of the ureAB sequences used in the analysis are summarized in Supplementary Table S4. The sequences were aligned by MAFFT version 7.49 and the tree was constructed using RAxML-NG version 1.1.0 with a GTR + G + I model and 1,000 bootstrap replicates. Numbers indicate bootstrap percentages, and the scale bar indicates the number of base substitutions per site. Open circles are 88 reference strains and nine strains analyzed in this study, and closed circles are 71 samples obtained from cats and dogs in Japan in previous studies (13, 14). Light blue, orange and green open circles indicate Helicobacter sp. strain NHP19-003T, NHP19-012T, NHP22-001 and NHP21-005T, respectively. The ureAB sequences in light blue, orange, and green subtrees were predicted to belong to the same species as Helicobacter sp. strain NHP19-003T, NHP19-012T, and NHP21-005T, respectively.

Table 4. Prevalence of gastric Helicobacter species among dogs and cats in Japan.
Mice infectionMice infection was performed using H. ailurogastricus ASB7T, H. suis NHP19-4004, NHP19-003T, NHP19-012T and NHP21-005T. Bacterial counts infecting the stomach were compared between groups and found that bacterial counts of H. suis NHP19-4004-infected mice were significantly higher compared to ASB7T, NHP19-003T, NHP19-012T and NHP21-005T-infected groups (Figure 8A). Counts of CD3(+)-T cells were significantly higher in NHP21-005T-infected stomach compared to control, H. ailurogastricus ASB7T, and H. suis NHP19-4004 in fundic mucosa (Figures 8B, 9), and compared to control and H. ailurogastricus ASB7T in pyloric mucosa (Figures 8B, 10). Counts of CD3(+)-T cells of NHP19-003T and NHP19-012T tended to be higher than control, H. ailurogastricus ASB7T, and H. suis NHP19-4004-infected mucosa without statistical significance (Figure 8B). Almost no CD19(+)-B cells were found in control and infected gastric mucosa (Figures 8D, 10) except the forestomach-glandular border (Figure 11). In the forestomach-glandular border, lymphocyte aggregation was found in H. suis NHP19-4004, NHP19-003T, NHP19-012T and NHP21-005T-infected mice and number of CD3(+)-T cells and CD19(+)-B cells were significantly higher in NHP19-012T-infected mice compared to control and H. ailurogastricus ASB7T-infected mice (Figure 8C).
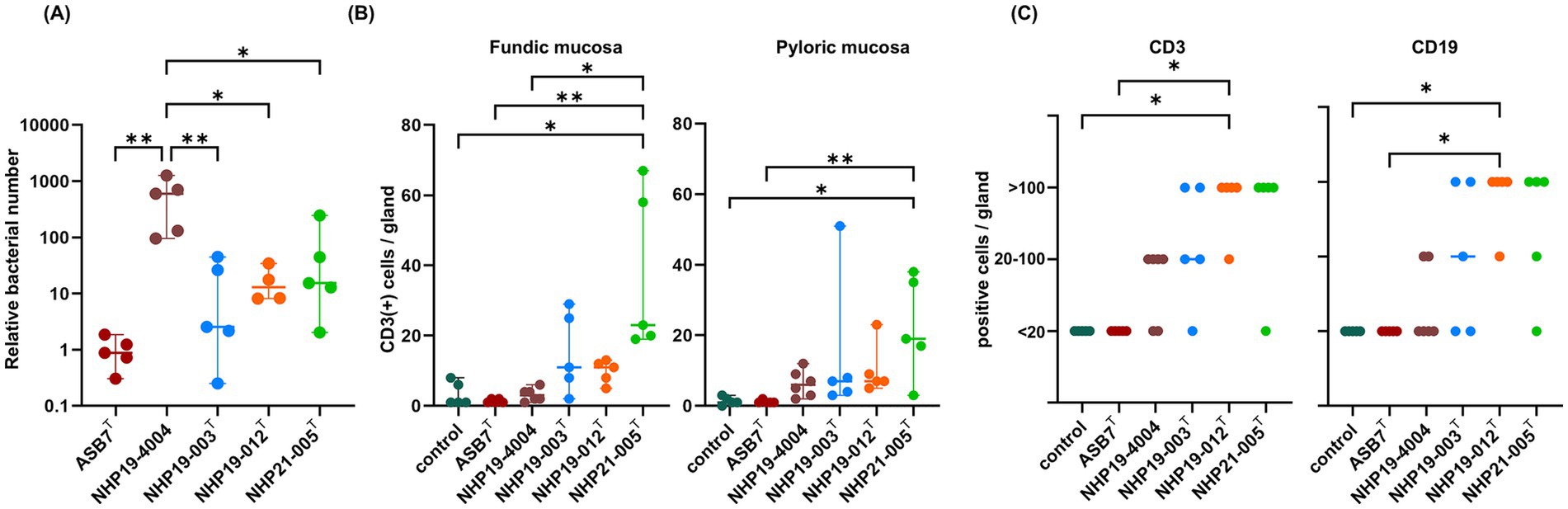
Figure 8. Effect of novel Helicobacter species infection in mouse stomachs. (A) Relative bacterial number of Helicobacter species in the mouse stomach. Bars indicate medians with 95% confidence interval. (B) Number of CD3(+) cells per glands in fundic and pyloric mucosa. (C) Number of CD3(+) cells and CD19(+) cells in the forestomach-glandular border. *P < 0.05, **P < 0.01.
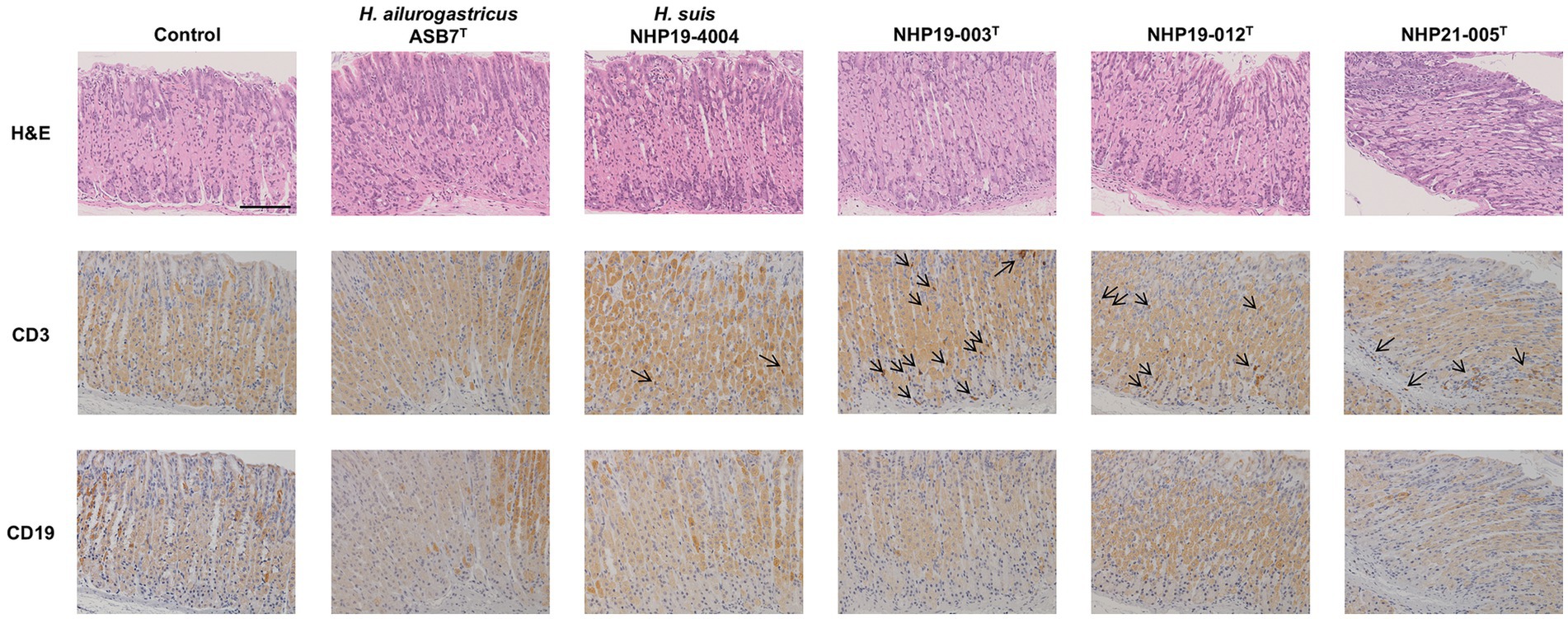
Figure 9. Effect of novel Helicobacter species infection in fundic mucosa of mouse stomachs. H&E, CD3, and CD19 staining in fundic mucosa of stomach sections of Helicobacter species-infected and control mice. CD3(+) lymphocytes (arrows) were observed in NHP19-003T, NHP19-012T, and NHP21-005T–infected mice, while most lymphocytes were mostly negative for CD19. Bar indicates 100 μm.
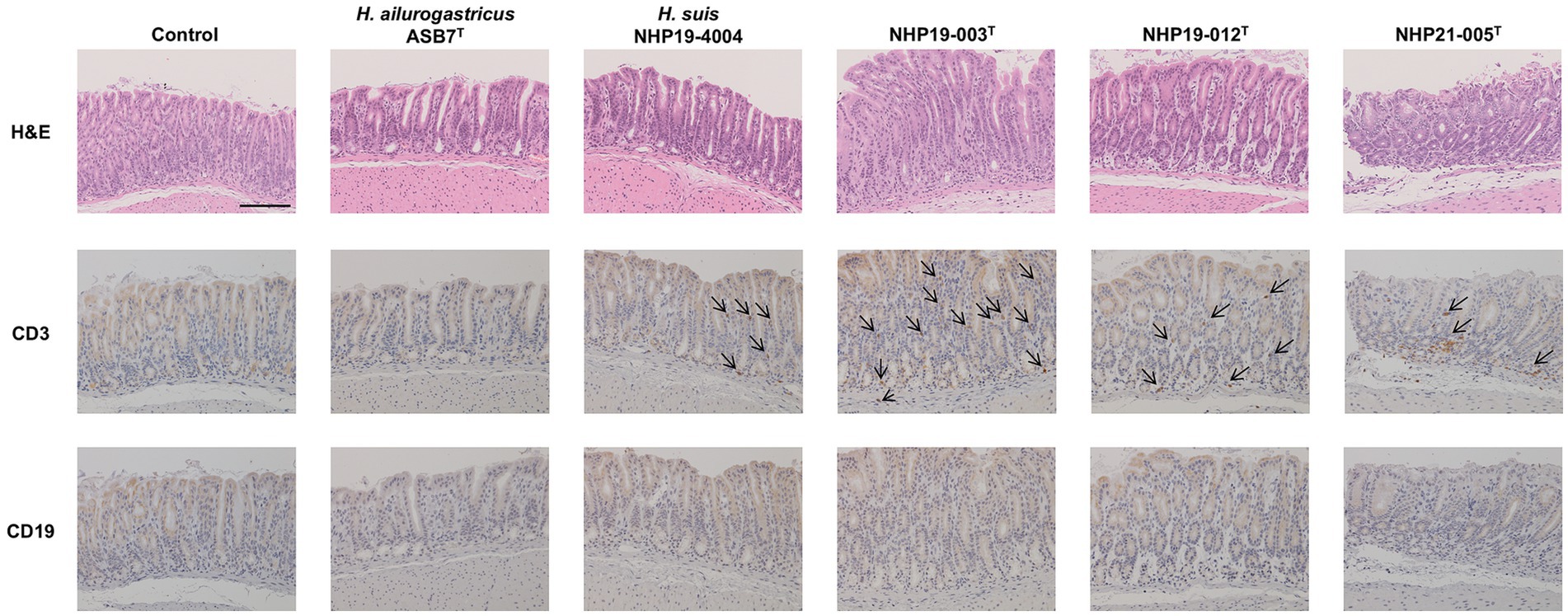
Figure 10. Effect of novel Helicobacter species infection in pyloric mucosa of mouse stomachs. H&E, CD3, and CD19 staining in pyloric (E) mucosa of stomach sections of Helicobacter species-infected and control mice. CD3(+) lymphocytes (arrows) were observed in NHP19-003T, NHP19-012T, and NHP21-005T–infected mice, while most lymphocytes were mostly negative for CD19. Bar indicates 100 μm.
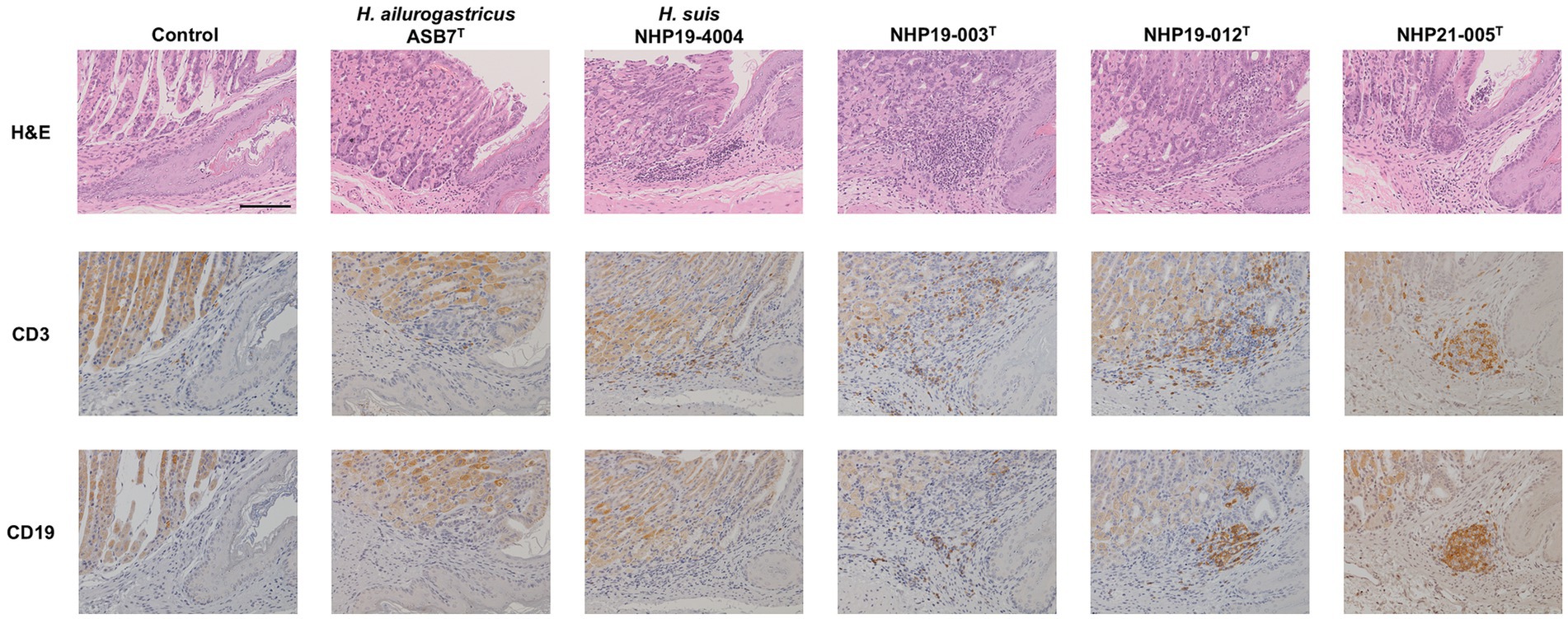
Figure 11. Effect of novel Helicobacter species infection in forestomach-glandular border of 689 mouse stomachs. H&E, CD3, and CD19 staining in the forestomach-glandular border of stomach sections from Helicobacter species-infected and control mice. Bar indicates 100 mm.
Discussion Genomic and pathogenetic characteristics of three novel Helicobacter speciesHelicobacter species are important causative agents of zoonotic diseases. In previous studies, it was suspected that dogs and cats might be infected with novel Helicobacter species. In this study, nine strains of Helicobacter spp. were isolated from gastric biopsy tissues of dogs and cats and identified through whole genome analysis. The nine strains include three known Helicobacter spp. H. ailurogastricus, H. heilmannii, and H. bizzozeronii. In addition, three strains NHP19-003T, NHP19-012T, and NHP21-005T were isolated in pure culture from the gastric biopsy of a dog and a cat, respectively, as novel Helicobacter species. Whole genome sequencing indicated that the ANI value was less than 95% identity between the genome of the three novel Helicobacter species and those of all other known species in the Helicobacter genus. The identity of 16S rRNA sequences between the genome of the three novel Helicobacter species and those of all other known species was more than 98.5%, consistent with previous reports showing that Helicobacter species cannot always be identified to species by 16S rRNA analysis alone (Dewhirst et al., 2005). Additionally, MALDI-TOF MS assay results showed that the peak at m/z approximately 13,000 pertaining to other gastric Helicobacter species was not detected in NHP19-003T, NHP19-012T and NHP21-005T. Based on these results, NHP19-003T, NHP19-012T and NHP21-005T are proposed as novel Helicobacter sp., named Helicobacter gastrocanis (NHP19-003T), Helicobacter gastrofelis (NHP19-012T) and Helicobacter felistomachi (NHP21-005T), respectively.
Whole genome sequencing revealed that H. gastrocanis sp. nov., H. gastrofelis sp. nov. and H. felistomachi sp. nov. strains had CGT, GGT, and VacA-like proteins, which have been reported as virulence factors in other Helicobacter spp. Among them, VacA-like protein, an autotransporter protein that transports the passenger domain of itself using the type V secretion system, is common to gastric Helicobacter species. H. pylori possess three autotransporter proteins other than VacA, ImaA, VlpC, and FaaA, which have been shown to contribute to colonization (Sause et al., 2012; Radin et al., 2013). H. heilmannii and H. ailurogastricus also have VacA-like proteins, the contribution of which to pathogenicity has been discussed (Baele et al., 2008). The VacA-like proteins of H. gastrocanis sp. nov., H. gastrofelis sp. nov. and H. felistomachi sp. nov. most resemble those of H. heilmannii and H. ailurogastricus and possess conserved domains among VacA-like proteins of ImaA, VlpC, and FaaA suggesting their contribution to colonization, although further analysis is needed to elucidate the functions of VacA-like proteins.
Three novel Helicobacter spp. were infected in mice. The results showed that inflammatory response in the stomach was induced after 1 month of infection. H. heilmannii is reported to cause mucous metaplasia in mice 9 weeks post-infection (Liu et al., 2014), while it is suggested in this study that H. heilmannii is not prevalent Helicobacter species in dogs and cats in Japan. On the other hand, H. ailurogastricus is prevalent in Japan; however, the pathogenicity of H. ailurogastricus has been reported to be less severe compared to H. heilmannii (Joosten et al., 2016). In this study, we compared the pathogenetic severity of three novel Helic
留言 (0)