Familial Mediterranean Fever (FMF) (MIM # 249100) is a hereditary autoinflammatory disorder predominantly affecting populations around the Mediterranean Sea including Arab and non-Arab countries. The disease is reportedly less common in other ethnic groups and populations, but it is becoming more prevalent worldwide due to global population movements (Lancieri et al., 2023).
FMF is mainly inherited in an autosomal recessive manner and caused by pathogenic variants in the Mediterranean Fever (MEFV) gene located on chromosome 16p13.3. This gene comprises 10 exons, encoding a 781-amino acid protein, named pyrin, that regulates inflammation. Mutations in MEFV lead to the loss of pyrin’s regulatory function, resulting in uncontrolled inflammation. Indeed, abnormal pyrin interacts with the inflammasome, a multiprotein complex responsible for activating pro-inflammatory cytokines, mainly interleukin-1β (IL-1β) and IL-18 (TUFAN and LACHMANN, 2020; Ibrahim et al., 2017; Ibrahim et al., 2018; Ibrahim et al., 2014). To date, around 400 MEFV variants have been identified (ClinVar, 2024; Infevers, 2024), with exon 10 and exon 2 being considered as hotspots for pathogenic variants (Bernot et al., 1998), and M694V being the most frequent mutation detected in FMF patients worldwide (Touitou, 2001).
FMF symptoms typically start before adulthood, with 90% of patients experiencing their first attack before the age of 20. The hallmark of FMF is recurrent, self-limiting febrile episodes accompanied by serositis. These attacks usually last between 1 and 3 days and manifest by symptoms like fever with chills (resolving within 24–72 h), abdominal pain, chest pain (pleural serositis), and joint pain (arthralgia or arthritis). Amyloidosis is the most severe complication of FMF, resulting from the deposition of serum amyloid A (SAA) protein in various organs, particularly in the kidneys. If not properly managed, it can lead to severe consequences including renal failure (Siligato et al., 2021). Interestingly, the wide variability in disease expression among individuals is primarily related to the allelic heterogeneity of MEFV, but is also influenced by other factors, including modifying genes and epigenetic events (Cazeneuve et al., 2000; Ibrahim et al., 2015; Chaaban et al., 2024a).
The diagnosis of FMF is primarily based on typical clinical findings in association with probable ethnicity, family history, and in some cases response to colchicine. Genetic testing is used to confirm the clinical diagnosis of FMF. It is very important for good clinical management, and to offer genetic counselling for FMF patients and their families, particularly in atypical cases or in populations with low disease prevalence (Manna and Rigante, 2019).
The gold standard treatment of FMF is the daily administration of colchicine, which significantly reduces the frequency and severity of attacks and prevents amyloidosis. This medication works by inhibiting microtubule polymerization, thereby reducing leukocyte chemotaxis and the inflammatory response. It is also thought to inhibit NF-κb signaling pathway and pyrin inflammasome assembly (Mezher et al., 2024). It is reported to be effective in 60%–75% of FMF patients, with partial response in an additional 10%–20%. The standard dosage varies from 0.5 to 2.0 mg/day, and it can be adjusted based on patient response and tolerance (Ozen et al., 2016; Ozturk et al., 2011). For patients unresponsive to colchicine or those experiencing severe side effects, alternative treatments include biologic agents targeting interleukin-1 (IL-1) such anakinra and canakinumab that are approved by the EMA (European Medicines Agency) and the FDA (Food and Drug Administration), respectively (Eroglu et al., 2015; Chaaban et al., 2024b).
In the Arab world, conducting FMF-related research is minimal, counting for only 3.80% of the total FMF-related publications worldwide (Masri et al., 2022). Even though Lebanon ranks first in FMF articles’ contribution among Arab countries (after normalizing to average population size, GDP, and number of physicians), there is still a lot of aspects to be addressed (Assouad et al., 2021). In Lebanon, the investigation of FMF started in 2005 with the analysis of specific MEFV gene variants (M694V, M680I, V726A, M694I, and E148Q) in a small cohort of patients (Medlej-Hashim et al., 2005; Sabbagh et al., 2008). This initial study identified M694V as the most prevalent variant, aligning with global patterns (Medlej-Hashim et al., 2005). Subsequent haplotype analysis across different Lebanese religious groups (Shiite, Sunni Muslims, and Christians) revealed that these variants originated from an ancient common ancestral population, where most MEFV mutations were already present with their associated haplotypes (Jalkh et al., 2008). Recently, a study done in Southern Lebanon examined 23 MEFV variants in 332 clinically diagnosed FMF patients. The findings supported the previous distribution of variant frequencies and highlighted the role of pseudo-dominant transmission of the disease in this region (El Roz et al., 2020). Despite providing valuable insights, these studies present limitations that are mainly related to the relatively sample size and the restricted number of tested MEFV variants, hence reducing the generalizability of the results. These limitations underscore the necessity to have a more comprehensive analysis of MEFV variants spectrum by looking at rare and newly identified variants and by analyzing those of uncertain significance (VUS) on a larger sample.
Accordingly, the current study is designed to analyze the molecular aspects of FMF and the spectrum of MEFV variants in the largest series of patients belonging to different geographic, ethnic, and religious groups across Lebanon. It also aims to explore the genotype-phenotype correlation of the most three common MEFV variants among affected individuals. The purpose is to establish a more accurate diagnosis of FMF, ultimately enabling improved management and treatment of the disease.
Materials and methodsStudy design and populationThis is a retrospective study on MEFV gene testing between 2006 and December 2023. A total of 3,167 clinically diagnosed FMF patients were referred, by either general physicians or specialists, to the Jacques Loiselet Center for Medical Genetics and Genomics (CGGM) at Saint Joseph University of Beirut (USJ) in Lebanon for MEFV sequencing. The major reported clinical symptoms were fever, abdominal pain, joint pain, thoracic pain and diarrhea. This study was conducted according to the Declaration of Helsinki and approved by the Ethical Committee at Saint Joseph University of Beirut and Hotel Dieu de France in Beirut, Lebanon (CEHDF-2315). An informed consent was obtained from all the participants prior to enrollment.
MEFV sequencingA blood sample was collected from all participants in EDTA tubes. DNA was extracted either by salting out method or using the QIAamp DNA® Kit (Qiagen). DNA quantity and quality were assessed by spectrophotometry using the Nanodrop ND-1000.
PCR reactions were performed using specific sets of primers (Forward and Reverse) for each exon. Exon 10 was sequenced first, followed by exons 2, 3, and 5 if needed. The remaining exons are tested upon recommendation to search for the second MEFV variant in heterozygous individuals. Then, PCR products were run on 1% agarose gel for verification. Afterwards, PCR products were purified and then sequenced using the Big Dye Terminator v1.1 Cycle sequencing kit (Applied Biosystems) under standard conditions. Sequencing reactions were purified using Sephadex G50 (Amersham Pharmacia Biotech) and loaded into the ABI 3130 or 3,500 Sequencer for capillary electrophoresis. Obtained electropherograms were analyzed and compared to the reference sequences using ChromasPro software (Technelysium).
ResultsAmong the 3,167 patients tested for MEFV variants, a total of 1,480 (46.73%) patients showed positive results, presenting at least one MEFV variant. Out of the 1,480 patients presenting variations in the MEFV gene, 48.38% were females and 51.62% were males, with an M/F sex ratio equal to 1.03 in the “heterozygous” group and 1.12 in patients carrying two variants (homozygous or compound heterozygous). In 64.88% of cases (n = 728), the age at testing was before 20 years old, with a mean age and a median age of 19 years and 12 years, respectively, in both groups (Figure 1). Of the 1,480 patients presenting MEFV variants, 628 patients (42.43%) presented two mutated alleles, with 177 (28,18%) being homozygous and 451 (71,82%) compound heterozygous (Table 1). The remaining 852 patients (57.57%) showed only one mutated allele and were classified as heterozygous. Interestingly, our findings revealed a significantly greater percentage of family history of FMF among patients carrying two MEFV variants (43.5%) compared to those with one variant (33.4%) (p = 0.0026).
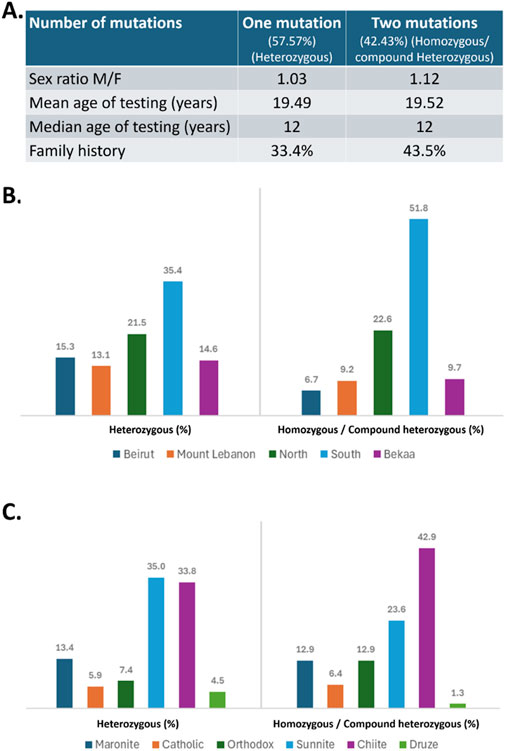
Figure 1. (A) Demographic characteristics of the studied population. (B) Geographic distribution (in %) of patients with one mutation (heterozygous) or two mutations (homozygous/compound heterozygous) across different regions of Lebanon. (C) Religious distribution (in %) in both mutation groups. Chi square analysis revealed a significant greater percentage of family history among patients carrying two mutations compared to those with one mutation (p = 0.0026).
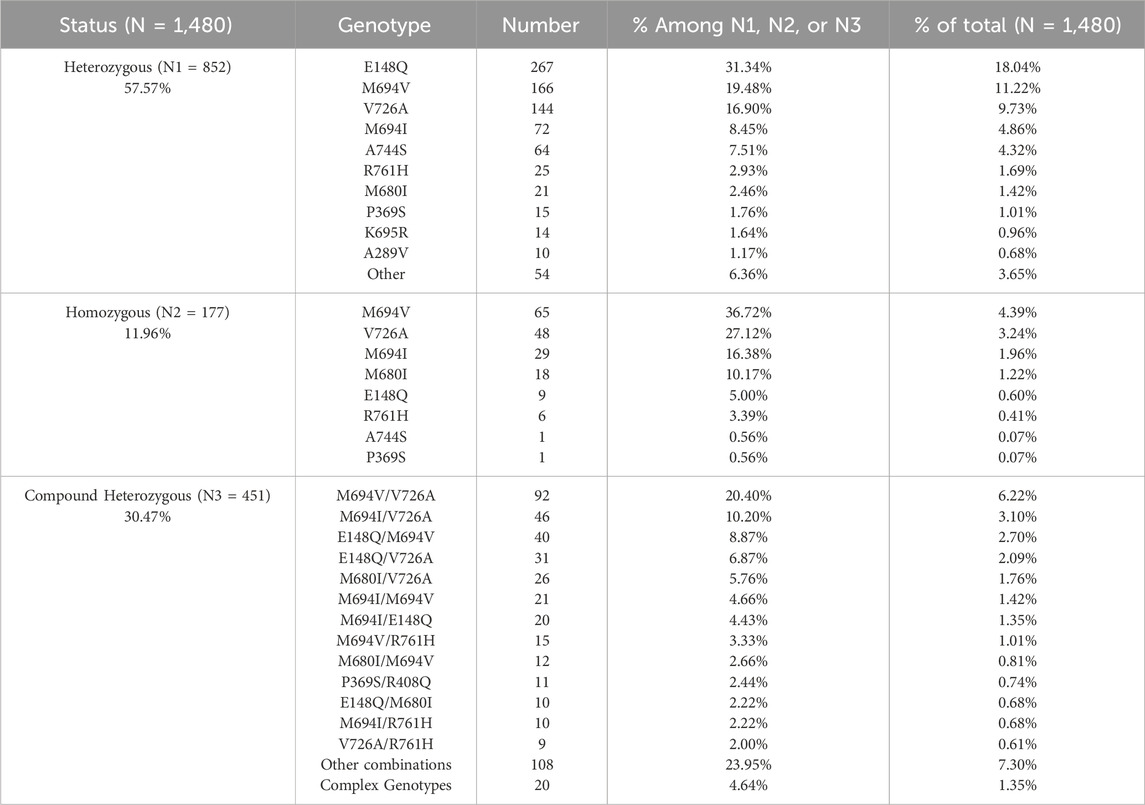
Table 1. Frequency of the identified MEFV variants among heterozygous, homozygous, and compound heterozygous patients.
On the other hand, most of the patients, heterozygous or homozygous/compound heterozygous, originated from South Lebanon and North Lebanon, with fewer cases from other regions including Beirut, Mount Lebanon, and Bekaa (Figure 1). A more detailed analysis of religious distribution revealed that the majority of the patients were Muslim Shiite or Sunni, while Druze patients were the minority (Figure 1).
In this study, we identified 48 different variations in the MEFV gene among 1,480 Lebanese patients. The distribution of the most prevalent variants and their allele frequencies are represented in Table 2. M694V and V726A were the most commonly detected variants in the studied population, accounting each for 28.98% of the cases, followed by E148Q (27.83%) and M694I (13.98%). M680I, A744S, R761H, and P369S accounted for 6.42%, 5.54%, 4.93%, and 3.04% respectively. On the other hand, M694V presented the highest allele frequency (16.57%), followed by V726A (15.99%), E148Q (14.22%), M694I (7.91%), M680I (3.79%), A744S (2.78%), R761H (2.65%), and finally P369S (1.54%). Interestingly, a significant number of patients (N = 178 or 12%) showed the presence of other variants that are considered rare or not very frequent.
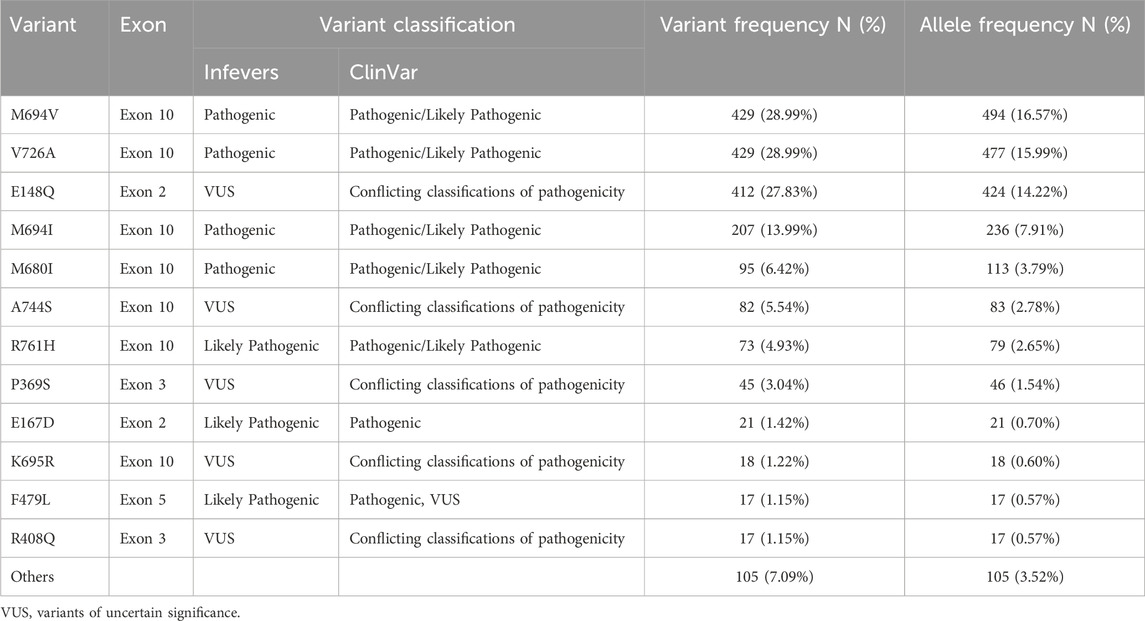
Table 2. Distribution of MEFV variants and allele frequencies among participants (N = 1,480).
Moreover, our results showed that E148Q was the most frequent variant in heterozygous patients accounting for 31.34% of cases (N = 267), while in homozygotes, the most detected variant was M694V (N = 65 or 36.72%). Furthermore, M694V/V726A was the most common genotype in compound heterozygous patients accounting for 20.40% (N = 92) of cases. The frequency of the variants among the total patients (N = 1,480) is represented in Table 2.
The most commonly reported symptoms among affected individuals were fever (78%), abdominal pain (88%), joint pain (65%), and thoracic pain (46%). Other clinical signs included diarrhea, constipation, vomiting, sensitivity to cold, and sensitivity to food. Amyloidosis was recorded in 5% of patients. Interestingly, the genotype-phenotype correlation analysis of the three most common variants revealed that patients carrying the M694V or V726A mutations, either in homozygous or compound heterozygous states, exhibited a more severe phenotype compared to those with the E148Q homozygous genotype (Figure 2). Indeed, a lower percentage of patients in the latter group reported experiencing the typical symptoms of FMF, such as fever, abdominal pain, and thoracic pain. More notably, none of the patients with the E148Q/E148Q genotype had amyloidosis.
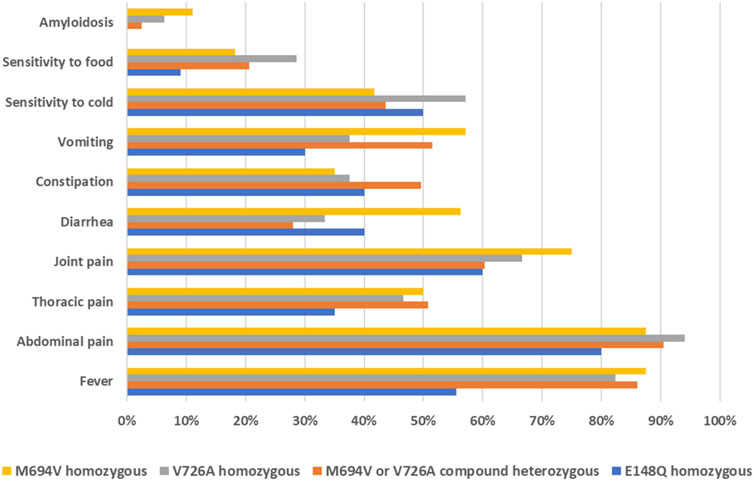
Figure 2. Distribution of FMF clinical manifestations based on the genotypes of affected individuals.
DiscussionThis study, the largest of its kind in Lebanon and the second in the Arab World, discusses the variant and allele frequencies of the MEFV gene among clinically diagnosed FMF patients. Our cohort of patients showed a high frequency of pathogenic/likely pathogenic variants in exons 10, 2, 3, and 5 of the MEFV gene. These findings endorse the standard approach utilized at our center for the molecular diagnosis of FMF, which involves initially sequencing exon 10, followed by exons 2, 3, and 5; the remaining exons are sequenced upon request if the results from the earlier exons are negative. Benign and likely benign variants were not reported or included in this study.
The findings of this study showed that two-third of the participants were tested under the age of 20 years, which is in line with previously published data (Sohar et al., 1967; Keskindemirci et al., 2014; Yasar Bilge et al., 2018). Moreover, the proportion of infants under 1 year of age was 2.77% (N = 41) which is comparable to that observed in the previous study performed in Southern Lebanon (2.45%) (El Roz et al., 2020). In fact, diagnosis in infants at such a young age is reported to be challenging, and this is due to their lack of verbal skills that are necessary for establishing an accurate diagnosis (Keskindemirci et al., 2014). On another hand, the overall male to female ratio in our study was 1.03 and 1.12, in heterozygous and genetically confirmed patients, respectively, indicating a comparable prevalence of FMF between the two sexes. Although data about the prevalence of FMF is still elusive, most of the reports indicate that FMF affects equally males and females (Mansour et al., 2019; Jalkh et al., 2008; Pradhan et al., 2017; Duruöz et al., 2021; Gumus, 2018). Interestingly, comparison of MEFV frequencies with respect to the different geographical regions and ethnicities revealed that the most affected patients were from South or North Lebanon and Muslims (Shiites and Sunni). These findings align with previous reports and are most likely attributed to the high rate of consanguinity among these particular communities. Indeed, previous research conducted by Barbour and Salameh in Lebanon indicated that non-Christian individuals and those from South or North Lebanon presented high consanguinity rates, potentially because of traditional practices and social norms prevalent in these communities (Barbour and Salameh, 2009).
Additionally, family history rates of FMF were higher in the group presenting two MEFV mutated alleles, underscoring the importance of genetic screening in families with a known history of FMF, in order to identify at-risk individuals and enable earlier intervention for a better disease management.
Considering the significant variation in the frequency distribution of MEFV variants among different populations, our purpose was to compare the prevalence of the seven most common MEFV variants identified in this study (M694V, V726A, E148Q, M694I, M680I, A744S, and R761H) with those observed in other populations across the Mediterranean Sea, encompassing both Arab and non-Arab countries (Table 3). Notably, our study reveals several differences when compared to previous research conducted in Lebanon, especially concerning the frequency of the V726A variant. In fact, all prior studies from Lebanon (Medlej-Hashim et al., 2005; Sabbagh et al., 2008; Jalkh et al., 2008; El Roz et al., 2020) identified M694V as the sole most frequent MEFV variant. However, our current study shows for the first time a high prevalence of the V726A in the Lebanese population, similar to that of M694V. Such difference might be attributed to the size of the studied population and the characteristics of the sample (geographic localization, age, sex, etc.). Indeed, the Lebanese population is known to be diverse in terms of ethnicity, religious groups, and regional distribution. Unlike previous studies that involved smaller sample size (Medlej-Hashim et al., 2005; Sabbagh et al., 2008; Jalkh et al., 2008) or focused on specific regions in Lebanon (El Roz et al., 2020), our research included patients from all over Lebanon and belonging to different religious groups.
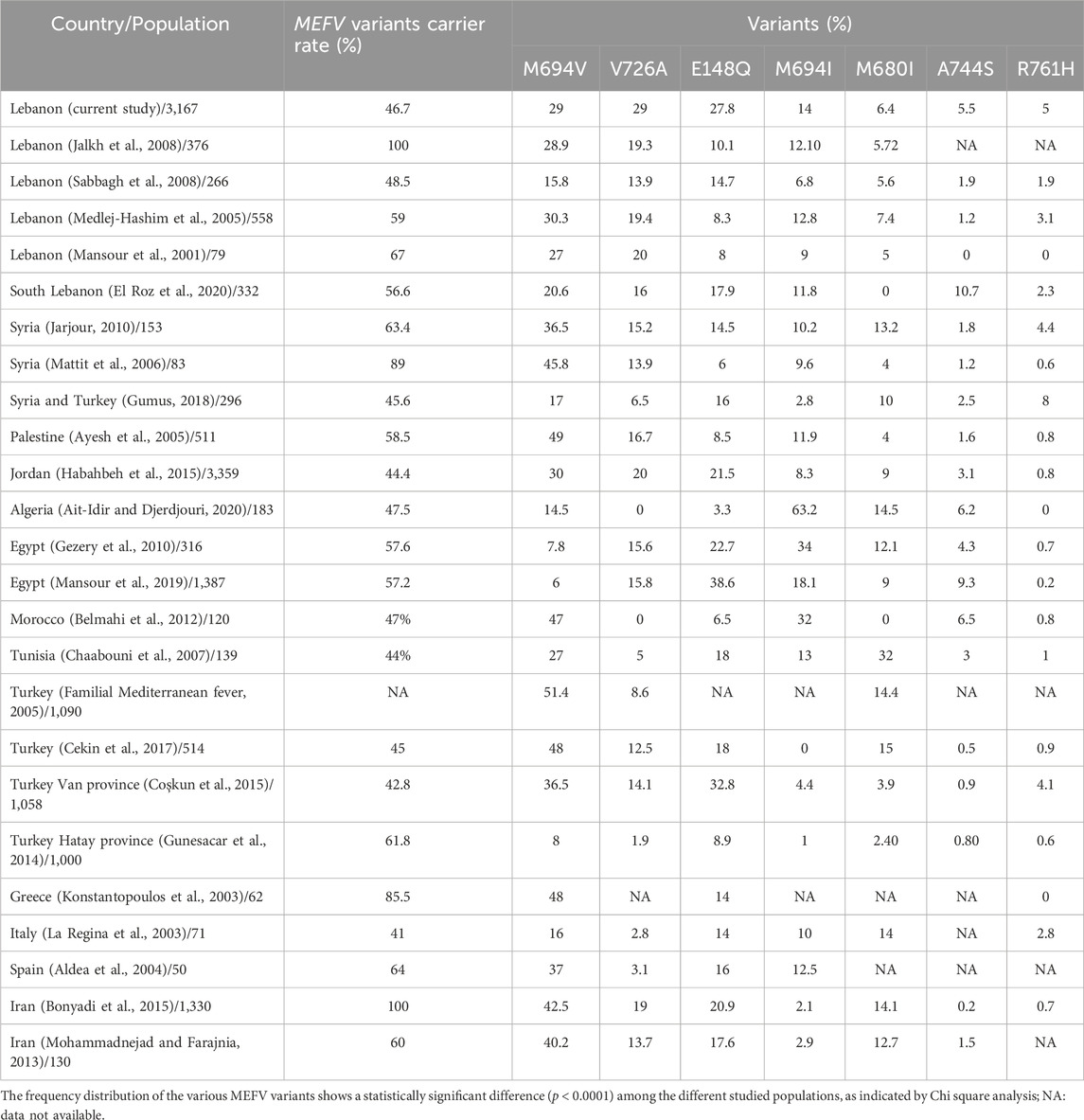
Table 3. Distribution and frequency of the seven most common MEFV variants among several populations, including the current study.
Research conducted in other Arab countries, such as Syria (Gumus, 2018; Jarjour, 2010; Mattit et al., 2006), Palestine (Ayesh et al., 2005), Jordan (Habahbeh et al., 2015), and Morocco (Belmahi et al., 2012) indicates that, similar to Lebanon, M694V is the most prevalent variant in these populations, while the frequency of other variants vary. In fact, it has been noted that Arabs exhibit a very diverse mutational pattern influenced by geographic origin (Touitou, 2001). For instance, oriental Arabs tend to have a higher prevalence of V726A compared to Arabs from North Africa; our study further supports this observation by recording elevated rates of V726A in Lebanon for the first time. Additionally, a 2001 study suggested that M694V and V726A likely originated in the middle East region, as their prevalence among Oriental Jews falls between that of Jews from North Africa and the Ashkenazi. On another hand, if we compare the results of Arab populations (Mansour et al., 2019; Belmahi et al., 2012; Gezery et al., 2010; Ait-Idir and Djerdjouri, 2020; Chaabouni et al., 2007) with those from our study and Arab Asian populations, we observe clear differences in the prevalence of the two variants M694I and E148Q. Indeed, M694I and E148Q are highly prevalent in Algeria (49.7%) (Ait-Idir and Djerdjouri, 2020) and Egypt (38.6%) (Mansour et al., 2019), respectively. Furthermore, M680I appears to be a particularly common frequent variant within the Tunisian population, with a notable allele frequency of 32%, which is not seen in any other Arab or non-Arab populations.
Finally, our findings align with those from studies conducted in non-Arab countries situated around or near the Mediterranean sea, including Greece (Konstantopoulos et al., 2003), Italy (La Regina et al., 2003), Spain (Aldea et al., 2004), Iran (Bonyadi et al., 2015; Mohammadnejad and Farajnia, 2013), and Turkey (Familial Mediterranean fever, 2005; Coşkun et al., 2015; Cekin et al., 2017), where M694V is also the most frequent variant, with frequencies sometimes surpassing 35%. Specifically in Turkey, where the prevalence of FMF is known to be the highest (Familial Mediterranean fever, 2005), literature has shown that the allele frequency of M694V can exceed 50% in certain regions. Nevertheless, it is important to note that in Hatay Province, located in the Mediterranean region of Turkey, R202Q emerged as the most common variant (21.35%) in a sample of 1,000 clinically diagnosed patients, followed by E148Q (8.85%), while M694V ranked third at 7.95%. These findings underscore that the differences in variants frequencies can be attributed to the specific sub-populations studied as well as the geographic and socio-demographic characteristics of the participants; a conclusion that is consistent with the diverse studies conducted in Lebanon.
The analysis of the distribution of clinical manifestations based on the genotypes of the affected individuals revealed a more severe phenotype among those carrying the M694V or V726A variants. This was evidenced by the higher frequency of typical FMF symptoms, namely, fever, abdominal pain, and thoracic pain, in these patients compared to individuals with the homozygous E148Q genotype. These findings corroborate earlier studies indicating that M694V has the highest penetrance and is associated with the most severe phenotype of the disease. On the contrary, the clinical significance of E148Q is contentious, with ongoing debates regarding whether it represents a pathogenic variant with low penetrance, or merely a benign polymorphism (El Roz et al., 2020). Our genotype-phenotype correlation findings underscore the vital role of molecular diagnosis in FMF. Beyond confirming the clinical diagnosis, molecular insights are crucial for understanding symptom onset and disease severity, thereby enabling more effective management and personalized care for patients.
In conclusion, this study represents the largest analysis of MEFV gene variants in a Lebanese population, providing valuable insights into the molecular aspects of FMF. For the first time, we report a high prevalence of the V726A variant in the Lebanese population, which differs from previous reports from Lebanon and other regional studies. V726A, along with M694V, was associated with the most severe phenotype of the disease, in contrast to E148Q. Our results enhance our understanding of the epidemiology, genetics, and clinical presentation of FMF in Lebanon and contribute to the broader global knowledge of this disease. Additionally, they emphasize the significant role of molecular diagnosis and genotype-phenotype correlation studies in improving the clinical management of FMF patients, and in providing genetic counseling for affected patients. Further studies exploring less common variants are needed to better understand the molecular spectrum of FMF in Lebanon.
Data availability statementThe datasets presented in this article are not readily available because of ethical constraints. Study participants did not provide consent for publicly sharing all the data. Requests to access the datasets should be directed to the corresponding author, Dr Alain Chebly, at YWxhaW4uY2hlYmx5QHVzai5lZHUubGI=.
Ethics statementThe studies involving humans were approved by Ethical committee at Hotel Dieu de France Hospital and Saint Joseph University of Beirut. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributionsRF: Data curation, Formal Analysis, Investigation, Software, Visualization, Writing–original draft. J-NI: Conceptualization, Data curation, Supervision, Visualization, Writing–original draft, Writing–review and editing. NS: Data curation, Formal Analysis, Investigation, Project administration, Software, Writing–original draft. RM: Data curation, Formal Analysis, Investigation, Software, Writing–original draft. GH: Data curation, Formal Analysis, Investigation, Project administration, Software, Writing–original draft. CA: Data curation, Formal Analysis, Investigation, Project administration, Writing–original draft. TY: Data curation, Formal Analysis, Investigation, Software, Writing–original draft. AC: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a grant from the research council at Saint Joseph University of Beirut (Grant no. FM451).
AcknowledgmentsWe would like to thank all the patients who participated in this study.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAldea, A., Calafell, F., Aróstegui, J. I., Lao, O., Rius, J., Plaza, S., et al. (2004). The west side story: MEFV haplotype in Spanish FMF patients and controls, and evidence of high LD and a recombination “hot-spot” at the MEFV locus. Hum. Mutat. 23 (4), 399. doi:10.1002/humu.9229
PubMed Abstract | CrossRef Full Text | Google Scholar
Assouad, E., El Hage, S., Safi, S., El Kareh, A., Mokled, E., and Salameh, P. (2021). Familial Mediterranean fever research activity in the Arab world: the need for regional and international collaborations. East Mediterr. Health J. Rev. Sante Mediterr. Orient Al-Majallah Al-Sihhiyah Li-Sharq al-mutawassit. 27 (10), 984–992. doi:10.26719/emhj.21.036
PubMed Abstract | CrossRef Full Text | Google Scholar
Ayesh, S. K., Nassar, S. M., Al-Sharef, W. A., Abu-Libdeh, B. Y., and Darwish, H. M. (2005). Genetic screening of familial Mediterranean fever mutations in the Palestinian population. Saudi Med. J. 26 (5), 732–737.
PubMed Abstract | Google Scholar
Belmahi, L., Cherkaoui, I. J., Hama, I., and Sefiani, A. (2012). MEFV mutations in Moroccan patients suffering from familial Mediterranean Fever. Rheumatol. Int. 32 (4), 981–984. doi:10.1007/s00296-010-1732-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Bernot, A., da Silva, C., Petit, J. L., Cruaud, C., Caloustian, C., Castet, V., et al. (1998). Non-Founder mutations in the MEFV gene establish this gene as the cause of familial mediterranean fever (FMF). Hum. Mol. Genet. 7 (8), 1317–1325. doi:10.1093/hmg/7.8.1317
PubMed Abstract | CrossRef Full Text | Google Scholar
Bonyadi, M. J., Gerami, S. M. N., Somi, M. H., and Dastgiri, S. (2015). MEFV mutations in Northwest of Iran: a cross sectional study. Iran. J. Basic Med. Sci. 18 (1), 53–57.
PubMed Abstract | Google Scholar
Cazeneuve, C., Ajrapetyan, H., Papin, S., Roudot-Thoraval, F., Geneviève, D., Mndjoyan, E., et al. (2000). Identification of MEFV-independent modifying genetic factors for familial Mediterranean fever. Am. J. Hum. Genet. 67 (5), 1136–1143. doi:10.1016/S0002-9297(07)62944-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Cekin, N., Akyurek, M. E., Pinarbasi, E., and Ozen, F. (2017). MEFV mutations and their relation to major clinical symptoms of Familial Mediterranean Fever. Gene 626, 9–13. doi:10.1016/j.gene.2017.05.013
PubMed Abstract | CrossRef Full Text | Google Scholar
Chaaban, A., Salman, Z., Karam, L., Kobeissy, P. H., and Ibrahim, J. N. (2024a). Updates on the role of epigenetics in familial mediterranean fever (FMF). Orphanet J. Rare Dis. 19 (1), 90. doi:10.1186/s13023-024-03098-w
PubMed Abstract | CrossRef Full Text | Google Scholar
Chaaban, A., Yassine, H., Hammoud, R., Kanaan, R., Karam, L., and Ibrahim, J. N. (2024b). A narrative review on the role of cytokines in the pathogenesis and treatment of familial Mediterranean fever: an emphasis on pediatric cases. Front. Pediatr. 12, 1421353. doi:10.3389/fped.2024.1421353
PubMed Abstract | CrossRef Full Text | Google Scholar
Chaabouni, H. B., Ksantini, M., M’rad, R., Kharrat, M., Chaabouni, M., Maazoul, F., et al. (2007). MEFV mutations in Tunisian patients suffering from familial mediterranean fever. Semin. Arthritis Rheum. 36 (6), 397–401. doi:10.1016/j.semarthrit.2006.12.004
PubMed Abstract | CrossRef Full Text | Google Scholar
Coşkun, S., Ustyol, L., Bayram, Y., Selçuk Bektaş, M., Gulsen, S., Çim, A., et al. (2015). The spectrum of MEFV gene mutations and genotypes in Van province, the eastern region of Turkey, and report of a novel mutation (R361T). Gene 562 (1), 128–131. doi:10.1016/j.gene.2015.02.059
PubMed Abstract | CrossRef Full Text | Google Scholar
Duruöz, M. T., Ozer, A., Gezer, H. H., Melikoglu, M. A., Hizmetli, S., Baklacioglu, H. S., et al. (2021). Pos1354 gender differences in clinical features and burden in patients with familial mediterranean fever: preliminary report. Ann. Rheum. Dis. 80 (Suppl. 1), 959.2–60. doi:10.1136/annrheumdis-2021-eular.2317
CrossRef Full Text | Google Scholar
El Roz, A., Ghssein, G., Khalaf, B., Fardoun, T., and Ibrahim, J. N. (2020). Spectrum of MEFV variants and genotypes among clinically diagnosed FMF patients from southern Lebanon. Med. Sci. 8 (3), 35. doi:10.3390/medsci8030035
PubMed Abstract | CrossRef Full Text | Google Scholar
Eroglu, F. K., Beşbaş, N., Topaloglu, R., and Ozen, S. (2015). Treatment of colchicine-resistant Familial Mediterranean fever in children and adolescents. Rheumatol. Int. 35 (10), 1733–1737. doi:10.1007/s00296-015-3293-2
PubMed Abstract | CrossRef Full Text | Google Scholar
Gezery, D. A. E., Abou-Zeid, A. A., Hashad, D. I., and El-Sayegh, H. K. (2010). MEFV gene mutations in Egyptian patients with familial mediterranean fever. Genet. Test. Mol. Biomarkers 14, 263–268. doi:10.1089/gtmb.2009.0180
PubMed Abstract | CrossRef Full Text | Google Scholar
Gumus, E. (2018). The frequency of MEFV gene mutations and genotypes in sanliurfa province, south-eastern region of Turkey, after the Syrian civil war by using next generation sequencing and report of a novel exon 4 mutation (I423T). J. Clin. Med. 7 (5), 105. doi:10.3390/jcm7050105
PubMed Abstract | CrossRef Full Text | Google Scholar
Gunesacar, R., Celik, M. M., Arica, V., Elmacioglu, S., and Ozturk, O. H. (2014). Frequency of MEFV gene mutations in Hatay province, Mediterranean region of Turkey and report of a novel missense mutation (I247V). Gene 546 (2), 195–199. doi:10.1016/j.gene.2014.06.019
PubMed Abstract | CrossRef Full Text | Google Scholar
Habahbeh, L. A., al, H. M., Zaben, S. F. A., Al-Momani, A., Khasawneh, R., Mallouh, M. abu, et al. (2015). Genetic profile of patients with familial mediterranean fever (FMF): single center experience at king hussein medical center (KHMC). Med. Arch. 69 (6), 417–420. doi:10.5455/medarh.2015.69.417-420
PubMed Abstract | CrossRef Full Text | Google Scholar
Ibrahim, J. N., Chouery, E., Lecron, J. C., Mégarbané, A., and Medlej-Hashim, M. (2015). Study of the association of IL-1β and IL-1RA gene polymorphisms with occurrence and severity of Familial Mediterranean fever. Eur. J. Med. Genet. 58 (12), 668–673. doi:10.1016/j.ejmg.2015.11.007
PubMed Abstract | CrossRef Full Text | Google Scholar
Ibrahim, J. N., Jéru, I., Lecron, J. C., and Medlej-Hashim, M. (2017). Cytokine signatures in hereditary fever syndromes (HFS). Cytokine Growth Factor Rev. 33, 19–34. doi:10.1016/j.cytogfr.2016.11.001
PubMed Abstract | CrossRef Full Text | Google Scholar
Ibrahim, J. N., Jounblat, R., Delwail, A., Abou-Ghoch, J., Salem, N., Chouery, E., et al. (2014). Ex vivo PBMC cytokine profile in familial Mediterranean fever patients: involvement of IL-1β, IL-1α and Th17-associated cytokines and decrease of Th1 and Th2 cytokines. Cytokine 69 (2), 248–254. doi:10.1016/j.cyto.2014.06.012
PubMed Abstract | CrossRef Full Text | Google Scholar
Ibrahim, J. N., Jounblat, R., Jalkh, N., Abou Ghoch, J., Al Hageh, C., Chouery, E., et al. (2018). RAC1 expression and role in IL-1β production and oxidative stress generation in familial Mediterranean fever (FMF) patients. Eur. Cytokine Netw. 29 (4), 127–135. doi:10.1684/ecn.2018.0416
PubMed Abstract | CrossRef Full Text | Google Scholar
Jalkh, N., Génin, E., Chouery, E., Delague, V., Medlej-Hashim, M., Idrac, C. A., et al. (2008). Familial Mediterranean Fever in Lebanon: founder effects for different MEFV mutations. Ann. Hum. Genet. 72 (Pt 1), 41–47. doi:10.1111/j.1469-1809.2007.00386.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Jarjour, R. A. (2010). Familial Mediterranean fever in Syrian patients: MEFV gene mutations and genotype–phenotype correlation. Mol. Biol. Rep. 37 (1), 1–5. doi:10.1007/s11033-009-9475-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Keskindemirci, G., Aktay Ayaz, N., Aldemir, E., Aydoğmuş, Ç., Aydoğan, G., and Kavuncuoğlu, S. (2014). Familial mediterranean fever: diagnosing as early as 3 Months of age. Case Rep. Pediatr. 2014 (1), 296479. doi:10.1155/2014/296479
PubMed Abstract | CrossRef Full Text | Google Scholar
Konstantopoulos, K., Kanta, A., Deltas, C., Atamian, V., Mavrogianni, D., Tzioufas, A., et al. (2003). Familial Mediterranean fever associated pyrin mutations in Greece. Ann. Rheum. Dis. 62 (5), 479–481. doi:10.1136/ard.62.5.479
PubMed Abstract | CrossRef Full Text | Google Scholar
Lancieri, M., Bustaffa, M., Palmeri, S., Prigione, I., Penco, F., Papa, R., et al. (2023). An update on familial mediterranean fever. Int. J. Mol. Sci. 24 (11), 9584. doi:10.3390/ijms24119584
PubMed Abstract | CrossRef Full Text | Google Scholar
La Regina, M., Nucera, G., Diaco, M., Procopio, A., Gasbarrini, G., Notarnicola, C., et al. (2003). Familial Mediterranean fever is no longer a rare disease in Italy. Eur. J. Hum. Genet. EJHG 11 (1), 50–56. doi:10.1038/sj.ejhg.5200916
PubMed Abstract | CrossRef Full Text | Google Scholar
Manna, R., and Rigante, D. (2019). Familial mediterranean fever: assessing the overall clinical impact and formulating treatment plans. Mediterr. J. Hematol. Infect. Dis. 11 (1), e2019027. doi:10.4084/MJHID.2019.027
PubMed Abstract | CrossRef Full Text | Google Scholar
Mansour, A. R., El-Shayeb, A., El Habachi, N., Khodair, M. A., Elwazzan, D., Abdeen, N., et al. (2019). Molecular patterns of MEFV gene mutations in Egyptian patients with familial mediterranean fever: a retrospective cohort study. Int. J. Inflamm. 2019, 2578760. doi:10.1155/2019/2578760
PubMed Abstract | CrossRef Full Text | Google Scholar
Mansour, I., Delague, V., Cazeneuve, C., Dodé, C., Chouery, E., Pêcheux, C., et al. (2001). Familial Mediterranean fever in Lebanon: mutation spectrum, evidence for cases in Maronites, Greek orthodoxes, Greek catholics, Syriacs and Chiites and for an association between amyloidosis and M694V and M694I mutations. Eur. J. Hum. Genet. 9 (1), 51–55. doi:10.1038/sj.ejhg.5200574
PubMed Abstract | CrossRef Full Text | Google Scholar
Masri, D. E., Alsaayed, B., Masri, J. E., Zreika, B., Chanbour, H., and Salameh, P. (2022). Contribution of Arab countries to familial mediterranean fever research: a PubMed-based bibliometric analysis. Rheumatol. Int. 42 (1), 95–100. doi:10.1007/s00296-021-04852-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Mattit, H., Joma, M., Al-Cheikh, S., El-Khateeb, M., Medlej-Hashim, M., Salem, N., et al. (2006). Familial Mediterranean fever in the Syrian population: gene mutation frequencies, carrier rates and phenotype–genotype correlation. Eur. J. Med. Genet. 49 (6), 481–486. doi:10.1016/j.ejmg.2006.03.002
PubMed Abstract | CrossRef Full Text | Google Scholar
Medlej-Hashim, M., Serre, J. L., Corbani, S., Saab, O., Jalkh, N., Delague, V., et al. (2005). Familial Mediterranean fever (FMF) in Lebanon and Jordan: a population genetics study and report of three novel mutations. Eur. J. Med. Genet. 48 (4), 412–420. doi:10.1016/j.ejmg.2005.05.010
PubMed Abstract | CrossRef Full Text | Google Scholar
Mezher, N., Mroweh, O., Karam, L., Ibrahim, J. N., and Kobeissy, P. H. (2024). Experimental models in familial mediterranean fever (FMF): insights into pathophysiology and therapeutic strategies. Exp. Mol. Pathol. 135, 104883. doi:10.1016/j.yexmp.2024.104883
PubMed Abstract | CrossRef Full Text | Google Scholar
Mohammadnejad, L., and Farajnia, S. (2013). Mediterranean Fever gene analysis in the azeri turk population with familial mediterranean Fever: evidence for new mutations associated with disease. Cell J. 15 (2), 152–159.
PubMed Abstract | Google Scholar
Ozen, S., Demirkaya, E., Erer, B., Livneh, A., Ben-Chetrit, E., Giancane, G., et al. (2016). EULAR recommendations for the management of familial Mediterranean fever. Ann. Rheum. Dis. 75 (4), 644–651. doi:10.1136/annrheumdis-2015-208690
PubMed Abstract | CrossRef Full Text | Google Scholar
Ozturk, MA, Kanbay, M, Kasapoglu, B, et al. Therapeutic approach to familial Mediterranean fever: a review update [published correction appears in Clin Exp Rheumatol. 2012 Jan-Feb;30(1):146]. Clin Exp Rheumatol. 2011;29(4 Suppl 67):S77-S86.
Pradhan, R., Kumar, R., Shekhar, S., Rai, N., Ambashtha, A., Banerjee, J., et al. (2017). Longevity and healthy ageing genes FOXO3A and SIRT3: serum protein marker and new road map to burst oxidative stress by Withania somnifera. Exp. Gerontol. 95, 9–15. doi:10.1016/j.exger.2017.05.013
留言 (0)