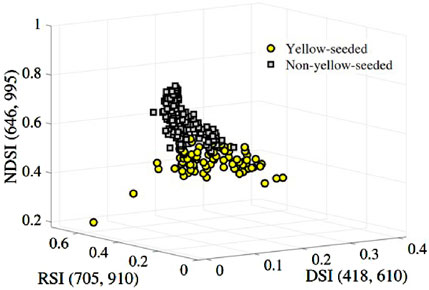
Figure 7. Two-classification of the yellow and non-yellow-seeded samples based on the feature combination .
Now, we hope to find a segmentation hyperplane to separate the above two types of samples. To do so, firstly, we input the remaining 150 samples including the spectral index combination and the corresponding labeled Y = 0/1 to train the Logit-R model. The trained discriminant function is
h=11+exp−3.3589RSI+4.3883DSI−5.1165NDSI.Then, inputting the three-spectral indexes combination of the 150 remaining samples into the trained model. Figure 7 depicts the sample distribution of four categories: yellow-seeded (yellow dots), non-yellow-seeded (grey squares), misclassified yellow-seeded samples (red dots), and misclassified non-yellow-seeded (red squares). As seen in Figure 8, there are only one yellow sample is misclassified and 9 non-yellow samples are misclassified. The RA of yellow-seeded and non-yellow-seeded is 97.78% and 91.43%, respectively, and the average RA is 93.33%.
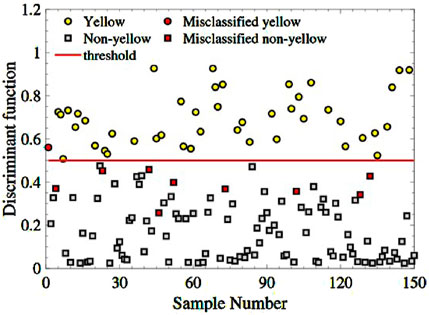
Figure 8. The classification of the yellow and non-yellow-seeded samples based on the feature combination .
3.2.3 Predication results with machine learning methodsIn this section, we employ two machine learning techniques: random forest Breiman (2001) and support vector classifier (SVC) Pisner and Schnyer (2020) to perform a classification task aimed at identifying the yellow seed. Differing from the preceding feature selection methods, we enhance our feature pool by integrating the original hyperspectral reflectances, consisting of nine spectral indexes calculated according to Table 3, and 23 trilateral parameters. This augmentation results in a total of 633 features.
To extract pivotal attributes and eliminate redundant data, we employ lasso-penalized logistic regression with the R package ‘glmnet’ (Friedman et al., 2010). The parameter λ, which balances the penalty and loss terms, is fine-tuned to maximize the “auc” index. Upon tuning, λ is set at 0.0062. Consequently, we identified 10 key features out of the initial 633. These features include the original hyperspectral values at wavelengths 394, 415, 416, 417, and 418, NDSI(568,988) (a spectral index), as well as Yep, Dymin, SDr, and ske (trilateral parameters).
According to these selected ten features, we conduct random forest and support vector classifier algorithms via R packages ‘rpart’ (Therneau et al., 2015) and ‘e1071’ (Meyer et al., 2024), respectively, by default settings with different training sizes (M: 150–240). The prediction results in test sets for the recognition task are reported in Table 5.
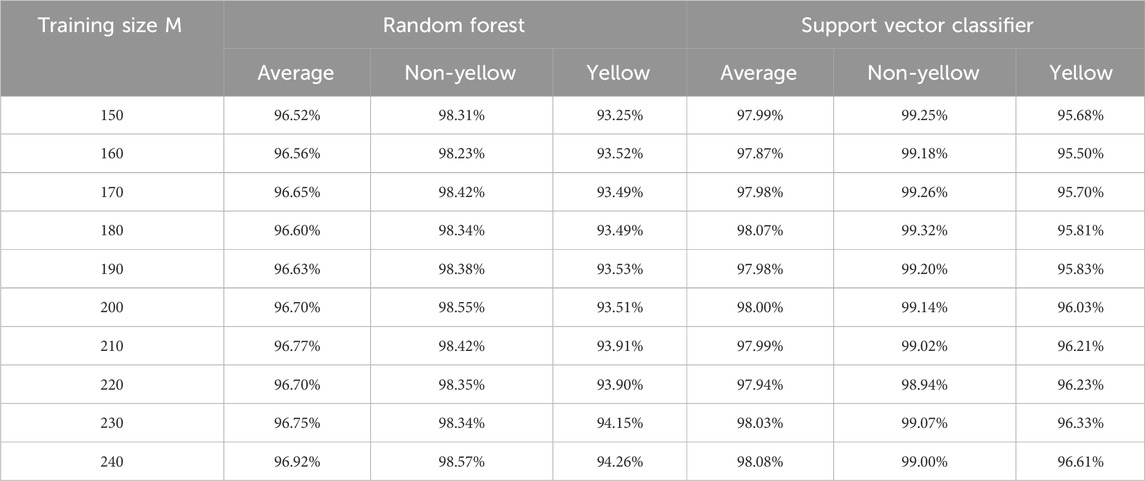
Table 5. Results of recognition accuracies with machine learning methods.
In comparing the recognition accuracies achieved by random forest and SVC methods, several key observations emerge. Regarding average accuracy, random forest demonstrates results ranging from approximately 96.52%–96.92%, while SVC exhibits a slightly higher range of around 97.87%–98.08%. Both methods exhibit an increase in accuracy with larger training sizes, with random forest’s accuracy gradually ascending and SVC’s accuracy following a similar trend, albeit with minor fluctuations.
In terms of category-specific accuracies, random forest achieves accuracy rates between approximately 93.25% and 94.26% in the yellow category, whereas SVC yields higher accuracy within the range of 95.68%–96.61%. For the non-yellow category, random forest consistently attains commendable accuracy, hovering between roughly 98.23% and 98.57%, whereas SVC shows even greater accuracy, ranging from approximately 98.94%–99.32%, generally surpassing random forest.
The consistency of performance across training sizes and categories is notable in the random forest’s case, where stability is observed. On the other hand, SVC displays slight performance variations, particularly noticeable within the yellow category. In the broader context of comparison, SVC emerges as the more favorable option, showcasing superior performance across most categories and training sizes. While random forest performs admirably in the non-yellow category, it falls short of SVC’s accuracy levels in the yellow category. This analysis underscores the nuanced strengths of each method and the importance of considering the specific problem context when selecting an appropriate machine learning approach.
3.3 Discussion of the proposed recognition methodsUp to this point, the task of recognizing yellow-seeded varieties has been effectively accomplished through the application of hyperspectral technology. Now, we would like to delve into the details of the four proposed models.
Beginning with the PLSR-based model, our approach involves identifying yellow-seeded varieties by predicting the RGB values through three essential trilateral parameters obtained from hyperspectral imaging of rapeseed, along with three significant spectral indices. During the process of predicting each R/G/B channel, we extract several noteworthy spectral features that contribute to enhancing the model’s interpretability. It’s important to note that the success of this method relies heavily on the accuracy of RGB calibration. Moving on to the Logit-R model, our strategy revolves around determining yellow-seeded or non-yellow-seeded categorization based on generating probabilities. However, one potential challenge of this model lies in dealing with imbalanced sample data. To address this, when data imbalance is encountered, it’s essential to consider adjusting classification thresholds to ensure accurate results. The optimal hyperspectral feature chosen for both the aforementioned models is determined through a thorough correlation analysis between the R/G/B values and the 23 trilateral parameters and spectral indices derived from a complete band combination. It’s worth mentioning that this approach might potentially omit some information from the original spectral reflectance data. Diverging from the two aforementioned methods, the machine learning models operate differently. In this case, we initially conduct feature dimensionality reduction from a total of 633 features, encompassing all 23 trilateral parameters, 9 spectral indices, and reflectance data from 601 original bands. Subsequently, we select ten key features to input into the random forest and SVC models.
All four models demonstrate high average accuracy rates, showcasing relatively similar performance differences ranging from 93% to 98%. This consistency highlights the feasibility of the framework that combines spectral features with intelligent models for accurately identifying yellow-seeded B. napus varieties. Considering factors like ease of operation and comprehensive utilization of information, the SVC model is recommended as an optimal choice for the task of identifying yellow-seeded varieties.
4 Discussions and conclusionRemote sensing technology, recognized as an essential national strategy, finds extensive application across both military and civilian domains. It facilitates the efficient acquisition of spectral data, enabling tasks like land classification and parameter inversion that are challenging for vision-based systems. Hyperspectral imaging technology, a near-Earth remote sensing tool, forms the basis for this advancement. As an innovative method of photoelectric detection and recognition, it integrates spectroscopy with optical imaging, offering a non-destructive and highly efficient alternative to traditional empirical and lab-based approaches for discerning the color of rapeseed seeds. Leveraging the rich spectral and image data inherent to rapeseed samples, this technology holds great promise for agricultural applications.
Comparing yellow seeds with black and brown seeds in Brassica napus reveals that yellow seeds have a thinner seed coat, higher oil content, and better quality. They also have higher protein content in the cake, lower cellulose and polyphenol levels, and higher economic value. Breeding yellow-seed varieties has become a key goal in rapeseed breeding worldwide. However, the complex seed color and inconsistent standards in current identification methods pose challenges. Most researchers use the naked eye or RGB color systems for seed color identification. However, the inconsistent phenotypic color and environmental influences make RGB methods unstable. In contrast, hyperspectral technology, which detects internal seed quality, is less affected by surface color, providing more stable results.
In this study, we introduce four intelligent models carefully designed to distinguish yellow-seeded rapeseed, as depicted in the model flowchart in Figure 9. The first two models, PLSR and Logit-R, synergize spectral indices with hyperspectral trilateral parameters. This process begins with extracting three spectral indices and 23 trilateral parameters. Through correlation analyses across the R, G, and B color channels of rapeseed seeds, we determine the optimal combinations of these spectral indices and trilateral parameters.
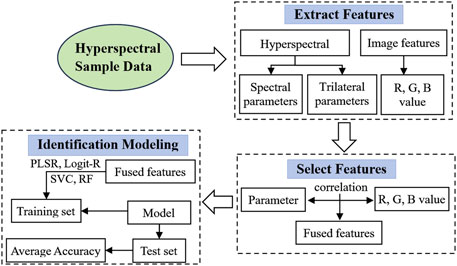
Figure 9. Flowchart of identification modeling.
The PLSR model leverages six features derived from three spectral indices and three trilateral parameters, achieving an impressive recognition accuracy (RA) between 92.32% and 96.55% in differentiating yellow from non-yellow seeds. The Logit-R model, which prioritizes the three spectral indices combined with the R channel, achieves a remarkable RA of 98%.
Additionally, we employ two machine learning models-random forest and SVC-to tackle the identification task. Beyond the 23 trilateral parameters and nine optimal spectral indices, we include the original 601 spectral reflectance values in the feature set. Using lasso-penalized logistic regression, we identify ten key features, which serve as input for the random forest and SVC models, achieving an average RA of approximately 98%, with SVC slightly outperforming random forest.
We emphasize that the proposed identification framework for yellow-seeded rapeseed, which integrates classical statistical methods and advanced machine learning tools, demonstrates robust generalizability. This framework is not limited to rapeseed classification but holds significant potential for application to seed classification and identification tasks across a wide range of other crops. By combining hyperspectral feature extraction with predictive modeling techniques, it provides a versatile approach that can adapt to various seed types, accommodating their unique physical and spectral characteristics. This generalizability makes it a valuable tool for advancing precision agriculture and improving the efficiency of crop breeding programs. Additionally, the framework’s ability to extract internal quality information and analyze large-scale data through machine learning models makes it adaptable for various agricultural tasks, including crop variety identification, stress detection, and quality assessment across different agricultural production systems. By customizing the spectral features and models for specific crops, this framework can be effectively extended to other agricultural systems, enhancing precision farming and crop management in diverse contexts.
This study identifies several limitations and proposes future research directions. It suggests integrating machine vision with machine learning for rapeseed color recognition as a cost-effective alternative to hyperspectral feature fusion, which remains expensive. Machine vision, efficient for non-destructive small-target color recognition, contrasts with hyperspectral remote sensing, which excels in large-area identification. A promising approach involves combining hyperspectral remote sensing with intelligent models, establishing a key paradigm for agricultural monitoring. Future work will focus on integrating machine vision and hyperspectral remote sensing to enhance rapeseed color recognition across broader areas.
Second, this study is limited by the small sample size and narrow color range, based on two B. napus varieties from a single field trial in Changsha (2020–2021). As an initial exploration of hyperspectral technology and machine learning for yellow-seeded rapeseed identification, it provides valuable insights but requires expansion. Future research will include more rapeseed varieties and account for environmental factors like temperature, humidity, light, and altitude by incorporating multi-year, multi-location data for a comprehensive analysis of seed color variability.
Data availability statementThe raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributionsFL: Conceptualization, Formal Analysis, Methodology, Writing–original draft. FW: Funding acquisition, Methodology, Supervision, Writing–review and editing. ZZ: Validation, Writing–review and editing. LC: Software, Visualization, Writing–review and editing. JW: Formal Analysis, Software, Supervision, Writing–review and editing. Y-GW: Formal Analysis, Software, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported partially by the National Natural Science Foundation of China (Grant No. 12471489), the Key Research Project of the Department of Education of Hunan Province (CN) (Grant No. 22A0135), the Excellent Young Research Project of the Department of Education of Hunan Province (CN) (Grant No. 24B1085), the “Chunhui” Program Collaborative Scientific Research Project (202202004), and the Australian Research Council project (DP160104292).
AcknowledgmentsWe would like to thank the Oil Institute of Hunan Agricultural University for providing two experimental rapeseed varieties of Xiangyou 708 and Xiangyou 710. The author wishes to thank the reviewers and the handling editor for their constructive comments and suggestions, which led to a great improvement in the presentation of this work.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAuger, B., Marnet, N., Gautier, V., Maia-Grondard, A., Leprince, F., Renard, M., et al. (2010). A detailed survey of seed coat flavonoids in developing seeds of brassica napus l. J. Agric. Food Chem. 58, 6246–6256. doi:10.1021/jf903619v
PubMed Abstract | CrossRef Full Text | Google Scholar
Baetzel, R., Lühs, W., Badani, A.-G., and Friedt, W. (2003). Development of segregating populations in the breeding of yellow-seeded winter rapeseed (brassica napus l.). Proc. 11th Int. Rapeseed Congr. 1, 238–242.
Bai, Z., Tian, J., Hu, X., Sun, T., Luo, H., and Huang, D. (2022). A back-propagation neural network model using hyperspectral imaging applied to variety nondestructive detection of cereal. J. Food Process Eng. 45, e13973. doi:10.1111/jfpe.13973
CrossRef Full Text | Google Scholar
Broeckx, J., Vanmaercke, M., Duchateau, R., and Poesen, J. (2018). A data-based landslide susceptibility map of africa. Earth-Science Rev. 185, 102–121. doi:10.1016/j.earscirev.2018.05.002
CrossRef Full Text | Google Scholar
Bu, Y., Jiang, X., Tian, J., Hu, X., Han, L., Huang, D., et al. (2023). Rapid nondestructive detecting of sorghum varieties based on hyperspectral imaging and convolutional neural network. J. Sci. Food Agric. 103, 3970–3983. doi:10.1002/jsfa.12344
PubMed Abstract | CrossRef Full Text | Google Scholar
Chen, C., Xiao, L., Zhao, Z., and Du, D. (2015). Research progress in seed coat color of yellow-seeded rapeseed. J. Henan Agric. Sci. 44, 1–6. doi:10.15933/j.cnki.1004-3268.2015.09.001
CrossRef Full Text | Google Scholar
Friedman, J., Hastie, T., and Tibshirani, R. (2010). Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 (1), 1–22. doi:10.18637/jss.v033.i01
PubMed Abstract | CrossRef Full Text | Google Scholar
Guo, L., Yu, Y., Yu, H., Tang, Y., Li, J., Du, Y., et al. (2019). Rapid quantitative analysis of adulterated rice with partial least squares regression using hyperspectral imaging system. J. Sci. Food Agric. 99, 5558–5564. doi:10.1002/jsfa.9824
PubMed Abstract | CrossRef Full Text | Google Scholar
He, S.-F., Zhou, Q., and Wang, F. (2022). Local wavelet packet decomposition of soil hyperspectral for som estimation. Infrared Phys. & Technol. 125, 104285. doi:10.1016/j.infrared.2022.104285
CrossRef Full Text | Google Scholar
Hong, Y., Liu, Y., Chen, Y., Liu, Y., Yu, L., Liu, Y., et al. (2019). Application of fractional-order derivative in the quantitative estimation of soil organic matter content through visible and near-infrared spectroscopy. Geoderma 337, 758–769. doi:10.1016/j.geoderma.2018.10.025
CrossRef Full Text | Google Scholar
Jiang, S., Wang, F., Shen, L., and Liao, G. (2018). Local detrended fluctuation analysis for spectral red-edge parameters extraction. Nonlinear Dyn. 93, 995–1008. doi:10.1007/s11071-018-4241-y
CrossRef Full Text | Google Scholar
Jiang, S., Wang, F., Shen, L., Liao, G., and Wang, L. (2017). Extracting sensitive spectrum bands of rapeseed using multiscale multifractal detrended fluctuation analysis. J. Appl. Phys. 121. doi:10.1063/1.4978308
CrossRef Full Text | Google Scholar
Liu, H. (1992). Studies on the inheritance of yellow-seeded brassica napus l. Acta Agron. Sin. (China). doi:10.3321/j.issn:0496-3490.1992.04.001
CrossRef Full Text | Google Scholar
Li, J., Chen, L., Tang, Z., Zhang, X., and Yan, S. (2001). “Genetic study and commercial application of the yellow-seeded rapeseed (brassica napus l.),” in Proceedings of the international symposium on rapeseed science (New York: Science Press), 19–23.
Li, J., Li, Q., Wang, F., and Liu, F. (2022). Hyperspectral redundancy detection and modeling with local hurst exponent. Phys. A Stat. Mech. its Appl. 592, 126830. doi:10.1016/j.physa.2021.126830
CrossRef Full Text | Google Scholar
Li, X., Chen, L., Hong, M., Zhang, Y., Zu, F., Wen, J., et al. (2012). A large insertion in bhlh transcription factor brtt8 resulting in yellow seed coat in brassica rapa. PLoS One 7, e44145. doi:10.1371/journal.pone.0044145
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y., Liu, X. L., Li, J., Yin, J., and Xu, X. (2012). Construction of near-infrared reflectance spectroscopy model for seed color of rapeseed. Chin. J. Oil Crop Sci. 34.
Liang, J., Wang, Y., Shi, Y., Huang, X., Li, Z., Zhang, X., et al. (2023). Non-destructive discrimination of homochromatic foreign materials in cut tobacco based on vis-nir hyperspectral imaging. J. Sci. Food Agric. 103, 4545–4552. doi:10.1002/jsfa.12528
PubMed Abstract | CrossRef Full Text | Google Scholar
Lin, Y., Deng, X., Li, X., and Ma, E. (2014). Comparison of multinomial logistic regression and logistic regression: which is more efficient in allocating land use? Front. Earth Sci. 8, 512–523. doi:10.1007/s11707-014-0426-y
CrossRef Full Text | Google Scholar
Liu, F., Wang, F., Liao, G., Lu, X., and Yang, J. (2021). Prediction of oleic acid content of rapeseed using hyperspectral technique. Appl. Sci. 11, 5726. doi:10.3390/app11125726
CrossRef Full Text | Google Scholar
Liu, F., Wang, F., Wang, X., Liao, G., Zhang, Z., Yang, Y., et al. (2022). Rapeseed variety recognition based on hyperspectral feature fusion. Agronomy 12, 2350. doi:10.3390/agronomy12102350
CrossRef Full Text | Google Scholar
Liu, X., Tu, J., Chen, B., and Fu, T. (2005). Identification and inheritance of a partially dominant gene for yellow seed colour in brassica napus. Plant Breed. 124, 9–12. doi:10.1111/j.1439-0523.2004.01051.x
CrossRef Full Text | Google Scholar
Meacham-Hensold, K., Montes, C. M., Wu, J., Guan, K., Fu, P., Ainsworth, E. A., et al. (2019). High-throughput field phenotyping using hyperspectral reflectance and partial least squares regression (plsr) reveals genetic modifications to photosynthetic capacity. Remote Sens. Environ. 231, 111176. doi:10.1016/j.rse.2019.04.029
PubMed Abstract | CrossRef Full Text | Google Scholar
Meyer, D., Dimitriadou, E., Hornik, K., Weingessel, A., Leisch, F., and Chang, C.-C. (2024). Misc functions of the department of statistics (e1071), tu wien. R. Package 1, 7–16. doi:10.32614/CRAN.package.e1071
CrossRef Full Text | Google Scholar
Munshi, T., Zuidgeest, M., Brussel, M., and van Maarseveen, M. (2014). Logistic regression and cellular automata-based modelling of retail, commercial and residential development in the city of ahmedabad, India. Cities 39, 68–86. doi:10.1016/j.cities.2014.02.007
CrossRef Full Text | Google Scholar
Peng, Z., Lin, S., Zhang, B., Wei, Z., Liu, L., Han, N., et al. (2020). Winter wheat canopy water content monitoring based on spectral transforms and “three-edge”’ parameters. Agric. Water Manag. 240, 106306. doi:10.1016/j.agwat.2020.106306
CrossRef Full Text | Google Scholar
Petisco, C., García-Criado, B., Vázquez-de Aldana, B. R., De Haro, A., and García-Ciudad, A. (2010). Measurement of quality parameters in intact seeds of brassica species using visible and near-infrared spectroscopy. Industrial Crops Prod. 32, 139–146. doi:10.1016/j.indcrop.2010.04.003
CrossRef Full Text | Google Scholar
Pisner, D. A., and Schnyer, D. M. (2020). “Support vector machine,” in Machine learning (Elsevier), 101–121.
Qu, C., Fu, F., Lu, K., Zhang, K., Wang, R., Xu, X., et al. (2013). Differential accumulation of phenolic compounds and expression of related genes in black-and yellow-seeded brassica napus. J. Exp. Bot. 64, 2885–2898. doi:10.1093/jxb/ert148
PubMed Abstract | CrossRef Full Text | Google Scholar
Sen, R., Sharma, S., Kaur, G., and Banga, S. S. (2018). Near-infrared reflectance spectroscopy calibrations for assessment of oil, phenols, glucosinolates and fatty acid content in the intact seeds of oilseed brassica species. J. Sci. Food Agric. 98, 4050–4057. doi:10.1002/jsfa.8919
PubMed Abstract | CrossRef Full Text | Google Scholar
Sibanda, M., Mutanga, O., Dube, T., Odindi, J., and Mafongoya, P. L. (2019). The utility of the upcoming HyspIRI’s simulated spectral settings in detecting maize gray leafy spot in relation to sentinel-2 MSI, VENµS, and landsat 8 OLI sensors. Agronomy 9, 846. doi:10.3390/agronomy9120846
CrossRef Full Text | Google Scholar
Somers, D. J., Rakow, G., Prabhu, V. K., and Friesen, K. R. (2001). Identification of a major gene and rapd markers for yellow seed coat colour in brassica napus. Genome 44, 1077–1082. doi:10.1139/g01-097
PubMed Abstract | CrossRef Full Text | Google Scholar
Tańska, M., Rotkiewicz, D., Kozirok, W., and Konopka, I. (2005). Measurement of the geometrical features and surface color of rapeseeds using digital image analysis. Food Res. Int. 38, 741–750. doi:10.1016/j.foodres.2005.01.008
CrossRef Full Text | Google Scholar
Wei, Y., Li, X., Pan, X., and Li, L. (2020). Nondestructive classification of soybean seed varieties by hyperspectral imaging and ensemble machine learning algorithms. Sensors 20, 6980. doi:10.3390/s20236980
PubMed Abstract | CrossRef Full Text | Google Scholar
Yang, F., Ye, S., Ma, X., Chen, Y., Yi, B., Ma, C., et al. (2021). Resynthesis of yellow-seeded brassica napus and comparative metabonomic analysis of differently colored seed coats. Mol. Plant Breed., 1–14. doi:10.13271/j.mpb.022.003979
CrossRef Full Text | Google Scholar
Ye, Q., Wang, Y., Zhou, S., Cheng, X., and Jia, J. (2018). Color discrimination based on hyperspectral imaging method. Spectrosc. Spectr. Analysis 38, 3310–3314. doi:10.3964/j.issn.1000-0593(2018)10-3310-05
CrossRef Full Text | Google Scholar
Zhang, J., Huang, Y., Li, Z., Liu, P., and Yuan, L. (2017). Noise-resistant spectral features for retrieving foliar chemical parameters. IEEE J. Sel. Top. Appl. Earth Observations Remote Sens. 10, 5369–5380. doi:10.1109/jstars.2017.2713039
CrossRef Full Text | Google Scholar
Zhang, L., Sun, H., Rao, Z., and Ji, H. (2020). Hyperspectral imaging technology combined with deep forest model to identify frost-damaged rice seeds. Spectrochimica Acta Part A Mol. Biomol. Spectrosc. 229, 117973. doi:10.1016/j.saa.2019.117973
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, T., Wei, W., Zhao, B., Wang, R., Li, M., Yang, L., et al. (2018). A reliable methodology for determining seed viability by using hyperspectral data from two sides of wheat seeds. Sensors 18, 813. doi:10.3390/s18030813
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, X., Pang, Z., Chen, L., Yin, J., and Li, J. (2006). Seed color detection by computer technology in rapeseed. Chin. J. Oil Crop Sci. 28, 11. doi:10.3321/j.issn:1007-9084.2006.01.003
留言 (0)