Low-grade glioma (LGG) is a primary brain tumor originating from glial cells (Weller et al., 2024), accounting for 7.6% of all primary brain tumors and 31.8% of gliomas. The median overall survival ranges from 5.6 to 13.3 years, depending on tumor histopathological characteristics, molecular phenotype, and growth rate (Li et al., 2021a). The optimal treatment strategy for LGG remains controversial, with current approaches including surgical resection, radiotherapy, chemotherapy, targeted therapy, and immunotherapy (van den Bent et al., 2023). Although LGG patients have slightly better survival rates than those with high-grade (WHO grade III or IV) gliomas (Aiman et al., 2024), the infiltrative nature of gliomas makes LGG prone to drug resistance and recurrence after treatment (Fukuya et al., 2019), with potential progression to high-grade gliomas, significantly shortening survival time (Tom et al., 2019). Following local dissemination, LGG tumor cells exhibit high heterogeneity (Nicholson and Fine, 2021), leading to greater variations in patient survival rates and times. This cellular diversity and heterogeneity in LGG are considered primary factors in tumor recurrence and malignant transformation (Ye et al., 2024; Gittleman et al., 2019). Therefore, understanding the cellular mechanisms underlying LGG development is crucial for clarifying its progression and developing new effective therapeutic targets to extend patient survival.
Histone modification is a crucial epigenetic regulatory mechanism encompassing various forms, including methylation, acetylation, phosphorylation, adenylation, ubiquitination, and ADP-ribosylation (Millán-Zambrano et al., 2022). With rapid advances in molecular biology, the World Health Organization substantially updated its diagnostic criteria for LGG in 2021, transitioning from traditional histological diagnosis to an integrated diagnostic system incorporating molecular markers (Figarella-Branger et al., 2022). This shift is prominently reflected in the WHO CNS5 classification system, which establishes IDH mutation and 1p/19q codeletion status as core molecular markers for adult-type diffuse low-grade glioma classification, fully reflecting the molecular heterogeneity of LGG.
Currently, epigenetic alterations (including histone methylation, DNA methylation, and histone acetylation) are increasingly being applied in brain tumor research (Han et al., 2024). While histone modifications have been extensively studied in high-grade gliomas (HGG), such as proteomics combined with other multi-omics revealing the central role of PTPN11 signaling in high-grade gliomas (Lowe et al., 2019a), and histone H3 mutations promoting diffuse glioma development through chromatin dysregulation (Lowe et al., 2019b), related research in LGG remains relatively scarce. Given the common cellular origins between LGG and HGG (Network et al., 2015), these findings may hold significant implications for LGG as well. Considering that tumor cell heterogeneity is a key factor in LGG recurrence and malignant transformation, conducting more extensive and comprehensive studies on histone modifications is crucial. This not only helps deepen our understanding of LGG’s molecular pathogenesis but also provides new insights for developing personalized treatment strategies for highly heterogeneous LGG.
In our study, we conducted a series of complex bioinformatics analyses, utilizing high-throughput sequencing and proteomics technologies to monitor genome-wide histone modification dynamics, while employing diverse machine learning frameworks and big data to perform comprehensive systematic analysis and identification of histone modifications and related multi-omics features in LGG. By integrating genomics, transcriptomics, and proteomics data to establish machine learning models, we comprehensively revealed key molecules and pathways controlling LGG development and treatment response. This cross-omics integrated analysis approach not only deepens our understanding of LGG epigenetic regulation but also identifies new biomarkers and potential therapeutic targets, potentially providing new directions for LGG treatment research.
2 Materials and methodsThe research workflow is shown in Figure 1.
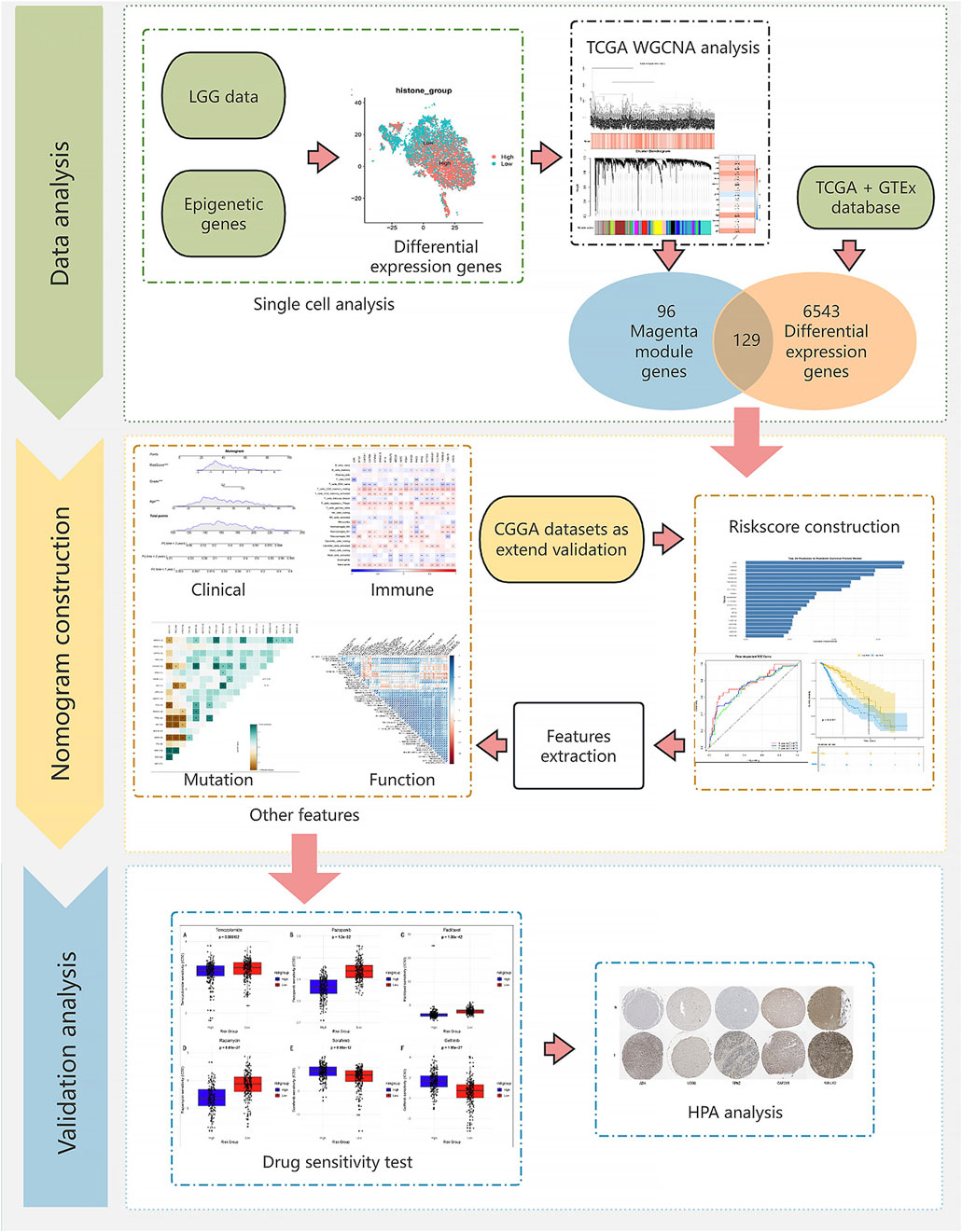
Figure 1. Flow chart.
2.1 Data sourceThis study integrated multiple independent datasets to construct and validate a prognostic model for low-grade glioma. Initially, we established a histone modification gene set based on Füllgrabe et al. (2011)’s research and the GeneCards database (https://www.genecards.org/) (relevance score >20). Subsequently, single-cell transcriptome data from 4 LGG samples (GSE182109) were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) for further single-cell analysis. TCGA-LGG (513 samples) and GTEx (2,642 normal control samples) datasets were merged after batch effect removal to calculate differentially expressed genes in LGG. Additionally, TCGA-LGG served as the training set, while two independent cohorts from the Chinese Glioma Genome Atlas (CGGA) database (https://www.cgga.org.cn/), containing 693 and 325 GBM samples respectively with clinical and survival information, were used as external validation sets.
2.2 Single cell analysis2.2.1 Data preprocessing and quality controlSingle-cell RNA sequencing data were analyzed using Seurat (v5.0.0) (Abdelfattah et al., 2022). Initially, raw data in 10X Genomics format were processed and preliminarily filtered, requiring each gene to be expressed in at least three cells and each cell to express at least 250 genes. Subsequently, the expression proportions of mitochondrial genes (prefixed with MT-) and ribosomal genes [prefixed with RP (SL)] were calculated for each cell. To ensure data quality, we filtered out the following cells: 1) cells expressing <100 or >7,500 genes; 2) cells with mitochondrial gene expression proportion >25%; 3) cells with total RNA counts <1,000.
2.2.2 Data integration and annotationFiltered data were normalized using the “LogNormalize” method (Luo et al., 2019) with a scale factor of 10,000. The “vst” method was used to select 2,000 highly variable genes for subsequent analysis. To eliminate batch effects, the Harmony algorithm was employed to integrate data from multiple samples (Petegrosso et al., 2020; Korsunsky et al., 2019). Subsequently, PCA dimensionality reduction was performed, selecting the top 30 principal components for further analysis. Using these principal components, UMAP and t-SNE dimensionality reduction visualizations were performed separately. Cell clustering was conducted using a graph-based clustering algorithm (Petegrosso et al., 2020), with the optimal resolution parameter of 0.2 determined through the clustree package. Cell types were automatically annotated using the SingleR package (Aran et al., 2019) in conjunction with the Human Primary Cell Atlas database (https://www.humancellatlas.org/) and scMayomap package (Yang et al., 2023). Additionally, manual verification was performed based on characteristic gene expression (Hassn Mesrati and Behrooz, 2020), including: OLIG2 and MBP (oligodendrocyte markers), CD44 and SOX2 (tumor cell markers), PDGFRB (pericyte marker), FOLR2, AIF1, and CD68 (macrophage markers), CD8A, CD3E, and CCL5 (T cell markers).
2.2.3 Histone score analysisBased on the predefined histone gene set, the ssGSEA algorithm was used to calculate histone scores for each cell (Chen et al., 2022). For LGG cell subgroups, cells were divided into high and low expression groups based on the median histone score, and differential expression analysis was performed (logFC threshold of 0.5, minimum expression proportion of 0.35) to identify functional pathways associated with histone expression.
2.3 Weighted gene co-expression network analysisWGCNA analysis was conducted based on the previously obtained histone-related differentially expressed gene set (Xu et al., 2023) to reveal gene co-expression relationships and their associations with phenotypes. During data preprocessing, gene expression data from the TCGA-LGG dataset were normalized, and quality was ensured by removing genes with zero standard deviation and samples with missing values. Subsequently, the goodSamplesGenes function was used for quality assessment, and outlier samples were detected using hierarchical clustering (Grabski et al., 2023). In network construction, the optimal soft threshold power = 5 was determined by analyzing the scale-free topology fit index and average connectivity under different soft thresholds. The blockwiseModules function was used to construct the co-expression network, setting the minimum module size to 50 genes, module merging similarity threshold to 0.15, and using unsigned network type. Through calculating Module Eigengenes (MEs) (Han et al., 2019), we analyzed the correlation between modules and histone scores, using Pearson correlation coefficients to evaluate module-trait relationships, and assessed correlation significance using Student’s t-test. The correlation intensity between modules and phenotypic features was visualized through heatmaps. Finally, in modules significantly correlated with histone scores, the biological significance was validated by analyzing the relationship between Module Membership and Gene Significance, with scatter plots demonstrating their positive correlation, further confirming these modules’ central role in the histone regulatory network.
2.4 Feature genes selectionDifferential expression analysis was performed using the TCGA-LGG dataset and GTEx dataset through DESeq2, with selection criteria of absolute logFC >1 and p-value <0.05. Feature genes were selected by intersecting the differentially expressed genes with module genes obtained from WGCNA.
2.5 Machine learning based prognosis signature constructionTo construct a reliable prognostic prediction model, this study used the TCGA-LGG dataset as the training set and CGGA325 and CGGA693 datasets as independent validation sets. Initially, all datasets underwent standardization and feature space consistency was ensured, with missing values eliminated through strict data quality control. During model construction, we systematically evaluated multiple machine learning algorithms and their combinations, including Random Survival Forest (RSF), Elastic Net (Enet), Stepwise Cox regression (StepCox), CoxBoost, Partial Least Squares Regression (plsRcox), Principal Component Regression (SuperPC), Gradient Boosting Machine (GBM), Survival Support Vector Machine (survival-SVM), Ridge regression, and Lasso regression as base models. To enhance prediction performance, we explored various combinations of these base models, such as RSF with CoxBoost and Lasso with GBM combinations. For the Elastic Net model, performance was optimized by adjusting the α parameter (0.1–0.9); for stepwise Cox regression, forward, backward, and bidirectional feature selection methods were employed. Model evaluation used C-index (Song et al., 2022) as the primary evaluation metric, assessing predictive ability and generalization performance through comprehensive performance on training and two independent validation sets. Finally, model performance across different datasets was visualized through heatmaps, and models were ranked based on average C-index values from validation sets to select the final prognostic prediction tool with optimal predictive efficacy and stability.
2.6 Optimal model performance validation and risk score constructionBased on the comprehensive evaluation results of machine learning models, the optimal prediction model underwent thorough performance validation and risk score system construction. Specifically, through feature importance analysis, 20 key features contributing most significantly to prognosis prediction were identified from the original features. Based on the model’s predictions, a risk score system was established, dividing patients into high and low-risk groups using the median score as the threshold, and Kaplan-Meier survival analysis was performed to observe survival differences between risk groups. Additionally, to evaluate the model’s time-dependent predictive ability, time-dependent ROC curves (Obuchowski and Bullen, 2018) and corresponding AUC values were calculated for 1-year, 3-year, and 5-year predictions.
2.7 Prognostic value analysis of clinical features and risk scoresThe prognostic value of clinical features and risk scores was evaluated through systematic survival analysis (Schober and Vetter, 2018). Initially, univariate Cox regression analysis assessed the impact of clinical features including age, gender, tumor type, grade, and risk score on patient prognosis. Subsequently, multivariate Cox regression analysis identified independent prognostic factors. Based on significant independent prognostic factors, an integrated nomogram prediction model (Gittleman et al., 2019) was constructed, and its prediction accuracy was verified through calibration curves for 1-year, 3-year, and 5-year survival predictions. Time-dependent C-index analysis was used to compare the predictive performance between the nomogram model and individual predictive factors, while decision curve analysis (DCA) evaluated the clinical net benefit of the model at different decision thresholds to validate this prediction tool’s value in clinical practice.
2.8 Enrichment analysisThis study explored molecular pathway differences between high and low risk score groups through systematic functional enrichment analysis (Canzler and Hackermüller, 2020). First, Gene Set Enrichment Analysis (GSEA) evaluated significantly enriched Hallmark pathways in the high-risk group. Subsequently, Gene Set Variation Analysis (GSVA) scored all samples, and limma differential analysis identified pathways with significantly different activities between high and low-risk groups. To verify the clinical relevance of key pathways, patients were divided into high and low pathway activity groups based on GSVA scores, with Kaplan-Meier survival analysis evaluating prognostic differences, and Cox proportional hazards regression model calculating hazard ratios (HR) and their 95% confidence intervals. For significantly correlated pathways, forest plots were generated to visualize their prognostic value. Finally, survival curves were verified for the six most significant pathways, comprehensively assessing these pathways’ potential roles in glioma development and progression.
2.9 Mutation analysis and heterogeneity assessmentThis study explored the association between tumor heterogeneity and risk scores through comprehensive analysis of somatic mutation data from the TCGA-LGG cohort. First, the Mutant-Allele Tumor Heterogeneity (MATH) score (Timar and Kashofer, 2020) was used to quantify tumor heterogeneity levels for each sample, comparing differences between high and low-risk groups. Distribution characteristics were visualized through violin plots, with statistical significance assessed using the Wilcoxon rank-sum test. Subsequently, patients were divided into high and low heterogeneity groups based on the median MATH score, with Kaplan-Meier survival analysis evaluating the relationship between tumor heterogeneity and prognosis. Further combining MATH scores with risk scores, patients were classified into four subgroups (high MATH/high risk, high MATH/low risk, low MATH/high risk, low MATH/low risk) to explore the joint predictive value of both indicators. Finally, maftools was used to analyze mutation characteristics of high and low-risk groups, with waterfall plots displaying distribution characteristics of top 20 mutated genes, while co-occurrence and mutual exclusivity analysis (Zang et al., 2023) revealed interaction patterns among key driver genes.
2.10 Immune analysisThis study conducted systematic analysis of the LGG tumor immune microenvironment using multiple algorithms. Initially, the ESTIMATE algorithm (Tennant et al., 2022) calculated stromal scores, immune scores, and overall scores for each sample, comparing differences between high and low-risk groups. Subsequently, immune-related pathways were scored using ssGSEA, with heatmaps visualizing differential patterns of immune pathway activity between risk groups. Further, the CIBERSORT algorithm (Chen et al., 2018) was employed to deconvolute the infiltration proportions of 22 immune cell types, with violin plots showing immune cell composition differences between high and low-risk groups. Additionally, ssGSEA analysis was performed using 28 immune cell characteristic gene sets, with box plots clearly displaying abundance differences of various immune cell types between risk groups. Finally, correlation analysis explored relationships between key gene expression and immune cell infiltration, as well as associations between risk scores and immune cell infiltration levels. Significant correlation patterns were displayed through heatmaps and correlation scatter plots, revealing potential connections between the risk score model and tumor immune microenvironment.
2.11 Drug sensitivity analysisSystematic drug sensitivity prediction analysis was performed on the TCGA-LGG cohort using the pRRophetic package (Yan et al., 2022). Initially, half maximal inhibitory concentration (IC50) values (Sebaugh, 2011) were predicted for all available drugs based on drug response data from the Cancer Genome Project (CGP) database. For each drug, drug sensitivity differences between high and low-risk groups were compared, with statistical significance assessed using the Wilcoxon rank-sum test. For drugs showing significant differences (P < 0.05), box plots were used to visually display drug sensitivity distribution characteristics across different risk groups.
2.12 Human protein atlas validationBased on previous analysis results, we selected the top five key genes with the highest weights for expression validation in the HPA database (https://www.proteinatlas.org/). Through immunohistochemical staining images, protein expression levels and distribution patterns of these key genes were visually demonstrated in normal brain tissue and glioma tissues of different grades. The validation results from the HPA database not only confirmed the differential expression characteristics of these genes in glioma development at the protein level but also provided histological evidence for understanding their potential roles in tumor progression.
2.13 Pan-cancer analysisTo comprehensively visualize the pan-cancer analysis results, we constructed a stratified forest plot integrating survival analysis outcomes across 29 cancer types, organized by eight major organ systems (Breast and Gynecologic System, Respiratory System, Digestive System, Urinary System, Nervous System, Endocrine System, Male Reproductive System, Hematologic System, and Others). In the forest plot, dot sizes represent -log10 (p-value), error bars indicate 95% confidence intervals, and color-coding displays risk levels (red for high-risk groups, cyan for low-risk groups, and gray for non-significant differences). A horizontal dashed line (HR = 1) serves as a reference, and hazard ratios are presented on a logarithmic scale to better illustrate the relative magnitude of risk differences.
3 Results3.1 Single-cell data reveals differential distribution of histone modificationsThrough dimensionality reduction clustering analysis and cell type annotation of LGG single-cell data (Figure 2A), we successfully identified five major cell subpopulations: SOX2 and OLIG2-expressing LGG tumor cells, CD68, AIF1, and FOLR2-expressing macrophages, MBP-expressing oligodendrocytes, PDGFRB-expressing pericytes, and CCL5, CD3E, CD8A, and CD44-expressing T cells. To validate the accuracy of cell type annotation, we constructed a dot plot displaying the expression patterns of marker genes for each cell type (Figure 2B).
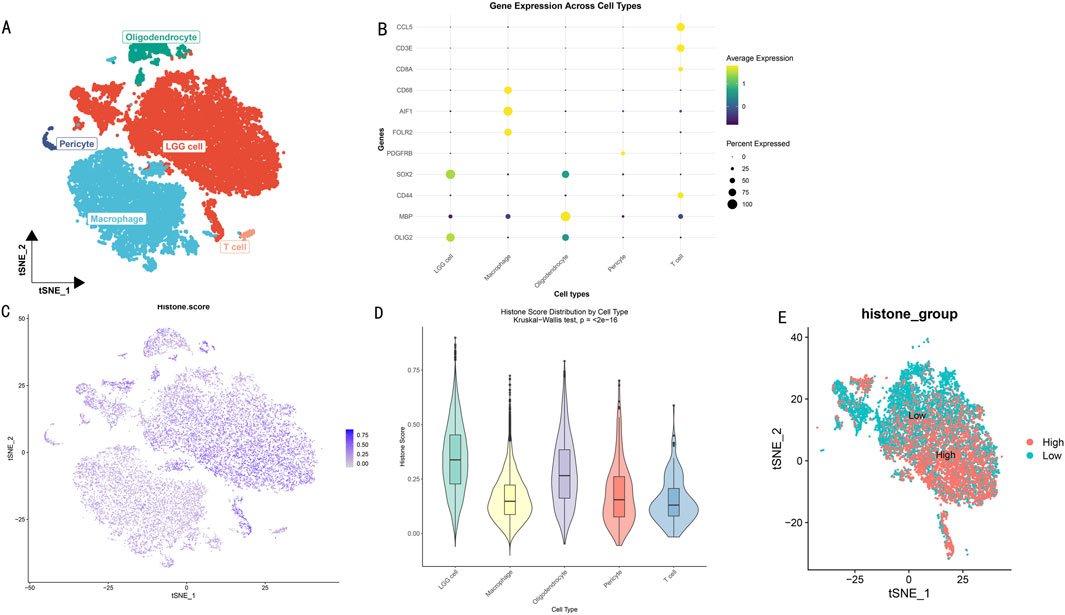
Figure 2. Histone Modification Lineage Analysis Based on Single-Cell Transcriptomics. (A) t-SNE dimensionality reduction showing spatial distribution of LGG cell subgroups. Different colors represent different cell subgroups. (B) Heatmap of cell subgroup-specific marker gene expression profiles. Rows represent genes, columns represent cells. (C) t-SNE plot showing distribution characteristics of histone modification scores across different cell subgroups. Color intensity indicates modification levels. (D) Box plot analysis of histone modification scores for five cell subgroups. (E) Bidirectional clustering analysis based on histone modification scores. t-SNE projection showing distribution patterns of high-score (red) and low-score (blue) cells.
To deeply explore the heterogeneity of histone modification levels among different cell types, we calculated and visualized histone modification scores for each cell subgroup (Figure 2C). Results showed significant differences in histone modification levels among different cell subgroups. Further statistical analysis (Figure 2D) revealed that LGG tumor cells and oligodendrocytes exhibited higher histone modification levels (scores >0.25). Based on the overall cellular histone modification levels (HMs), we separated LGG cells into high HMs and low HMs groups (Figure 2E). Differential expression analysis identified 5,278 differentially expressed genes (|logFC|>1, p < 0.05), with 3,638 genes upregulated in the high HMs group and 1,640 genes upregulated in the low HMs group.
3.2 HMs-related gene network analysisDifferential expression analysis revealed significant transcriptomic differences between high and low HMs groups. Volcano plot analysis showed numerous genes with significant differential expression (Figure 3A), suggesting these genes may participate in the histone modification regulatory network. To deeply analyze key regulatory genes, we visualized the top 50 up- and downregulated genes with the most significant differences in a circular plot (Figure 3B). To systematically identify co-expression modules related to histone modifications, we performed WGCNA analysis on differentially expressed genes. The hierarchical clustering dendrogram displayed gene co-expression relationships, while the bottom heatmap reflected HMs score variation patterns among samples (Figure 3C). Based on the dynamic tree-cutting algorithm, we ultimately identified 15 functional modules with significant co-expression characteristics (Figure 3D). Module-trait correlation analysis indicated that the magenta module (225 genes) showed the strongest positive correlation with histone modification scores (cor = 0.44, P < 0.005) (Figure 3E). Further module membership analysis revealed that genes in the magenta module showed significant positive correlation between Gene Significance (GS) and Module Membership (MM) (cor = 0.58, P < 1e-21) (Figure 3F), strongly suggesting this module plays a core role in the histone modification regulatory network.
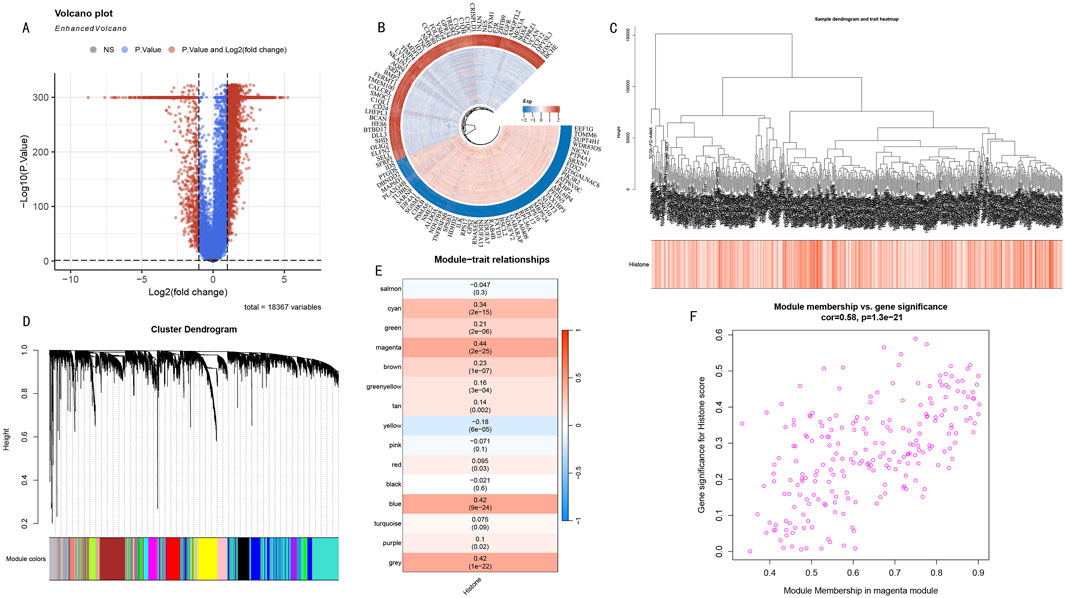
Figure 3. WGCNA Network Analysis Reveals Key Gene Modules in Histone Modification Regulation. (A) Volcano plot distribution of differentially expressed genes. Red and blue dots represent significantly upregulated and downregulated genes in the high HMs group, respectively. (B) Circular plot of top 50 most significantly up- and downregulated genes. Inner to outer rings show gene names, expression change direction, and statistical significance. (C) Hierarchical clustering dendrogram of differentially expressed genes. Upper part shows hierarchical clustering relationships among genes, lower heatmap shows distribution characteristics of sample HMs scores. Color intensity represents score levels. (D) WGCNA module identification results. Dendrogram shows gene clustering relationships, bottom colored bands represent 15 functional modules identified by dynamic tree-cutting algorithm. (E) Module-trait correlation heatmap. Each row represents a co-expression module, values and color intensity indicate Pearson correlation coefficients with histone modification scores. Magenta module shows strongest positive correlation (cor = 0.44, P < 0.005). (F) GS-MM scatter plot analysis of magenta module. X-axis: Module Membership; Y-axis: Gene Significance. Distribution trend (cor = 0.58, P < 1e-21) validates this module’s core position in histone modification regulatory network.
3.3 Construction of prognostic model using feature genesThrough integrative transcriptomic analysis of TCGA-LGG and GTEx datasets, we initially identified 6,672 LGG-related differentially expressed genes (DEGs), including 5,798 upregulated and 874 downregulated genes. Intersection analysis of these DEGs with previously determined magenta module genes yielded 129 LGG-specific histone modification-related genes (LGG-HMRgenes) (Figure 4A). Functional enrichment analysis revealed these LGG-HMRgenes were significantly enriched in pathways including heterocycle catabolic process, nucleobase-containing compound catabolic process, RNA catabolic process, and negative regulation of cellular macromolecule biosynthetic process (Figure 4B, P < 0.05). These results emphasize the regulatory role of histone modifications in LGG development and provide theoretical basis for developing targeted therapeutic strategies.
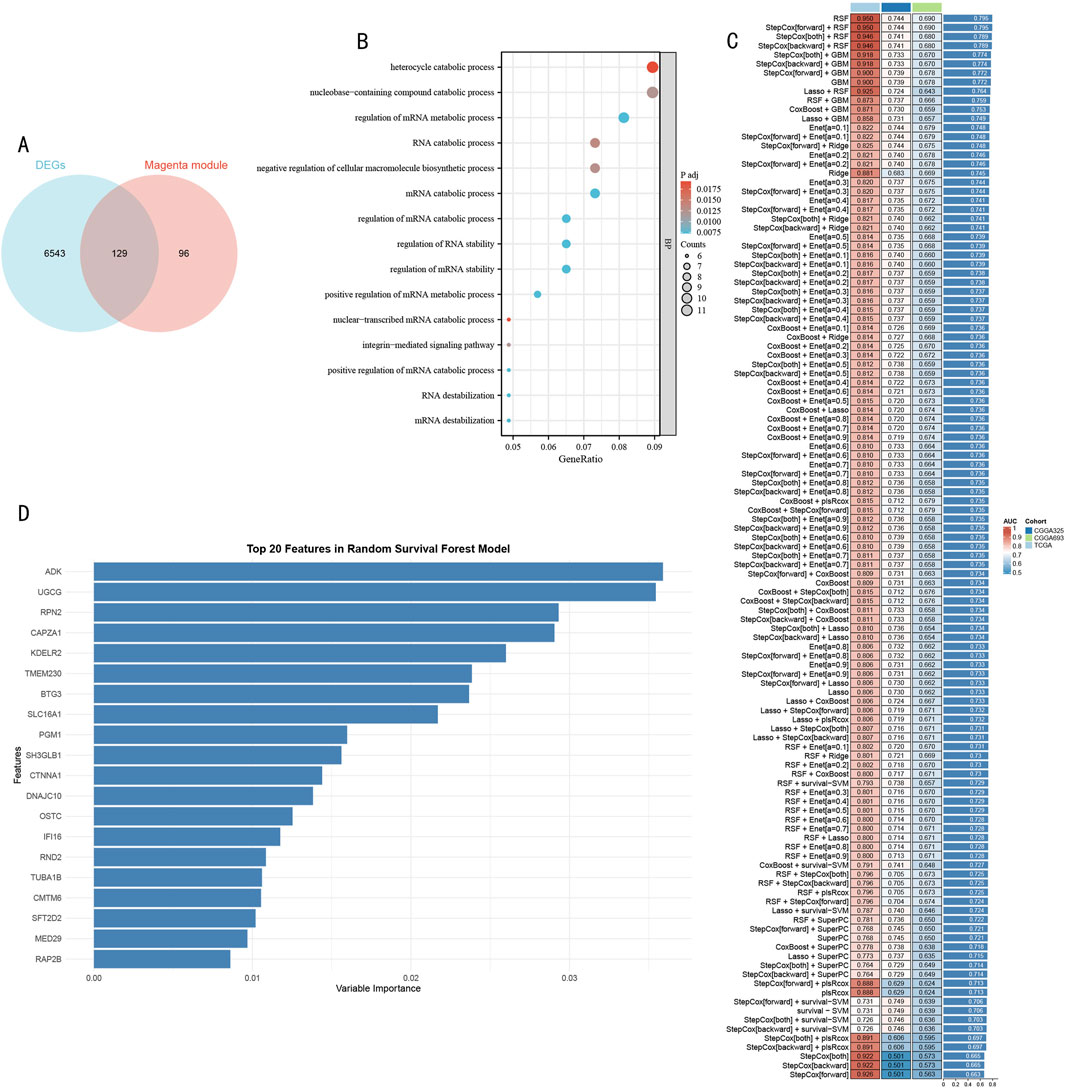
Figure 4. Systematic Identification and Model Construction of Histone Modification-Related Prognostic Markers (A) Venn diagram analysis of LGG DEGs and magenta module genes. (B) GO functional enrichment dot plot of LGG-HMRgenes. X-axis represents gene ratio, dot size represents number of enriched genes, color intensity represents statistical significance [-log10 (P-value)]. (C) Machine learning model performance evaluation heatmap. Rows represent different algorithms, columns represent validation datasets. Color scale indicates C-index values (red indicates higher prediction accuracy, blue indicates lower prediction accuracy). (D) Importance ranking plot of Top 20 feature genes identified by RSF model. X-axis represents feature importance scores, Y-axis represents gene symbols. Bar length reflects each gene’s contribution to prognostic prediction.
To construct a robust prognostic prediction model, we systematically evaluated the predictive performance of 105 machine learning algorithms using 129 HMR genes as feature inputs (Figure 4C). Through comprehensive comparison of C-index performance across validation sets, the Random Survival Forest (RSF) model demonstrated optimal predictive performance. Based on feature importance analysis of the RSF model, we further selected 20 core feature genes with the strongest predictive contributions (Figure 4D).
3.4 Multi-center validation and performance assessment of HMRS prognostic modelTo systematically evaluate the time-dependent predictive performance of the HMRS model, we first conducted time-dependent receiver operating characteristic (ROC) analysis in the TCGA-LGG cohort (513 samples). Results showed that the model demonstrated excellent discriminative ability in 1-year, 3-year, and 5-year survival predictions, with corresponding areas under the curve (AUC) reaching 0.77, 0.73, and 0.71 respectively (Figure 5A). After stratifying patients into high and low-risk groups based on the optimal cutoff value, Kaplan-Meier survival analysis revealed significant prognostic differences between the groups (log-rank test, P < 0.001) (Figure 5B).
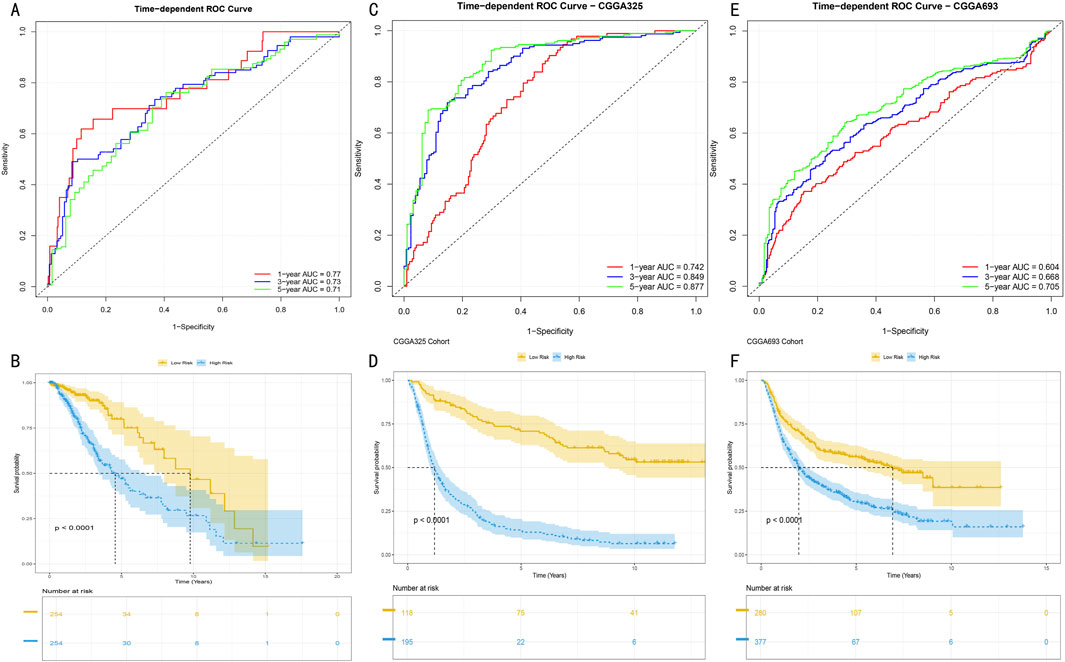
Figure 5. Predictive Performance and External Validation of HMRS Model (A) Time-dependent ROC curve analysis in TCGA training set. Red, blue, and green curves represent AUC values for 1-year, 3-year, and 5-year survival predictions. (B) Kaplan-Meier survival analysis based on HMRS scores in TCGA cohort. Yellow and blue curves represent high-risk group (n = x) and low-risk group (n = y) respectively. Shaded areas indicate 95% confidence intervals. (C, E) Time-dependent ROC curves in CGGA325 and CGGA693 validation sets. AUC values at various time points demonstrate the model’s stable predictive performance. (D, F) Survival stratification analysis in validation sets. Separation of survival curves validates the model’s prognostic discrimination ability (log-rank test, P < 0.001). Numbers at bottom indicate number at risk at each time point.
To validate the external applicability of the HMRS model, we conducted validation in two independent CGGA validation cohorts (CGGA325 and CGGA693). In the CGGA325 cohort (325 samples), the model demonstrated time-dependent prediction accuracy comparable to the training set (Figure 5C), with survival stratification differences showing statistical significance (P < 0.001) (Figure 5D). These results were further confirmed in the CGGA693 cohort (693 samples) (Figures 5E, F). Multi-center validation results confirmed that the HMRS model possesses robust prognostic prediction capability and broad clinical applicability.
3.5 Construction and evaluation of clinical variable-integrated prognostic modelTo systematically evaluate the prognostic value of clinical features and HMRS scores, we first conducted Cox proportional hazards regression analysis. Univariate analysis showed that age, WHO grade, and HMRS score were significant prognostic factors (Figure 6A, all P < 0.001). Multivariate analysis further confirmed the independent prognostic value of these three factors (Figure 6B). Based on these independent prognostic factors, we constructed an integrated nomogram prediction model. Calibration curve analysis showed that the model demonstrated excellent calibration in 1-year, 3-year, and 5-year survival predictions (Figure 6C).
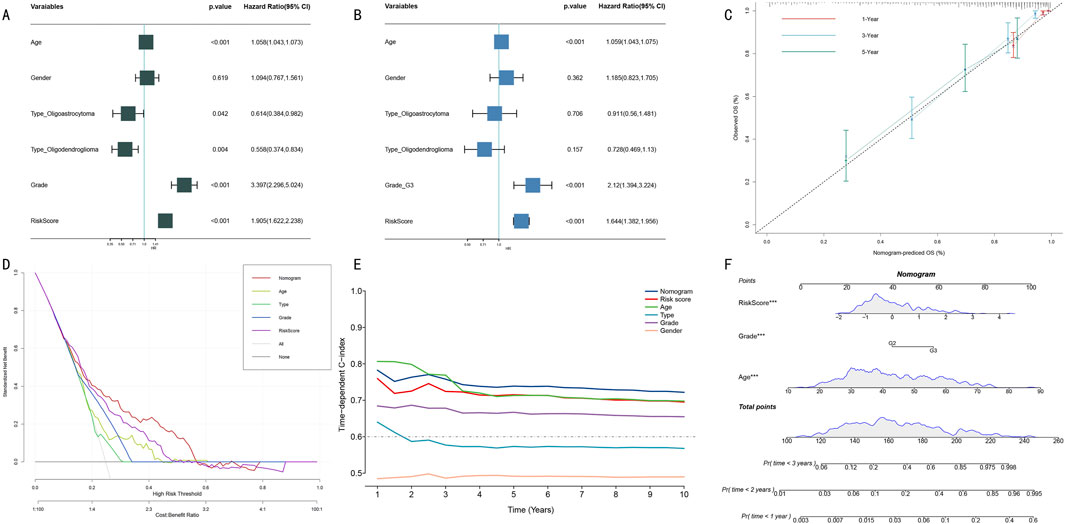
Figure 6. Construction and Performance Evaluation of Integrated Prognostic Prediction Model. (A) Forest plot of univariate Cox regression analysis. Shows hazard ratio (HR) and 95% confidence intervals for each clinical feature. (B) Forest plot of multivariate Cox regression analysis. Confirms independent prognostic factors (age, WHO grade, HMRS score). (C) Calibration curves for nomogram model. Shows consistency between predicted and actual survival probabilities at 1-year (red), 3-year (blue), and 5-year (green). Diagonal line represents perfect prediction. (D) DCA. Compares net benefit of different prediction strategies at various risk thresholds. (E) Dynamic comparison of time-dependent C-indices. Shows prediction accuracy of nomogram model (red) versus single prognostic factors at different follow-up time points. (F) Integrated prognostic prediction nomogram. Includes three independent prognostic factors: HMRS score, WHO grade, and age, for individualized prognosis prediction.
Clinical decision curve analysis (DCA) indicated that the integrated nomogram model demonstrated greater net benefit compared to single prognostic factors (Figure 6D). Dynamic analysis of time-dependent C-index showed that the nomogram model’s prediction accuracy (C-index >0.70) consistently outperformed single prognostic factors (Figure 6E). Notably, although age performed well in short-term (1–3 years) prediction, its long-term prediction stability was insufficient. Finally, we established a visual nomogram incorporating HMRS score, WHO grade, and age (Figure 6F), providing an intuitive quantitative tool for clinical prognostic assessment.
3.6 Systematic functional analysis of HMRS-related molecular mechanismsTo elucidate the molecular biological basis of HMRS prognostic stratification, we conducted systematic functional enrichment analysis between high and low-risk groups. GSEA showed that the high-risk group was significantly enriched in multiple cancer-related Hallmark pathways, including Allograft Rejection, E2F Targets, Interferon Gamma Response, MYC Targets V1, and TNFα Signaling via NFκB (Figure 7A, FDR <0.05).
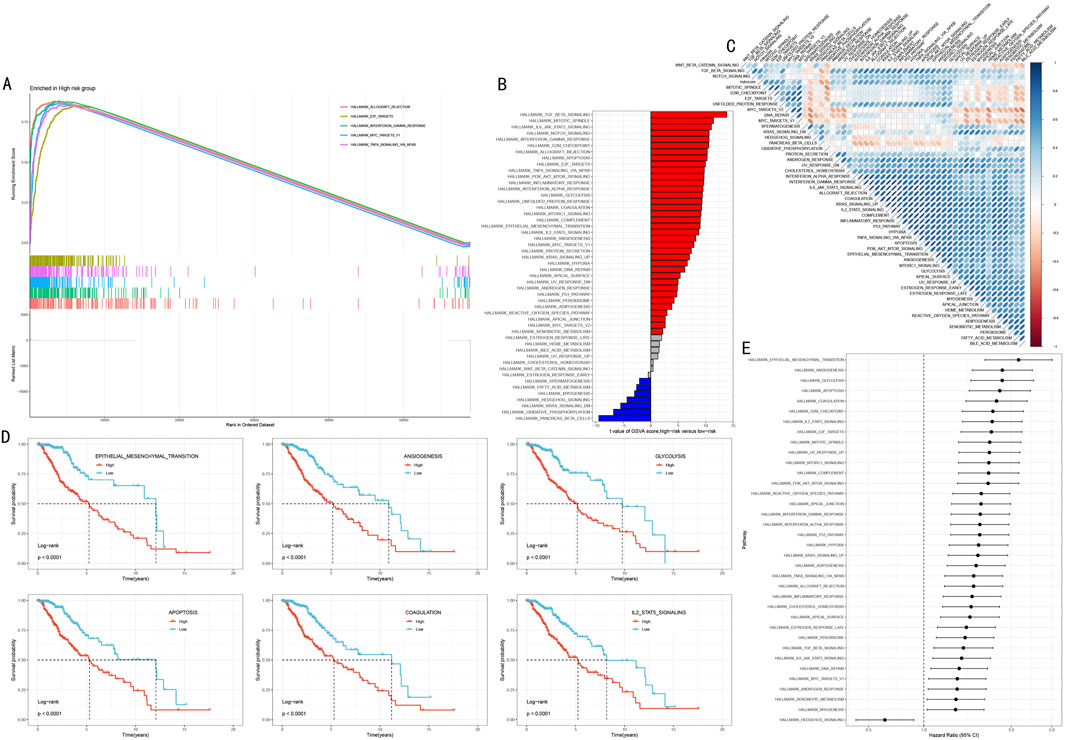
Figure 7. Multi-dimensional Functional Analysis of HMRS-Related Molecular Mechanisms. (A) GSEA waterfall plot showing five key pathways significantly enriched in high-risk group. Upper part shows enrichment plots, lower part shows gene expression heatmap. (B) Differential pathways revealed by GSVA analysis between high and low-risk groups. Red and blue indicate upregulated pathways in high-risk and low-risk groups respectively. (C) Correlation heatmap between HMRS scores and key pathway activities. Red and blue indicate positive and negative correlations respectively. (D) Survival analysis of six key pathways. Patients divided into high activity (red line) and low activity (blue line) groups based on pathway activity scores. (E) Forest plot of pathway hazard ratios. Shows degree of impact and 95% confidence intervals of each pathway on prognosis.
Gene Set Variation Analysis (GSVA) further revealed risk stratification-specific signaling pathway activity characteristics (Figure 7B). The high-risk group showed significant activation of TGF Beta Signaling, Mitotic Spindle, and IL6 JAK STAT3 Signaling; while the low-risk group was characterized by Pancreas Beta Cells, Oxidative Phosphorylation, and KRAS Signaling DN. Correlation analysis between HMRS scores and these pathway activity scores further validated these findings (Figure 7C, P < 0.05).
To assess the clinical prognostic significance of key pathways, we selected six most significant signaling pathways for survival analysis, including Epithelial Mesenchymal Transition, Angiogenesis, Glycolysis, Apoptosis, Coagulation, and IL2 STAT5 Signaling. Kaplan-Meier analysis showed that high activity in these pathways was significantly associated with poorer overall survival (Figure 7D, all P < 0.001). Hazard ratio (HR) analysis further confirmed these outcomes and found that the role of Hedgehog signaling pathway as the sole protect prognostic factor (Figure 7E).
3.7 Analysis of somatic mutation spectrum and tumor heterogeneityTo deeply understand the genomic characteristics of LGG patients, we conducted systematic analysis of histone modification gene mutation patterns and tumor heterogeneity. Using MATH (Mutant-Allele Tumor Heterogeneity) scores to quantify intratumoral heterogeneity levels, results showed significant difference between high score group and low score group (Figure 8A, p < 0.001). Survival analysis based on MATH scores indicated that low MATH scores were significantly associated with poorer prognosis (Figure 8B, p = 0.005). Further analysis integrating MATH scores with HMRS risk stratification showed that the “low risk + low MATH” subgroup had the most favorable prognosis (Figure 8C, p < 0.001).
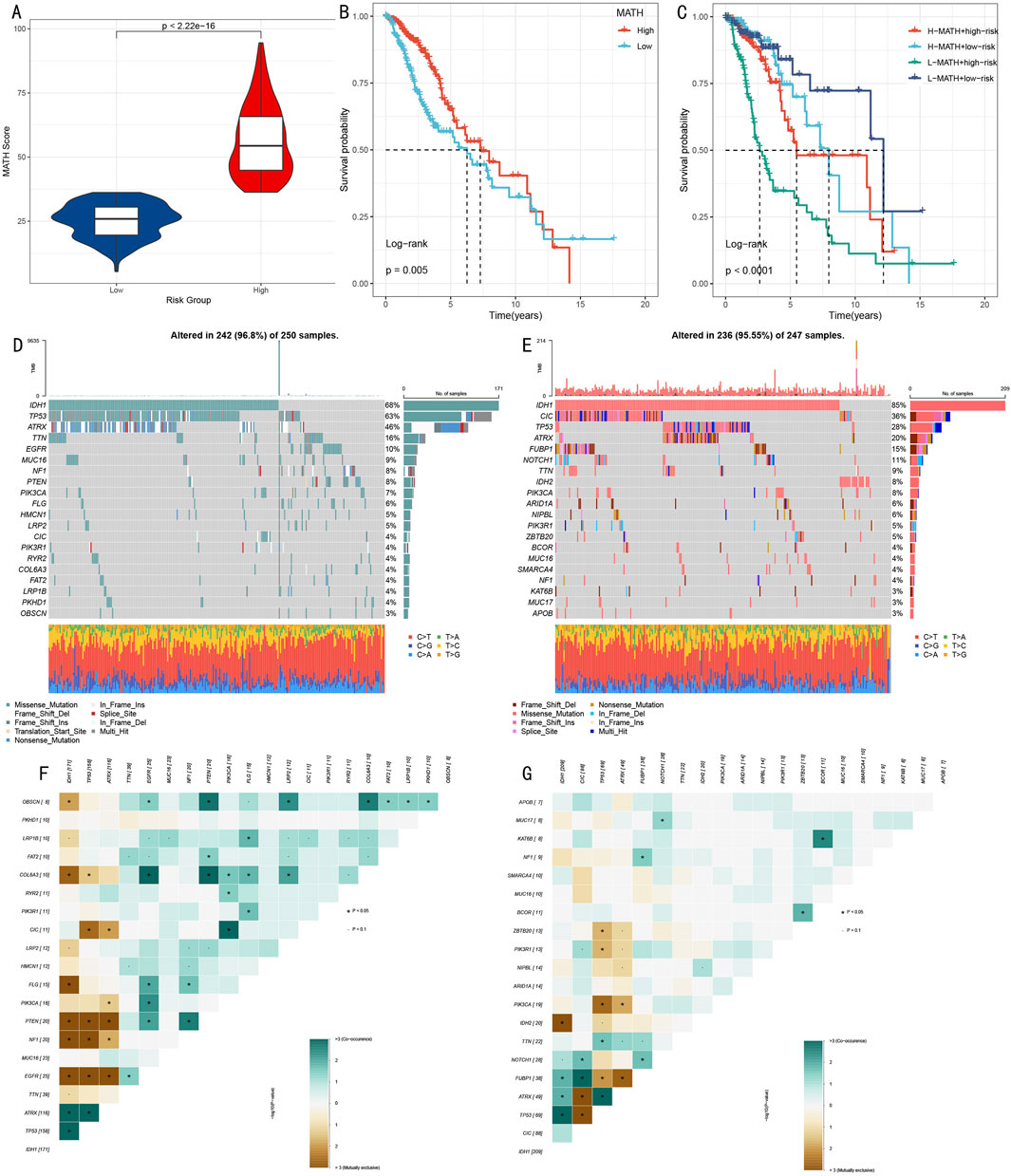
Figure 8. Multi-dimensional Analysis of Somatic Mutation Spectrum and Tumor Heterogeneity. (A) Box plot comparison of MATH scores between high and low score groups. (B) Kaplan-Meier survival analysis based on MATH scores. (C) Survival analysis combining MATH scores and risk scores. (D, E) Mutation landscape waterfall plots showing top 20 mutated genes in high-risk (D) and low-risk (E) groups. (F, G) Mutation gene co-occurrence/mutual exclusivity relationship heatmaps for high-risk (F) and low-risk (G) groups.
Through systematic analysis of mutation landscapes in high and low-risk groups (Figures 8D, E), we found: 1) TP53, as a key tumor suppressor gene, had a mutation frequency of 63% in the high-risk group, significantly higher than 28% in the low-risk group; 2) The transcription repressor CIC had a mutation frequency of 4% in the high-risk group compared to 36% in the low-risk group; 3) IDH1 mutations, associated with specific cytogenetic abnormalities and 1p/19q codeletion, showed mutation frequencies of 68% and 85% in high and low-risk groups respectively; 4) ATRX gene, involved in transcriptional regulation and chromatin remodeling, had mutation frequencies of 46% and 20% in high and low-risk groups respectively; 5) Additionally, characteristic mutations in the high-risk group included TTN (16%), while the low-risk group included FUBP1 (15%).
Mutation co-occurrence analysis (Figures 8F, G) revealed that in the high-risk group, TP53 mutations significantly co-occurred with IDH1 and ATRX. In the low-risk group, besides observing co-occurrence patterns of TP53, IDH1, and ATRX, significant mutation co-occurrence characteristics were also found between COL6A3 and PTEN.
3.8 Analysis of immune microenvironment characteristics and model associationsWe conducted multi-dimensional analysis of the immune microenvironment in high and low-risk groups. ESTIMATE algorithm assessment results showed (Figures 9A–C) that the high-risk group had significantly higher stromal scores, immune scores, and overall scores than the low-risk group (p < 0.001), suggesting more active immune responses and stromal components in the high-risk group.
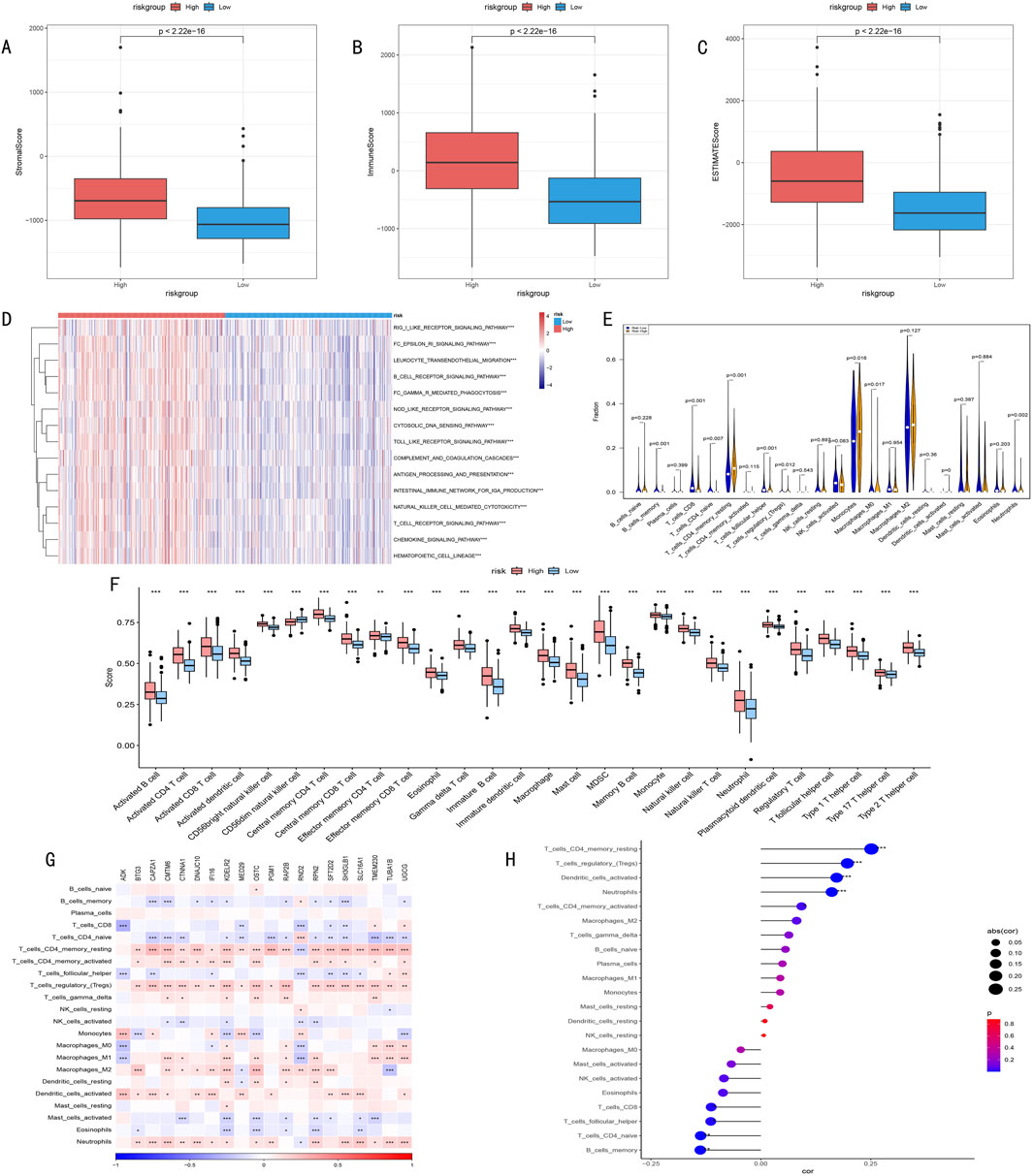
Figure 9. (A–C) ESTIMATE algorithm assessment of immune microenvironment differences between high and low-risk groups. (A) Stromal Score reflects tumor stromal components; (B) Immune Score quantifies immune cell infiltration levels; (C) ESTIMATE Score comprehensively characterizes tumor microenvironment features. (D) Heatmap of differential immune-related pathway activities between high and low-risk groups identified by ssGSEA algorithm, red indicates pathway upregulation, blue indicates pathway downregulation, color intensity represents degree of difference. (E) Violin plot of infiltration proportion differences of 22 immune cells between high and low-risk groups quantified by CIBERSORT algorithm. Shows distribution characteristics, density, and significant differences of each immune cell type. (F) Box plot of abundance differences of 28 characteristic gene-defined infiltrating immune cell types between high and low-risk groups. Box shows interquartile range, whiskers show 1.5 times interquartile range, outliers shown separately. (G) Correlation heatmap between risk score-related gene expression levels and various immune cell infiltration degrees. Red indicates positive correlation, blue indicates negative correlation, color intensity represents correlation strength. (H) Correlation scatter plot between risk scores and key immune cell infiltration levels. Point size represents absolute value of correlation coefficient, color indicates correlation direction and statistical significance.
Furthermore, ssGSEA algorithm analysis revealed all 15 significantly different immune-related pathways between high and low-risk groups (Figure 9D).
CIBERSORT algorithm analysis of immune cell infiltration characteristics showed (Figures 9E, F): 1) Memory CD4+ T cells were significantly higher in the high-risk group (P < 0.001); 2) Memory B cells, CD8+ T cells, and follicular helper T cells were more abundant in the low-risk group (P = 0.001); 3) Except for CD56dim NK cells, other immune cells generally showed higher expression levels in the high-risk group.
Correlation analysis between key gene expression and immune cell infiltration (Figure 9G) revealed that histone modification genes might participate in LGG progression by regulating immune cell infiltration. Correlation analysis between risk scores and immune cell infiltration (Figure 9H) indicated: 1) Significant positive correlations with memory CD4+ T cells, regulatory T cells, dendritic cells, and neutrophils; 2) Significant negative correlations with naive T cells and memory B cells. These findings suggest that HMRS can effectively quantify the immune status of LGG patients, reflecting significant immune landscape differences among patients with different risk levels.
3.9 Drug sensitivity analysisBased on the risk score model, we predicted sensitivity differences to common drugs between high and low-risk groups. Through comparison of IC50 values (Figure 10), significant response differences were found for the following drugs: Temozolomide (A), Pazopanib (B), Paclitaxel (C), Rapamycin (D), Sorafenib (E), and Gefitinib (F). These findings provide important references for risk stratification-based individualized medication.
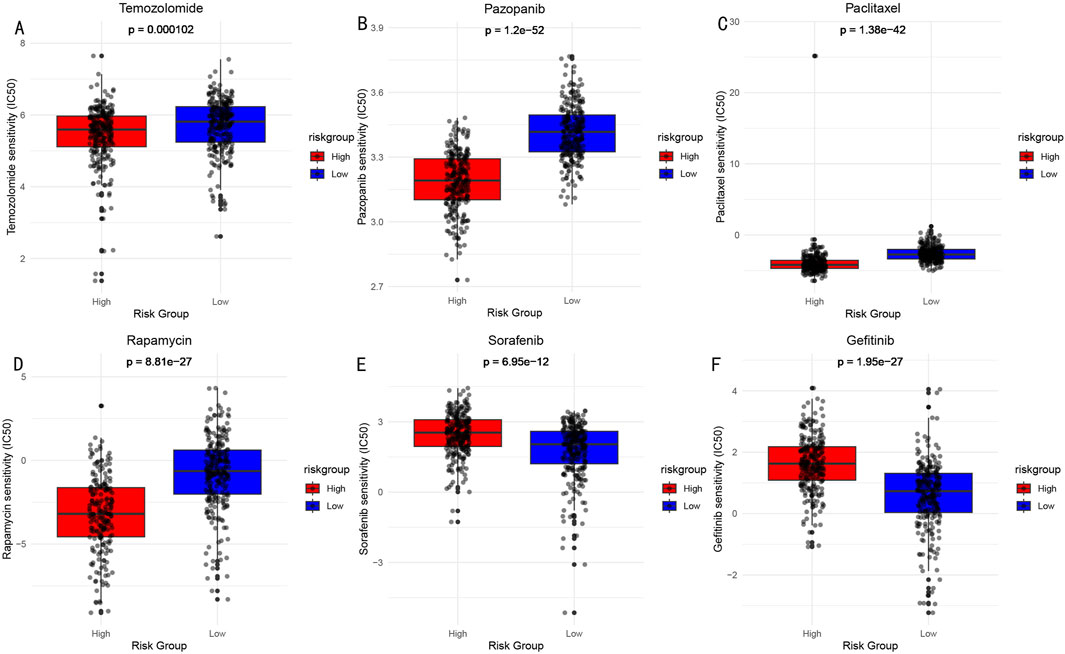
Figure 10. Drug Sensitivity Analysis (IC50 values) Between High and Low-risk Groups. (A) Temozolomide: DNA alkylating agent, widely used chemotherapy drug for brain glioma treatment. (B) Pazopanib: Multi-target tyrosine kinase inhibitor, used for treatment of various solid tumors. (C) Paclitaxel: Microtubule protein inhibitor, classic anti-tumor chemotherapy drug. (D) Rapamycin: mTOR pathway inhibitor, with immunosuppressive and anti-tumor effects. (E) Sorafenib: Multi-target tyrosine kinase inhibitor. (F) Gefitinib: EGFR tyrosine kinase inhibitor.
3.10 HPA validation analysisTo further validate the expression characteristics of key genes in the risk model, we selected five genes with the highest weight coefficients (ADK, UGCG, RPN2, CAPZA1, KDELR2) for HPA database immunohistochemical validation (Figure 11). Results showed that all these genes exhibited significantly high expression in LGG tissue.
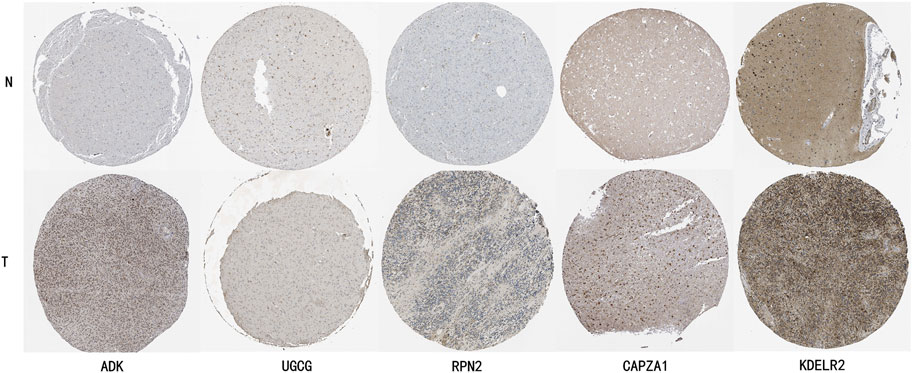
Figure 11. HPA Database Immunohistochemical Validation of Key Gene Expression Characteristics in LGG Tissue. ADK (Adenosine Kinase): Normal brain tissue shows light gray weak positive expression, while glioma tissue shows deep brown moderate to strong positive staining, mainly localized in cytoplasm. UGCG (UDP-Glucose Ceramide Glucosyltransferase): Normal brain tissue shows faint staining with almost no expression, glioma tissue shows obvious brown positive staining in cell membrane and cytoplasm. RPN2 (Ribophorin II): Normal brain tissue shows uniform light gray weak expression, glioma tissue shows uneven dark strong positive staining, mainly localized in endoplasmic reticulum. CAPZA1 (F-Actin-Capping Protein Subunit Alpha-1): Normal brain tissue shows weak expression, glioma tissue shows obvious brown moderate to strong positive staining, distributed in cytoplasm. KDELR2 (KDEL Endoplasmic Reticulum Protein Retention Receptor 2): Normal tissue shows light brown weak expression, glioma tissue shows obvious deep brown strong positive expression, mainly localized in Golgi apparatus.
3.11 Pan-cancer analysisOur pan-cancer analysis revealed distinct survival patterns across different cancer types and organ systems (Figure 12). Among the 29 cancer types analyzed, four cancer types demonstrated statistically significant associations with survival outcomes (p < 0.05). In the Urinary System, both Kidney Renal Clear Cell Carcinoma (KIRC, HR = 1.72, p = 0.00016) and Kidney Renal Papillary Cell Carcinoma (KIRP, HR = 2.87, p = 0.00077) showed significantly higher risk in the high-risk group. Within the Respiratory System, Lung Adenocarcinoma (LUAD) exhibited significantly poorer outcomes (HR = 1.57, p = 0.0014). Additionally, in the others category, Sarcoma (SARC) demonstrated significantly worse survival (HR = 1.59, p = 0.024).
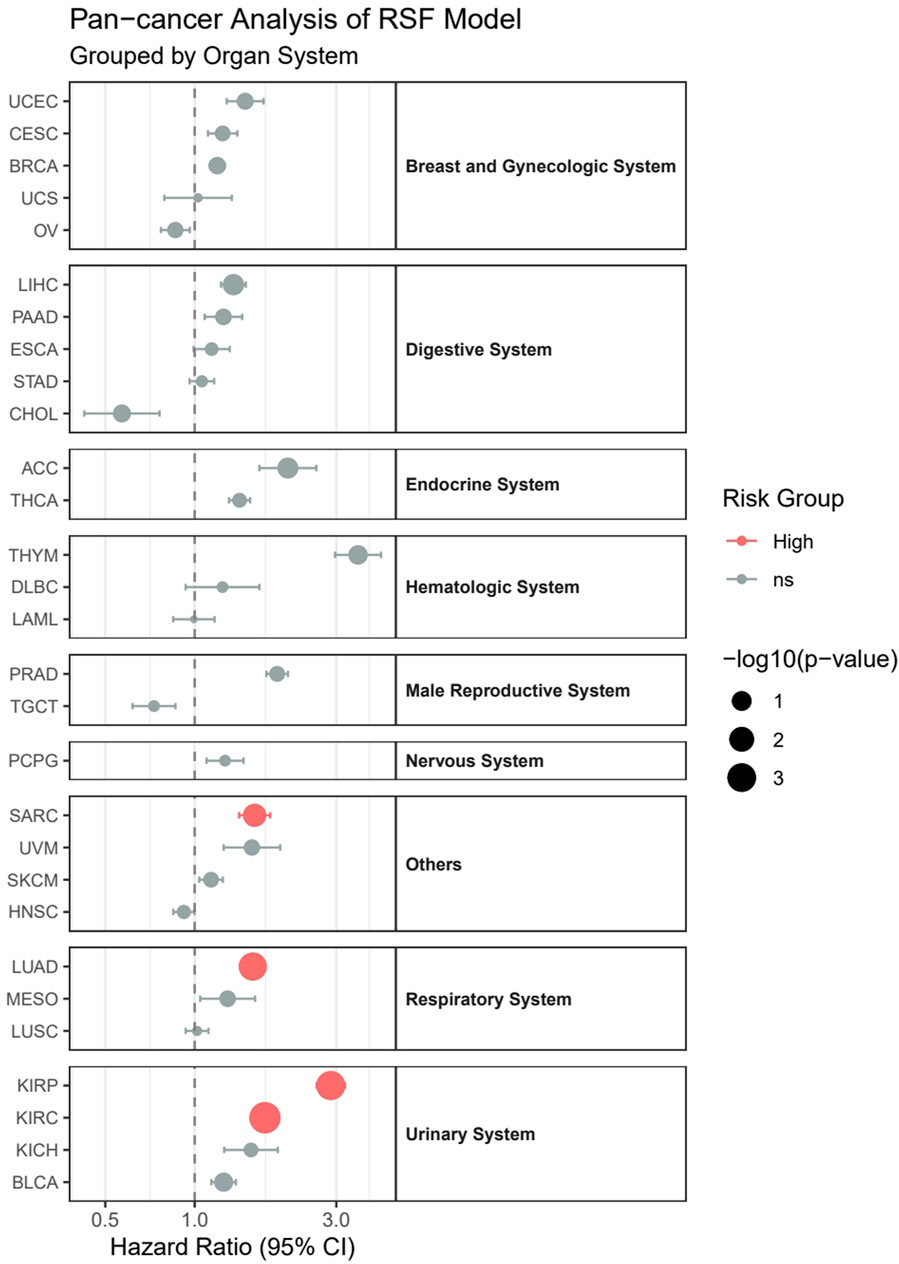
Figure 12. Pan-cancer analysis of survival outcomes across 29 cancer types stratified by organ systems.
Although not reaching statis
留言 (0)