Triple negative breast cancer (TNBC) is a subtype of breast cancer that lacks estrogen receptor (ER), progesterone receptor (PR), and HER2 expression (Lee et al., 2020). The majority of TNBC shows the expression of basal markers along with a smaller fraction that lacks the expression of basal markers and is called non-basal-like (Sahoo et al., 2024). Surgery, radiotherapy, chemotherapy, and immunotherapy are the treatment options for TNBC patients. However, TNBC treatment options are limited due to its unresponsiveness to hormone therapy and anti-HER2 therapy (Zagami and Carey, 2022; Agostinetto et al., 2022). TNBC is recognized for its heterogeneity, extremely aggressive onset as well as high occurrence of metastasis, highlighting an urgent need for targeted therapies.
Recently, ion channels have emerged as a potential target in cancer therapy (Capatina et al., 2022). Ion channels are imperative for maintaining the control of membrane potential, cell signaling, and the movement of ions necessary for cellular functions. Abnormal expression or function of these channels can lead to uncontrolled cell division, a primary hallmark of cancer (Capatina et al., 2022; Leanza et al., 2016). Voltage gated Cl− channels play a role in cell cycle regulation, cell proliferation, migration, and apoptosis in many cancer cells (Rao et al., 2015). However, not many groups have specifically investigated the expression and function of ligand-gated ion channel (LGIC) receptors in TNBC. The Cys-loop LGIC family of receptors are expressed as pentameric membrane-bound receptors of multiple subtypes which have unique pharmacological properties (Alexander et al., 2017). Moreover, amino acid transmitters such as GABA (gamma-amino butyric acid) which endogenously activate the GABA type A receptor (GABAAR) LGIC can also act as an energy source for cancer cells via direct conversion to TCA cycle intermediates (Ravasz et al., 2017). Activation of GABAAR mediates hyperpolarization via Cl− ion influx in the adult central nervous system (CNS), promoting inhibitory neurotransmission (Knoflach et al., 2016). GABAAR have been shown to be expressed in many peripheral cancers (Bhattacharya et al., 2021). We recently showed that pharmacological antagonism and genetic knockdown of GABAA β3 subunit decreases TNBC proliferation and migration via decreased cyclin D1 expression and increased p21 expression, combined with cell cycle arrest in the G0/G1 phase (Bundy et al., 2024). Similarly, the GABAAR π subunit is implicated in pancreatic cancer, and α3 GABAAR subunit shows overexpression in HER2+ breast cancer (Juvale et al., 2021; Gumireddy et al., 2016; Takehara et al., 2007). GABAAR π has been shown to stimulate breast cancer cell invasion through the ERK1/2 pathway (Sizemore et al., 2014) and it also interacts with EGFR and sustains EGFR expression in TNBC (Li et al., 2021). However, the functionality of such overexpressed GABAAR subunits in peripheral cancers outside the brain is understudied. With respect to brain cancers, benzodiazepine analogs have been shown to induce Cl− efflux from the medulloblastoma cells, depolarizing their mitochondria and inducing fission (Kallay et al., 2019). Little is known about the cell-surface expression and functional status of the upregulated GABAAR in cancers outside the brain, i.e. if they mediate Cl− ion flux. Understanding the mechanisms by which upregulated GABAAR function and mediate Cl− flux in TNBC could identify new approaches to target TNBC, since ion flux (e.g., Ca2+, K+) can alter tumor growth and metastasis (Prevarskaya et al., 2018). Cl− ions play a role in cell-cycle progression in gastric cancer; wherein low [Cl−] levels induce G1 cell cycle arrest and upregulation of p21 (Shiozaki et al., 2011). Therefore, we wanted to investigate whether GABAAR overexpressed in TNBC are expressed on the cell-surface and if they mediate Cl− flux. Elucidating cell-surface expression is important since membrane-bound receptors are easier to target with small molecules and antibody-based approaches. Understanding the nature of GABAAR mediated Cl− flux in TNBC can further help in mapping out how Cl− flux may contribute to increased proliferation and migration in TNBC.
To address these questions, we employed a panel of TNBC and non-tumorigenic MCF-10A cell lines. We employed surface biotinylation experiments to study GABAAR localization and N-(Ethoxycarbonylmethyl)-6-methyl quinolinium bromide (MQAE) fluorescence quench assays to assess function and direction of GABAAR-mediated Cl− flux. Our results indicate that α1 and β3 subunit containing GABAAR in TNBC cell lines are localized on the cell surface and are functionally active, mediatng GABA-mediated Cl− influx. Further, employing TNBC cells with GABAA β3 subunit knockdown, we show that GABAAR mediated Cl− influx is attenuated after knockdown. Moreover, GABAAR antagonist bicuculline (BC) decreased cell proliferation in TNBC cells.
MethodsCell cultureAll cell lines were obtained from ATCC. MCF-10A cells were cultured in mammary epithelial basal medium (MEBM) (Lonza, MD) supplemented with 5% horse serum (Invitrogen), 20 ng/mL epidermal growth factor (Lonza), 0.5 mg/mL hydrocortisone (Lonza), 10 μg/mL insulin (Sigma-Aldrich, MO), 100 ng/mL cholera toxin (Sigma-Aldrich), and 1% penicillin/streptomycin (ThermoFisher Scientific, MA). MDAMB231 cells were cultured in Dulbecco’s modified eagle medium (DMEM) (ThermoFisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (ThermoFisher Scientific), 1% sodium pyruvate (ThermoFisher Scientific), and 1% penicillin/streptomycin (ThermoFisher Scientific). BT-549 cells were cultured in RPMI-1640 growth medium (ATCC, VA) supplemented with 10% FBS, 0.023 IU/mL bovine insulin, and 1% penicillin/streptomycin. HCC1806 cells were cultured in RPMI-1640 growth medium supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were incubated at 37°C in a 5% CO2 incubator.
Cell proliferation MTS assay2.5 × 104 cells were plated on 96 well plates and incubated overnight at 37°C, 5% CO2. Cells were treated with GABAAR antagonist BC for 48 h to assess effects of pharmacological inhibition of GABAAR. 20 μL of CellTiter 96® AQueous One Solution Reagent containing a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] (Promega, WI) was added to each well, incubated at 37°C, 5% CO2. Absorbance was read on a plate reader at 490 nm.
Lentiviral Mediated Knockdown of GABAAR subunit in TNBC Epithelial Cells: As described earlier (Bundy et al., 2024), to knockdown the GABAA β3 subunit, TNBC cells (HCC1806 and BT 549) were cultured in appropriate complete medium until cells were 50% confluent. Medium was replaced with polybrene (5 μg/mL) containing medium to increase transduction efficiency. Cells were infected with transduction-ready scramble control shRNA lentiviral particles (#TR30021V, Origene, MD) or human GABAAR shRNA lentiviral particles (constructs # 2 and 3 targeting the GABAA β3 gene, #TL304428V, Origene) for 24 h at multiplicity of infection (MOI) of 5. Constructs 2 and 3 were chosen out of 4 unique constructs since they led to the highest GABAA β3 protein knockdown in HCC1806 and BT549 cells (Bundy et al., 2024). Stably transduced cells were selected with puromycin (1–2.5 μg/mL).
Cell-surface biotinylationThe protocol followed was based on a cell surface protein biotinylation protocol we employed previously (Bundy et al., 2024). Briefly, MCF-10 A and TNBC cell lines were incubated with membrane-impermeable biotin, lysed with lysis buffer. Biotinylated proteins were pulled down with neutrAvidin Ultralink beads and analyzed via western blotting. GAPDH was used as a loading control and as a cytosolic marker, thus GAPDH signal was only detected in the ‘input’ and not ‘pulldown’ samples. ABCB1 expression, also known as multidrug resistance 1 (MDR1), was used as a positive control to confirm detection of a membranous protein.
Western blottingWestern blotting was carried out as previously described (Bundy et al., 2024). Briefly, protein samples (20 μg) were separated by SDS-PAGE and transferred to a PVDF membrane for probing and blocked in TBS-Tween supplemented with 5% nonfat dry milk for 1 h at room temperature (RT). The membranes were incubated with these primary antibodies: GABAA β3 (1:1,000, #73–149, RRID: AB_2109585, Antibodies Inc., CA), GABAA ɑ1 (1:1,000, #75–136, RRID: AB_2108811, Antibodies Inc.), ABCB1 (ABCB1 (1:1,000, #12683, RRID: :AB_2715689 Cell Signaling) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:20,000, #10R-2932, RRID: AB_11199818, Fitzgerald, MA) antibody as the loading control. Each blot was incubated with the respective dilution of primary antibody overnight with the exception of GAPDH antibody which was incubated for 1 h. IRDye 680RD secondary antibodies (1:10,000, LI-COR BioSciences, NE) were used to visualize bound primary antibodies. The Odyssey CLx Imaging System (LI-COR BioSciences) was utilized for near-infrared fluorescent detection of proteins. Image Studio software on the Odyssey CLx was used to carry out densitometry analysis (LI-COR BioSciences).
Preparation of buffer for MQAE fluorescence assaysWe employed a Cl−free HEPES buffer containing 10 mM HEPES, 95 mM Sodium Nitrate, 2.5 mM Potassium Nitrate and 1.8 mM Calcium Nitrate since NO3− in the range between 0 and 100 mM does not quench MQAE fluorescence (Hiroshi Kaneko et al., 2002; Rocha-Gonzalez et al., 2008). This complete HEPES buffer was then adjusted to pH of 7.4 with 0.1 M sodium hydroxide and osmolarity of 310 mOsm/kg and 1 M sucrose, respectively. For double ionophore calibration experiments, the K+/H+ antiporter nigericin (5 μM) was added to this HEPES buffer to remove H+ and OH− gradients, and the Cl−/OH−antiporter tributyltin (10 μM) was added to equalize Cl− gradients; i.e. to ensure that the intracellular [Cl−] was equal to the extracellular [Cl−]. The choice of antiporter concentrations was guided by previously published studies (Krapf et al., 1988; Hiraoka et al., 2010).
MQAE loading2.5 × 105 cells/well were seeded into 24 well plates and left overnight to adhere. The next day, cells were washed with HEPES buffer three times. Cells were then loaded with 5 mM N-(Ethoxycarbonylmethyl)-6-methyl quinolinium bromide (MQAE), a halide-sensitive dye (ThermoFisher Scientific) in HEPES buffer for 90 min at 37°C and 5% CO2. Cells were washed again three times with HEPES buffer to remove extracellular MQAE and exposed to respective treatments and controls explained in the methods below (Cl− calibration and GABAAR mediated Cl− flux). Fluorescence of MQAE was read on a plate reader (BioTek Synergy Neo2 multimode microplate reader, ThermoFisher Scientific) at an excitation wavelength of 350 nm and emission wavelength of 460 nm.
Cl− calibrationAfter MQAE loading incubation and HEPES buffer washes, cells were exposed to various concentrations of potassium chloride (KCl) (0–80 mM) in HEPES buffer with double ionophores (5 μM nigericin and 10 μM tributyltin to equalize Cl− gradients) to assess MQAE fluorescence quenching. The emitted MQAE fluorescence intensity is inversely related to the [Cl−] of the MQAE-containing solution due to quenching by a collisional mechanism with a linear relation as described by the Stern Volmer equation: F0/Ft = 1 + Ksv [Cl−] (Krapf et al., 1988).
MQAE fluorescence assay to assess GABAAR-mediated intracellular Cl− changesBased on Cl− calibration experiments, cells were loaded with MQAE and incubated with HEPES buffer with 60 mM KCl. Cells were exposed to various concentrations of GABA (0–3,000 μM) in HEPES buffer with 60 mM KCl to assess fluorescence quenching. Relative Cl− flux was calculated by the Stern–Volmer equation, where F0 is the initial fluorescence before GABA treatment, and Ft is the final fluorescence after GABA treatment. EC50 values were determined by plotting a dose-response curve and interpolating the concentration at which the response reaches 50% of its maximum effect.
To assess pharmacological GABAAR inhibition of Cl− flux, respective wells were pre-treated with GABAAR antagonist 10 μM BC for 15 min to block receptor activity before 100 μM GABA ligand was added to the respective wells. Dosing of BC was chosen based on the cell viability data (Figure 4). Next, to assess pharmacological positive allosteric modulation of GABAAR, 7 μM diazepam was added to respective wells simultaneously with 5 μM GABA. Dosing for BC, diazepam and GABA was chosen based on well-established prior research on GABAAR in the CNS field (Wongsamitkul et al., 2017). Fluorescence quenching was assessed via the plate reader, and relative Cl− flux was calculated using the Stern–Volmer plots of fluorescent ratios versus concentration of the quencher, where F0 is the initial fluorescence before treatment, and Ft is the final fluorescence in the presence of the quencher (Motz et al., 2023).
MQAE fluorescence assay to assess GABAAR-mediated intracellular Cl− flux after GABAA β3 subunit knockdownHCC 1806 and BT 549 cells that have undergone GABAA β3 lentiviral mediated knockdown as described previously were employed (Bundy et al., 2024). Knockdown cells (Scramble control, GABAA β3 KD Constructs # 2 and 3) were washed with HEPES buffer, loaded with MQAE, washed with HEPES again, and then exposed to HEPES buffer with 60 mM KCl and various concentrations of GABA (100–1,000 uM). Fluorescence quenching was assessed as described above.
Cell viability assayCell viability experiments were performed as described in our previous study (Bundy et al., 2024). Cells were treated with BC at various concentrations (0–300 uM) for 48 h. As compared to Bicuculline Methiodide (BCM), BC does not have a quaternary ammonium charge and therefore, it is blood brain barrier permeable. In previous experiments (Bundy et al., 2024), BCM was used due to its blood brain barrier impermeability, making BCM suitable for future in vivo studies. We chose BC here in these experiments since BC is the parent compound and has a higher affinity for GABAAR and is better suited to study pharmacological inhibition of GABAAR (Johnston, 2013). Absorbance of the CellTiter 96® AQueous One Solution Reagent containing a tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3- carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] (Promega, WI) was read at 490 nm. IC50 values were determined by plotting a dose-response curve and interpolating the concentration at which the response is reduced by 50% compared to the control.
Statistical analysist-test, one-way ANOVA followed by a Tukey post-hoc test in GraphPad Prism 8.0 (Boston, MA) were employed as needed. p < 0.05 was considered significant.
ResultsBiotinylation suggests that β3 and α1 subunits of GABAAR are localized on the cell surfaceCell surface biotinylation assays were performed in MCF 10A cells and TNBC cells to confirm localization. Results confirm that the GABAA β3 and α1 subunits are located on the cell surface in MCF10A cells and TNBC cells indicating that the GABAAR is membranous (Figure 1). Consistent with western blotting results with whole cell lysates reported by us, GABAA α1 and β3 subunit expression is significantly higher in HCC 1806 and BT 549 cell lines as compared to MCF10A cell lines (Bundy et al., 2024).
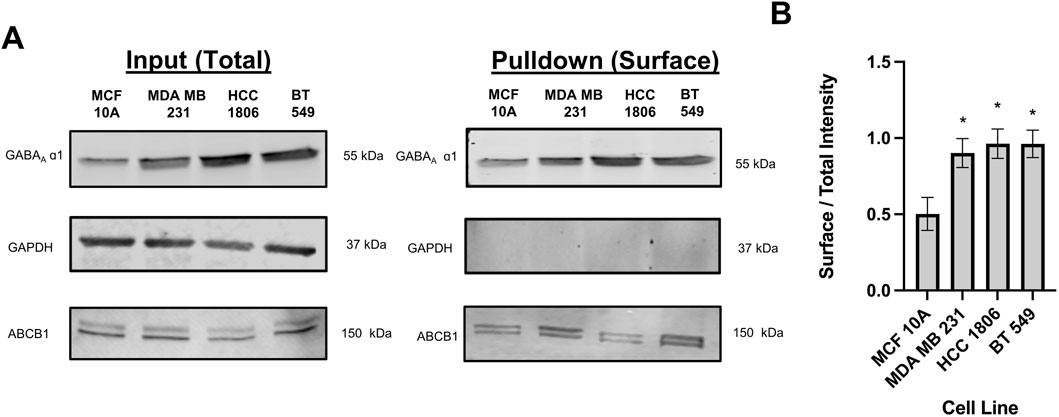
Figure 1. Biotinylated protein from TNBC cells and MCF10A cells show that GABAAR α1 subunit is localized on the cell surface. (A) GABAA α1 subunit protein expression in whole cell samples (input) versus biotinylated samples (pulldown) in TNBC cells and MCF10A cells showing that GABAAR α1 subunits are localized on the cell surface, n = 3. GAPDH is a cytosolic marker shown as loading control. ABCB1 protein expression was used as a positive control to confirm the detection of a membrane-bound protein in the samples. (B) Corresponding densitometry of the ratio of pulldown to input samples across MCF10A and TNBC cell lines. * represent significance compared to MCF10A. Data are presented as mean±SE, *p < 0.05 (ANOVA).
MQAE dye shows no significant change in fluorescence over time indicating no dye leakage during experimentsDye leakage experiments were done to test whether MQAE dye leakage was a contributing factor in the change in fluorescence. Results show that there is no detectable change in fluorescence over 30 min of time in MCF 10A cells and TNBC cells indicating that no detectable dye leakage occurs during this time and therefore leakage does not contribute to fluorescence changes in MQAE experiments (Figure 2A).
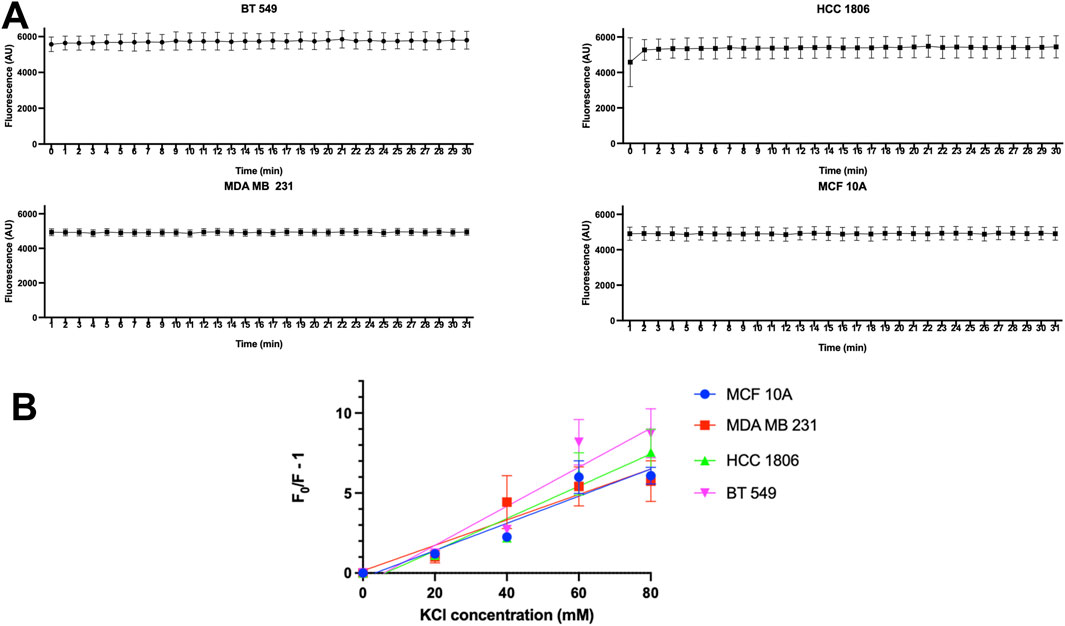
Figure 2. MQAE fluorescence intensity indicates no major dye leakage in TNBC cells and MCF10A cells and Stern–Volmer plots. (A) Fluorescent intensity of MQAE of MCF 10A, MDA MB 231, HCC 1806 and BT 549 cells over time, n = 3. (B) Stern–Volmer plots for MCF 10A, MDA MB 231, HCC 1806 and BT 549 cells indicate that the KCl concentration used as a constant in MQAE Cl-flux experiments is 60 mM, n = 3. Data are presented as mean±SE (ANOVA).
Relationship between MQAE fluorescence quench and [Cl−]i in TNBC and MCF10A cellsTo convert the fluorescence intensity into the [Cl−]i, the relationship between intracellular MQAE fluorescence and [Cl−]i was determined using the double ionophore technique. MCF10A and TNBC cells were exposed to various concentrations of KCl (0–80 mM) as the quencher of MQAE fluorescence. The Stern–Volmer plot showing the quench in fluorescence intensity against KCl concentrations is shown in Figure 2B, indicating a KSV of 5.56 M-1, 6.77 M-1, 7.32 M-1, 8.56 M-1 for MCF 10A, MDA MB 231, HCC 1806, and BT 549 cells, respectively. These Ksv values reflect Cl−sensitivity of MQAE and fall within the range of Ksv values (5–25 M-1) in previous studies with MQAE in neurons (Weilinger et al., 2022; Kaneko et al., 2004).
MCF 10A cells and TNBC cells exposed to GABA ligand show a concentration-dependent increase in Cl− ion influxNon-tumorigenic MCF 10A cells and TNBC cells were exposed to increasing concentrations of GABA to assess GABAAR function and the direction of ion flux when activating the receptor. Results indicate a significantly higher quench in fluorescence in TNBC cells as compared to MCF10A cells indicating that the GABAAR is functional and expressed at a higher level in TNBC cells. Additionally, there is an increase in intracellular Cl− concentration in all cell lines, indicating that the directionality of Cl− ions in the GABAAR is influx under these experimental conditions (Figure 3A). As shown by GABA dose-response curves, relative Cl− influx efficacy shows the rank of BT549>HCC1806>MDA MB231>MCF10A cells. These data are in agreement with surface biotinylation data where GABAA α1 and β3 show the highest expression in BT549 and HCC1806 cells as compared to MCF10A cells. Calculated EC50 values for GABA in MCF10A and 3 TNBC cell lines are shown in Figure 3B.
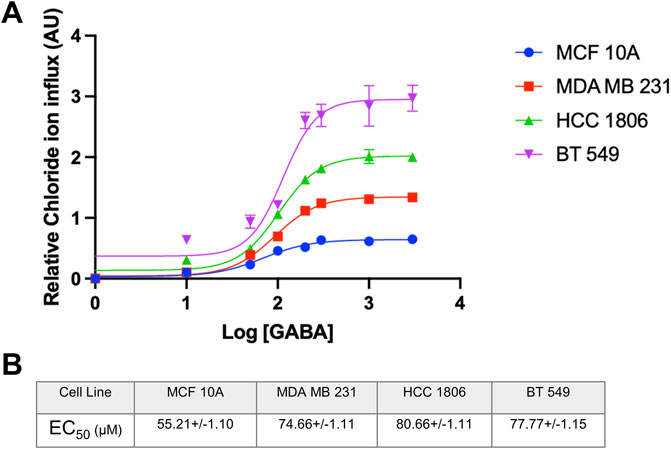
Figure 3. EC50 of GABA ligand concentration and relative Cl-concentration in TNBC cells and MCF 10A cells. (A) EC50 curve of GABA ligand concentration on MCF 10A, MDA MB 231, HCC 1806, and BT549 cells n = 3. (B) EC50 values of GABA (µM) interpolated from the curve for each cell line, respectively. Data are presented as mean±SE (ANOVA).
GABAAR-mediated intracellular Cl− flux is pharmacologically modulated by GABAAR ligandsIn order to characterize pharmacological properties, MCF10A and TNBC cells were exposed to well-established GABAAR modulators-the antagonist BC, and the positive allosteric modulator, Diazepam. When exposed to BC, Cl− influx decreased in BT 549 and HCC 1806 cells and showed no significant change in MCF 10A cells (Figure 4A). When exposed to Diazepam, MCF 10A cells and TNBC cells resulted in an increase in Cl− influx (Figure 4B). Moreover, BT 549 cells showed a significantly higher level of intracellular Cl− than MCF 10A and HCC 1806 cells.
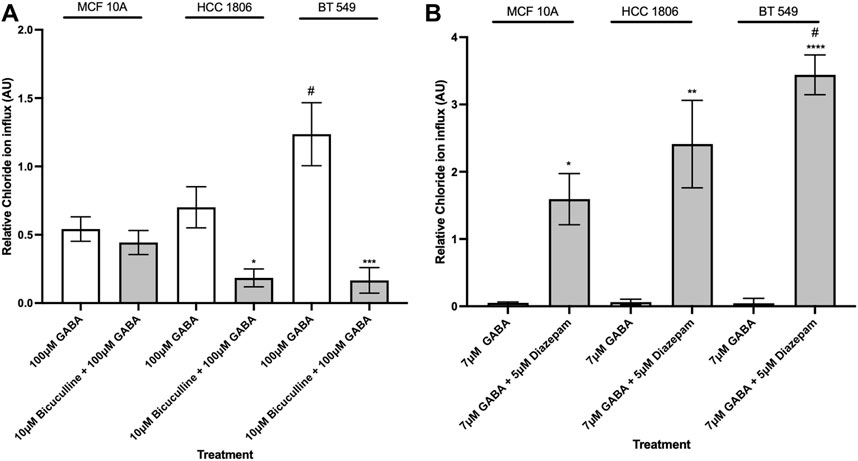
Figure 4. Intracellular Cl− in TNBC cells and MCF 10A cells decreases when exposed to GABAAR competitive antagonist and increases when exposed to GABAAR positive allosteric modulator. (A) Relative Cl− in TNBC cells and MCF 10A cells exposed to 100 µM GABA alone and in combination with 10 µM GABAAR competitive antagonist, Bicuculline, n = 3. (B) Relative Cl− in TNBC cells and MCF 10A cells exposed to 7 µM GABA alone and in combination with 5 µM GABAAR positive allosteric modulator, Diazepam, n = 3. * represent significance compared to 7 µM GABA alone within each cell line, respectively. # represent significance compared to the HCC 1806 cell line. Data are presented as mean±SE, *p < 0.05, **p < 0.01, ***p < 0.001,****p < 0.0001 (ANOVA).
GABA mediated Cl− influx is attenuated in GABAAβ3 subunit knockdown cellsTNBC cells that have undergone GABAAβ3 subunit knockdown were exposed to GABA at various concentrations. HCC 1806 cells with GABAAβ3 subunit knockdown show a significant decrease in relative intracellular Cl− when exposed to 300 μM and 1,000 µM GABA (Figure 5A). BT 549 cells with GABAAβ3 subunit knockdown also show a significant decrease in relative intracellular Cl− when exposed to 100 μM, 300 μM, 1,000 µM GABA (Figure 5B). In addition, in BT 549 cells, Construct 2 and 3 show a more significant reduction of intracellular Cl− than HCC 1806 knockdown cells.
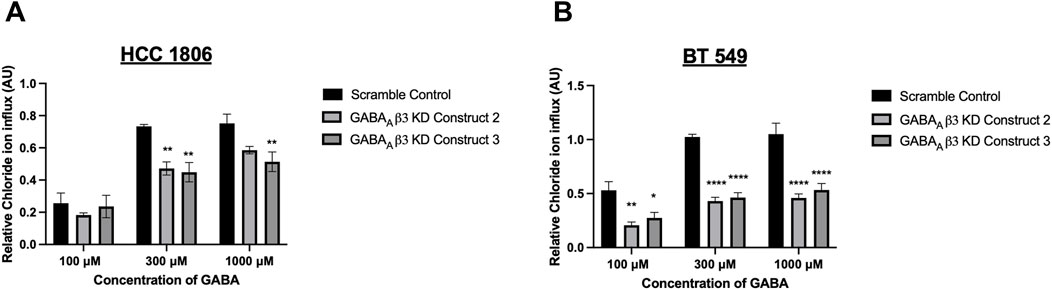
Figure 5. Relative GABAAR-mediated Cl− flux in TNBC cells decreases after GABAAβ3 subunit knockdown. (A) Relative Cl− in HCC 1806 cells that have undergone GABAAβ3 subunit knockdown (scramble and KD constructs 2,3) exposed to 100 μM, 300 μM, and 1,000 μM GABA, n = 3. (B) Relative Cl− in HCC 1806 cells that have undergone GABAAβ3 subunit knockdown (scramble and KD constructs 2,3) exposed to 100 μM, 300 μM, and 1,000 μM GABA, n = 3. * represent significance compared to the scramble control within concentration of GABA, respectively. Data are presented as mean±SE, *p < 0.05, **p < 0.01, ****p < 0.0001 (ANOVA).
GABAAR antagonist BC significantly decreases cell viability in TNBC cells as compared to MCF 10A cellsNon-tumorigenic MCF 10A cells and TNBC cells were exposed to GABAAR antagonist BC at various concentrations. Results indicate that there is a significant decrease in cell viability in all TNBC cell lines at 5 µM (Figure 6A) as compared to MCF10A cells. IC50 values suggest that TNBC cell lines are more sensitive to BC as compared to MCF 10A cells (Figures 6B,C). Morphology of the cells exposed to BC are in Supplementary Section.
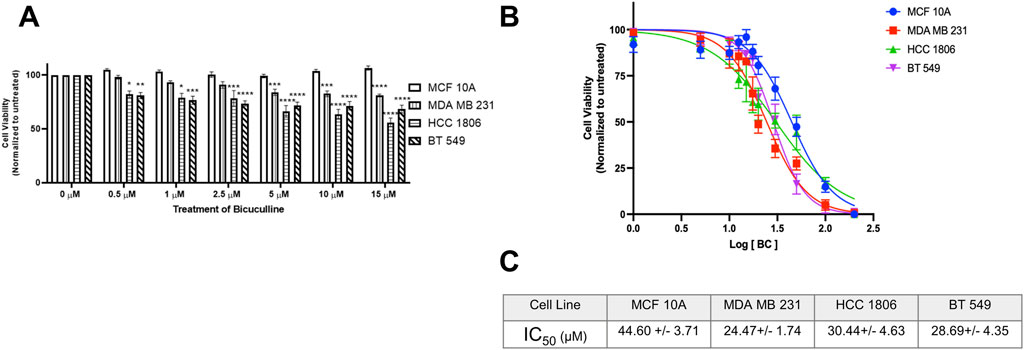
Figure 6. GABAAR competitive antagonist Bicuculline decreases cell proliferation in TNBC cells. (A) Cell viability of MCF 10A cells and TNBC cell lines exposed to Bicuculline for 48 h at various concentrations, n = 3. (B) Complete IC50 curve for Bicuculline in all TNBC cell lines (C) IC50 values of Bicuculline for all TNBC cell lines * represent significance compared to MCF10A within each concentration, respectively. Data are presented as mean±SE, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (ANOVA).
DiscussionOur previous study showed that GABAA β3 subunit plays a vital role in proliferation, migration and cell-cycle progression of TNBC cells (Bundy et al., 2024). To elucidate the mechanisms by which β3 subunit containing GABAAR mediate these effects in TNBC and to develop therapeutic approaches to target these GABAAR, confirming surface localization of GABAAR is imperative to deem the receptor as druggable in TNBC cells. Surface biotinylation experiments reported here confirm that GABAA α1 and β3 subunits, which are critical for forming the GABA-binding interface, are localized at the cell surface. We next investigated if these cell-surface GABAAR are functional and whether they mediate Cl− influx or efflux in TNBC cells. In the adult CNS, binding of GABA to GABAAR causes a conformational change, opening the ion channel, allowing Cl− ions to flow through (Lynagh and Pless, 2014). Our results suggest that GABA causes a concentration-dependent increase in Cl− influx leading to a fluorescence quench. This GABAAR-dependent Cl− influx occurs to a much greater degree in TNBC cells as compared to MCF10A cells. This functional rank efficacy for GABA observed here in TNBC vs. MCF10A cells also correlates with the rank order of α1 and β3 GABAA total protein levels (BT549>HCC1806>MDA MB231>MCF 10A) reported earlier by us (Bundy et al., 2024). The EC50 values for GABA obtained from our experiments are within the EC50 range of 6–106 μM established in literature from various expression systems (Hadingham et al., 1993; Karim et al., 2013; Baur and Sigel, 2003).
We next studied the modulation of GABAAR in cell lines using well-characterized GABAAR pharmacological modulators such as BC (GABAAR competitive antagonist) and Diazepam (GABAAR positive allosteric modulator). Diazepam is a classic benzodiazepine that binds to allosteric site at the α-γ interface of the GABAAR, causes a conformational change, enhancing GABA-mediated channel opening frequency and Cl− influx (Olsen, 2018). The competitive GABAAR antagonist, BC, binds to the α-β interface and inhibits the receptor, therefore inhibiting flow of Cl− ions (Johnston, 2013). The GABAAR-mediated Cl− influx was inhibited by BC and potentiated by diazepam, further supporting GABAAR function.
It is important to note that even though GABAAR activation typically results in Cl− influx in the adult CNS, this is not always the case, especially in brain cancers. For example, the α5 GABAA subunit is overexpressed in medulloblastoma, however the GABAAR in medulloblastoma show efflux of Cl− ions, contributing to mitochondrial depolarization, inducing mitochondrial fission and dysfunction (Kallay et al., 2019). On the other hand, little is known about GABAAR subunit composition and function in peripheral cancers where GABAAR overexpression has been detected (with α3, π GABAAR subunits). Many studies indicate the overexpression/knockdown of specific GABAAR subunits can affect cancer cells, but these studies do not investigate if these subunits form functional receptors (Gumireddy et al., 2016; Sizemore et al., 2014). Here, we show that in contrast to GABAAR in medulloblastoma, GABAAR overexpressed in TNBC cells mediate Cl− influx to a much higher extent than MCF10A cells. These observations allow us to further study how Cl− influx affects proliferation and migration of TNBC cells. These findings are also supported by results from TNBC cells that have undergone GABAAβ3 subunit knockdown which show a reduction in GABAAR-mediated Cl− influx as compared to cells treated with scramble control.
With respect to Cl− ions, intracellular Cl− accumulation via Na+, K+, 2Cl- (NKCC) cotransporter activity is implicated in glioma cells (Luo et al., 2020). It is also known that low intracellular Cl− can induce cell cycle arrest in the G1 phase in prostate cancer (Hiraoka et al., 2010). Additionally, several studies suggest that membrane hyperpolarization at the G1/S checkpoint is required for S phase initiation. For example, depolarizing the cell membrane halts G1/S progression in MCF-7 breast cells (Wonderlin et al., 1995). Therefore, we speculate that if Cl− ion influx is blocked via GABAAR inhibition or genetic knockdown, it could relatively depolarize the membrane potential of the TNBC cells, and halt cell cycle progression. Thus, our results show, for the first time, that GABAAR overexpressed in TNBC mediate Cl− influx. Moreover, pharmacological inhibition of this Cl− influx with BC also decreases TNBC cell viability, with TNBC cells showing a higher sensitivity to BC as compared to MCF10A cells. Therefore, decreasing intracellular Cl− may be a novel mechanism by which TNBC proliferation and migration can be controlled. Future studies will focus on employing patch clamp electrophysiology to elucidate the membrane potential, ECl (Chloride reversal potential) and Cl− influx kinetics in individual TNBC cells after GABAAR modulation. These studies will further guide the design of novel ligands that can target this membrane-bound LGIC that is upregulated in TNBC.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributionsJB: Conceptualization, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. YA: Methodology, Validation, Writing–review and editing. SW: Methodology, Writing–review and editing. JN: Methodology, Writing–review and editing. JS: Methodology, Writing–review and editing. IM: Conceptualization, Resources, Writing–review and editing. AS: Conceptualization, Funding acquisition, Methodology, Resources, Software, Supervision, Writing–original draft, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Pennsylvania Department of Health.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1449256/full#supplementary-material
ReferencesAgostinetto, E., Losurdo, A., Nader-Marta, G., Santoro, A., Punie, K., Barroso, R., et al. (2022). Progress and pitfalls in the use of immunotherapy for patients with triple negative breast cancer. Expert Opin. Investig. Drugs 31 (6), 567–591. doi:10.1080/13543784.2022.2049232
PubMed Abstract | CrossRef Full Text | Google Scholar
Alexander, S. P., Peters, J. A., Kelly, E., Marrion, N. V., Faccenda, E., Harding, S. D., et al. (2017). The concise guide to pharmacology 2017/18: ligand-gated ion channels. Br. J. Pharmacol. 174 (Suppl. 1), S130–S159. doi:10.1111/bph.13879
PubMed Abstract | CrossRef Full Text | Google Scholar
Bhattacharya, D., Gawali, V. S., Kallay, L., Toukam, D. K., Koehler, A., Stambrook, P., et al. (2021). Therapeutically leveraging GABA(A) receptors in cancer. Exp. Biol. Med. (Maywood) 246 (19), 2128–2135. doi:10.1177/15353702211032549
PubMed Abstract | CrossRef Full Text | Google Scholar
Bundy, J., Shaw, J., Hammel, M., Nguyen, J., Robbins, C., Mercier, I., et al. (2024). Role of β3 subunit of the GABA type A receptor in triple negative breast cancer proliferation, migration, and cell cycle progression. Cell Cycle 23 (4), 448–465. doi:10.1080/15384101.2024.2340912
PubMed Abstract | CrossRef Full Text | Google Scholar
Capatina, A. L., Lagos, D., and Brackenbury, W. J. (2022). Targeting ion channels for cancer treatment: current progress and future challenges. Rev. Physiol. Biochem. Pharmacol. 183, 1–43. doi:10.1007/112_2020_46
PubMed Abstract | CrossRef Full Text | Google Scholar
Gumireddy, K., Li, A., Kossenkov, A. V., Sakurai, M., Yan, J., Li, Y., et al. (2016). The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 7, 10715. doi:10.1038/ncomms10715
PubMed Abstract | CrossRef Full Text | Google Scholar
Hadingham, K. L., Wingrove, P. B., Wafford, K. A., Bain, C., Kemp, J. A., Palmer, K. J., et al. (1993). Role of the beta subunit in determining the pharmacology of human gamma-aminobutyric acid type A receptors. Mol. Pharmacol. 44 (6), 1211–1218.
PubMed Abstract | Google Scholar
Hiraoka, K., Miyazaki, H., Niisato, N., Iwasaki, Y., Kawauchi, A., Miki, T., et al. (2010). Chloride ion modulates cell proliferation of human androgen-independent prostatic cancer cell. Cell Physiol. Biochem. 25 (4-5), 379–388. doi:10.1159/000303042
PubMed Abstract | CrossRef Full Text | Google Scholar
Hiroshi Kaneko, I. P., Frings, S., and Gensch, T. (2002). “Determination of intracellular chloride concentration in dorsal root ganglion neurons by fluorescence lifetime imaging,” in Current topics in membranes (Academic Press), 167–189.
CrossRef Full Text | Google Scholar
Juvale, I. I. A., Hassan, Z., and Has, A. T. C. (2021). The emerging roles of pi subunit-containing GABA(A) receptors in different cancers. Int. J. Med. Sci. 18 (16), 3851–3860. doi:10.7150/ijms.60928
PubMed Abstract | CrossRef Full Text | Google Scholar
Kallay, L., Keskin, H., Ross, A., Rupji, M., Moody, O. A., Wang, X., et al. (2019). Modulating native GABA(A) receptors in medulloblastoma with positive allosteric benzodiazepine-derivatives induces cell death. J. Neurooncol 142 (3), 411–422. doi:10.1007/s11060-019-03115-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Kaneko, H., Putzier, I., Frings, S., Kaupp, U. B., and Gensch, T. (2004). Chloride accumulation in mammalian olfactory sensory neurons. J. Neurosci. 24 (36), 7931–7938. doi:10.1523/JNEUROSCI.2115-04.2004
PubMed Abstract | CrossRef Full Text | Google Scholar
Karim, N., Wellendorph, P., Absalom, N., Johnston, G. A. R., Hanrahan, J. R., and Chebib, M. (2013). Potency of GABA at human recombinant GABA(A) receptors expressed in Xenopus oocytes: a mini review. Amino Acids 44 (4), 1139–1149. doi:10.1007/s00726-012-1456-y
PubMed Abstract | CrossRef Full Text | Google Scholar
Knoflach, F., Hernandez, M. C., and Bertrand, D. (2016). GABAA receptor-mediated neurotransmission: not so simple after all. Biochem. Pharmacol. 115, 10–17. doi:10.1016/j.bcp.2016.03.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Krapf, R., Berry, C. A., and Verkman, A. S. (1988). Estimation of intracellular chloride activity in isolated perfused rabbit proximal convoluted tubules using a fluorescent indicator. Biophys. J. 53 (6), 955–962. doi:10.1016/S0006-3495(88)83176-X
PubMed Abstract | CrossRef Full Text | Google Scholar
Leanza, L., Managò, A., Zoratti, M., Gulbins, E., and Szabo, I. (2016). Pharmacological targeting of ion channels for cancer therapy: in vivo evidences. Biochim. Biophys. Acta 1863 (6 Pt B), 1385–1397. doi:10.1016/j.bbamcr.2015.11.032
PubMed Abstract | CrossRef Full Text | Google Scholar
Lee, Y. M., Oh, M. H., Go, J. H., Han, K., and Choi, S. Y. (2020). Molecular subtypes of triple-negative breast cancer: understanding of subtype categories and clinical implication. Genes Genomics 42 (12), 1381–1387. doi:10.1007/s13258-020-01014-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, X., Wang, H., Yang, X., Wang, X., Zhao, L., Zou, L., et al. (2021). GABRP sustains the stemness of triple-negative breast cancer cells through EGFR signaling. Cancer Lett. 514, 90–102. doi:10.1016/j.canlet.2021.04.028
PubMed Abstract | CrossRef Full Text | Google Scholar
Luo, L., Wang, J., Ding, D., Hasan, M. N., Yang, S. S., Lin, S. H., et al. (2020). Role of NKCC1 activity in glioma K(+) homeostasis and cell growth: new insights with the bumetanide-derivative STS66. Front. Physiol. 11, 911. doi:10.3389/fphys.2020.00911
PubMed Abstract | CrossRef Full Text | Google Scholar
Motz, R. N., Sun, A. C., Lehnherr, D., and Ruccolo, S. (2023). High-throughput determination of stern-volmer quenching constants for common photocatalysts and quenchers. ACS Org. Inorg. Au 3 (5), 266–273. doi:10.1021/acsorginorgau.3c00019
PubMed Abstract | CrossRef Full Text | Google Scholar
Ravasz, D., Kacso, G., Fodor, V., Horvath, K., Adam-Vizi, V., and Chinopoulos, C. (2017). Catabolism of GABA, succinic semialdehyde or gamma-hydroxybutyrate through the GABA shunt impair mitochondrial substrate-level phosphorylation. Neurochem. Int. 109, 41–53. doi:10.1016/j.neuint.2017.03.008
PubMed Abstract | CrossRef Full Text | Google Scholar
Rocha-Gonzalez, H. I., Mao, S., and Alvarez-Leefmans, F. J. (2008). Na+,K+,2Cl-cotransport and intracellular chloride regulation in rat primary sensory neurons: thermodynamic and kinetic aspects. J. Neurophysiol. 100 (1), 169–184. doi:10.1152/jn.01007.2007
PubMed Abstract | CrossRef Full Text | Google Scholar
Sahoo, S., Ramu, S., Nair, M. G., Pillai, M., San Juan, B. P., Milioli, H. Z., et al. (2024). Increased prevalence of hybrid epithelial/mesenchymal state and enhanced phenotypic heterogeneity in basal breast cancer. iScience 27 (7), 110116. doi:10.1016/j.isci.2024.110116
PubMed Abstract | CrossRef Full Text | Google Scholar
Shiozaki, A., Otsuji, E., and Marunaka, Y. (2011). Intracellular chloride regulates the G(1)/S cell cycle progression in gastric cancer cells. World J. Gastrointest. Oncol. 3 (8), 119–122. doi:10.4251/wjgo.v3.i8.119
PubMed Abstract | CrossRef Full Text | Google Scholar
Sizemore, G. M., Sizemore, S. T., Seachrist, D. D., and Keri, R. A. (2014). GABA(A) receptor pi (GABRP) stimulates basal-like breast cancer cell migration through activation of extracellular-regulated kinase 1/2 (ERK1/2). J. Biol. Chem. 289 (35), 24102–24113. doi:10.1074/jbc.M114.593582
PubMed Abstract | CrossRef Full Text | Google Scholar
Takehara, A., Hosokawa, M., Eguchi, H., Ohigashi, H., Ishikawa, O., Nakamura, Y., et al. (2007). Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res. 67 (20), 9704–9712. doi:10.1158/0008-5472.CAN-07-2099
PubMed Abstract | CrossRef Full Text | Google Scholar
Weilinger, N. L., Wicki-Stordeur, L. E., Groten, C. J., LeDue, J. M., Kahle, K. T., and MacVicar, B. A. (2022). KCC2 drives chloride microdomain formation in dendritic blebbing. Cell Rep. 41 (4), 111556. doi:10.1016/j.celrep.2022.111556
留言 (0)