Acute myeloid leukemia (AML) is one of the most common malignant diseases in the hematological system, characterized by the malignant proliferation of hematopoietic stem cells. It predominantly affects adults and is estimated to be the 11th most common cancer globally (Döhner et al., 2015; Sasaki et al., 2021). Despite significant advancements in AML treatment, including intensive chemotherapy, allogeneic stem cell transplantation, and targeted therapy, which have gradually become the primary treatment modalities and have achieved certain outcomes, the prognosis of AML remains poor, with a 5-year survival rate of less than 25% (Liu, 2021; Rm et al., 2019). Consequently, improving the prognosis of AML has become a key focus for many researchers. Since the European LeukemiaNet (ELN) introduced a method for predicting AML patient prognosis based on cellular and genetic abnormalities in 2017, numerous scholars have been dedicated to identifying indicators for monitoring AML prognosis (Liu, 2021; Döhner et al., 2017). In recent years, long non-coding RNA (lncRNA), an RNA molecule longer than 200 nucleotides that does not code for proteins, has gained attention due to its abnormal expression in various human diseases. LncRNAs regulate gene expression through mechanisms such as epigenetic modification, transcription, and post-transcriptional regulation, and play a role in the development of multiple diseases (Lu et al., 2024). Multiple studies have demonstrated that lncRNAs play a significant role in the pathogenesis of AML (7–31) and show substantial value in diagnosis, prognosis monitoring, and targeted therapy. However, the relationship between lncRNA dysregulation and AML prognosis remains controversial. Therefore, this study aims to use meta-analysis to investigate the evidence-based relationship between lncRNA dysregulation and AML prognosis, providing references for risk stratification and treatment of AML.
2 Materials and methods2.1 Document retrievalSearch the Chinese and English databases, including PubMed, Embase, Cochrane Library, China National Knowledge Infrastructure (CNKI), Wanfang Data Knowledge Service Platform, VIP Database, and China Biomedical Literature Service (SinoMed). Additionally, perform manual searches to obtain relevant literature to the greatest extent possible. The search period ranges from the establishment of the databases to 4 August 2024. The Chinese search terms include “lncRNA,” “acute myeloid leukemia,” and “prognosis.” For the English search terms, a combination of subject headings and free text terms is used, with subject headings including “lncRNA,” “acute myeloid leukemia,” and “prognosis.” An example of the PubMed search strategy is provided in Table 1.
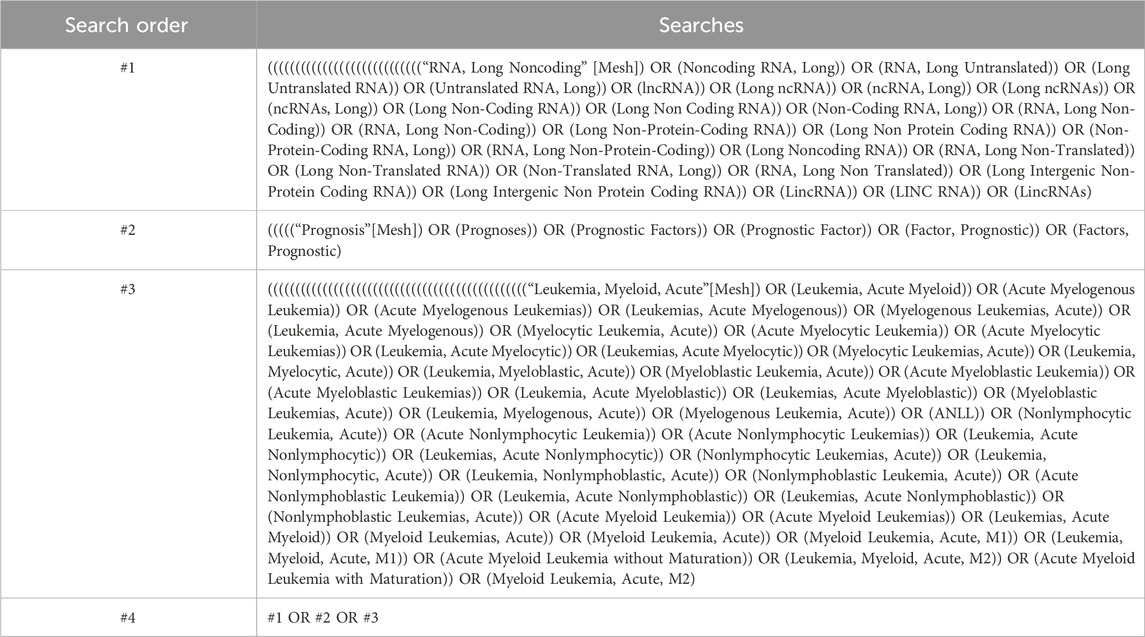
Table 1. Pubmed search strategy.
2.2 Criteria for inclusion and exclusion2.2.1 Inclusion criteria1. Study Subjects: Healthy individuals or patients with benign diseases as the control group, and patients with acute myeloid leukemia (AML) as the case group (age of study subjects ≥14 years).
2. Research Content: Studies examining the prognostic correlation of lncRNA in tissue specimens and AML.
3. Research Type: Cohort studies.
4. Outcome Indicators: Studies must report overall survival (OS), event-free survival (EFS), relapse-free survival (RFS), and disease-free survival (DFS). Hazard ratios (HR) and their 95% confidence intervals (CI) must be either directly extractable or indirectly extractable from survival curves, along with the type, expression level, and cutoff value of lncRNA.
2.2.2 Exclusion criteria1. Studies published repeatedly or in different languages.
2. Review articles, conference papers, dissertations, and other non-primary research articles.
3. Studies with incomplete data or where the full text is unavailable.
4. Research content not related to the topic.
5. Studies where outcome indicators do not meet the specified criteria or cannot be extracted.
2.3 Screening of literature and extraction of dataThe retrieved literature was independently screened and included by two researchers in strict adherence to the inclusion and exclusion criteria. In the event of disagreement, a third researcher would arbitrate the decision. The data to be extracted included: ① authors, publication year ② study region ③ sample size ④ detection method ⑤ type, expression level, and cutoff value of lncRNA ⑥ source of samples ⑦ disease type and stage ⑧ follow-up time ⑨ outcome indicators ⑩ source of HR extraction, etc.
2.4 Evaluation of literature qualityFor the included cohort studies, the authors will employ the Newcastle-Ottawa Scale (NOS) to assess the quality of the literature. Studies scoring ≥6 on the NOS will be considered high-quality and deemed suitable for inclusion in the meta-analysis.
2.5 Statistical approachesMeta-analysis was conducted using Stata16SE software, and the results were presented in a forest plot. The relationship between lncRNA and the prognosis of AML patients was assessed using the combined effect size (HR) and its corresponding 95% CI values. To evaluate the heterogeneity of the studies, the authors used the Q test and the I2 statistic. If the test results indicated I2 < 50% and P > 0.05, it suggested low heterogeneity, and a fixed-effects model was applied; if the test results showed I2 > 50% and P ≤ 0.05, it indicated substantial heterogeneity, and a random-effects model was used. Sensitivity analysis was performed for studies with significant heterogeneity, and subgroup analysis was conducted to explore the sources of heterogeneity. When the number of studies included in the outcome measure exceeded 10, Begg’s test and Egger’s test were employed to assess publication bias.
3 Results3.1 Results of literature screeningAfter conducting the search in strict accordance with the inclusion and exclusion criteria, 826 Chinese and English articles were retrieved. After eliminating 283 duplicates, 62 articles were initially selected based on the abstracts and titles. Finally, 25 articles were included after reviewing the full texts (Figure 1).
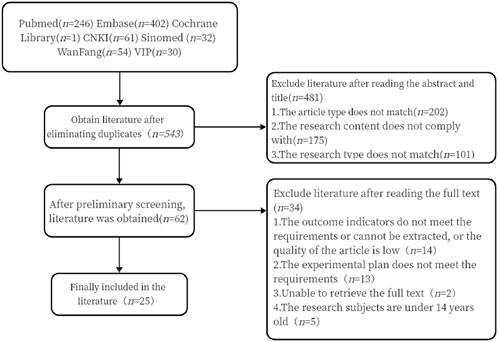
Figure 1. Literature screening process diagram
3.2 The basic characteristics of the included literature and the quality evaluation of NOSA total of 25 articles were included, with the research period spanning from 2015 to 2024. The research regions included China (20 articles), Egypt (3 articles), and Iran (2 articles). A total of 2,372 patients diagnosed with AML were involved in these studies. The studies examined 22 types of lncRNAs, and the detection method used in all studies was qRT-PCR. The NOS scores of all studies were ≥6 points, indicating their suitability for inclusion in the meta-analysis. The specific details are presented in Table 2.
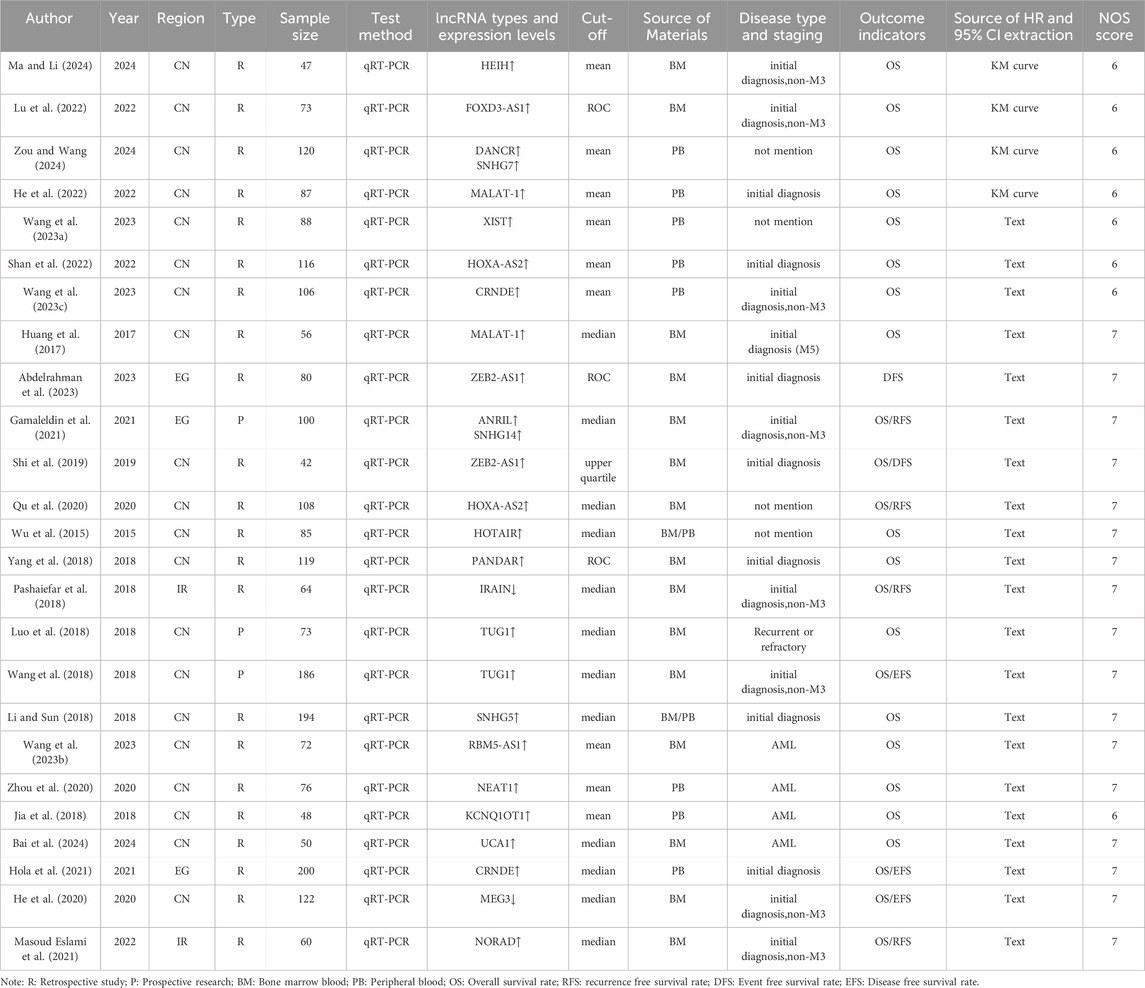
Table 2. Basic characteristics of included literature.
3.3 The outcomes of the meta-analysis3.3.1 The correlation between aberrant lncRNA expression and overall survival (univariate analysis) in AML patients17 studies have utilized univariate analysis to investigate the relationship between aberrant lncRNA expression and overall survival (OS) in AML patients. Heterogeneity analysis (I2 = 0.0%, P > 0.05) indicated no significant heterogeneity, leading to the use of a fixed-effects model for the meta-analysis. The results (Figure 2, HR = 2.46, 95% CI 2.11–2.88, P < 0.001) demonstrated that the aberrant expression of lncRNAs, including high expression of DANCR, SNHG7, HEIH, FOXD3-AS1, MALAT-1, PANDAR, TUG1, ZEB2-AS1, NEAT1, ANRIL, SNHG14, SNHG5, CRNDE, NORAD, RBM5-AS1, and low expression of IRAIN, was significantly associated with reduced OS in AML patients, with a statistically significant difference.

Figure 2. Forest plot of the relationship between abnormal expression of lncRNA and OS (univariate) in AML patients.
3.3.2 The correlation between aberrant lncRNA expression and overall survival (multivariate) in AML patientsThe 19 included studies utilized multivariate analysis to investigate the relationship between aberrant lncRNA expression and overall survival (OS) in AML patients. Heterogeneity analysis revealed significant heterogeneity (I2 = 53.1%, P < 0.05), prompting the use of a random-effects model for the meta-analysis. The meta-analysis results (Figure 3) showed that AML patients with aberrant lncRNA expression (high expression of XIST, HOXA-AS2, CRNDE, MALAT-1, ANRIL, SNHG14, ZEB2-AS1, HOTAIR, PANDAR, TUG1, SNHG5, RBM5-AS1, NEAT1, KCNQ1OT1, UCA1, CRNDE, NORAD, etc., and low expression of MEG3, IRAIN) had a shorter OS, indicating a poor prognosis. The difference was statistically significant (HR = 2.21, 95% CI 1.82–2.70, P < 0.001). Additionally, we summarized all the factors included in the multivariate analysis of each study with OS as the outcome variable to facilitate the exploration of potential sources of heterogeneity or error in the meta-analysis (Table 3).
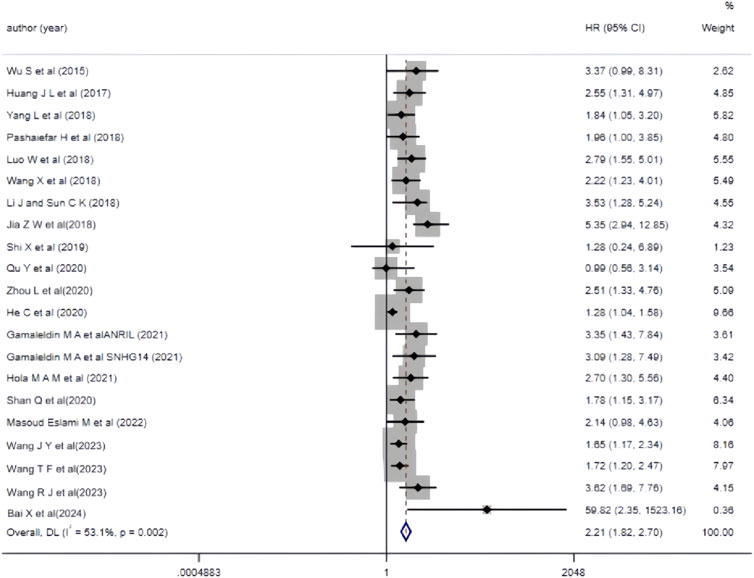
Figure 3. Forest plot of the relationship between abnormal expression of lncRNA and OS (multifactorial) in AML patients.
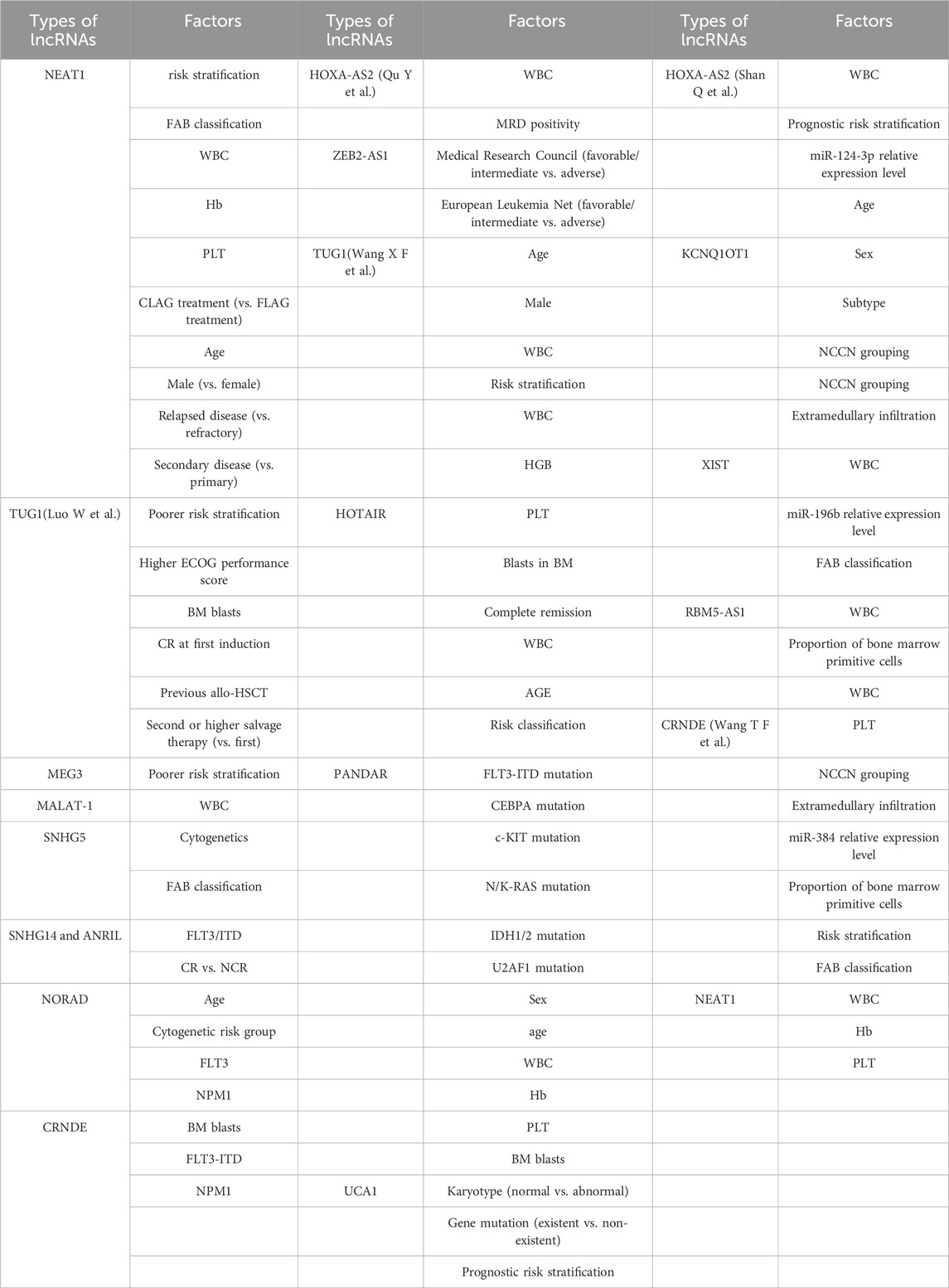
Table 3. Summary of factors in multiple factor analysis of different studies.
Furthermore, we conducted subgroup analyses based on study region, sample size (number of cases), source of material, and disease type to explore the sources of heterogeneity. The results (Table 4) indicated that each subgroup analysis consistently demonstrated that aberrant lncRNA expression was associated with a reduced OS.
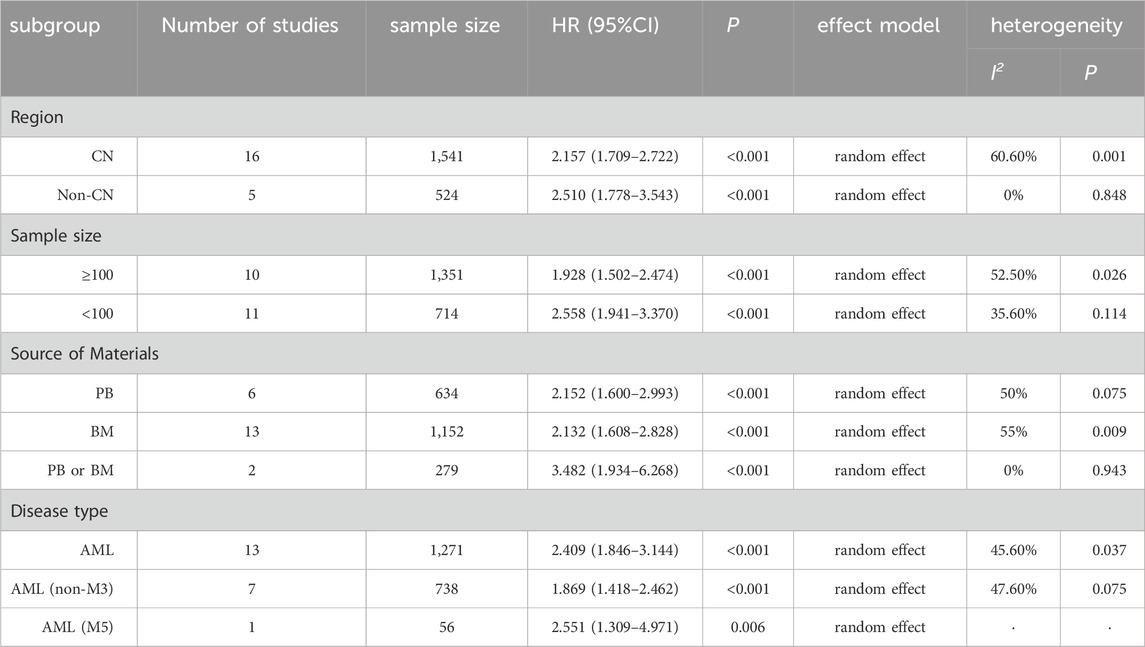
Table 4. Subgroup analysis results.
3.3.3 The correlation between aberrant lncRNA expression and DFS, EFS, and RFS (multivariate variables) in AML patients2 studies reported the correlation between aberrant lncRNA expression and DFS in AML patients. Heterogeneity analysis indicated I2 = 16.9% and P > 0.05, leading to the use of a fixed-effects model for analysis. The results (Figure 4) demonstrated that high expression of lncRNA ZEB2-AS1 was associated with a decreased DFS in AML patients (HR = 2.390, 95%CI 1.037–5.507, P = 0.041), with a statistically significant difference.
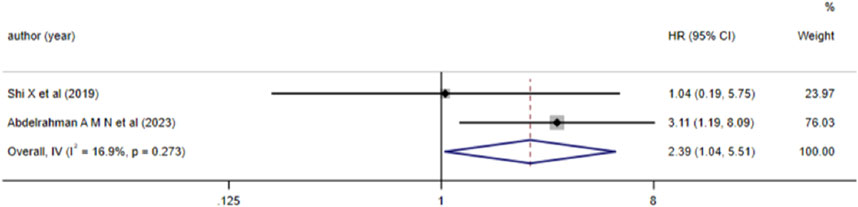
Figure 4. Forest plot of the relationship between lncRNA expression levels and DFS in AML patients.
3 studies analyzed the relationship between lncRNA and AML prognosis, with EFS as the outcome measure. Since the results of Hola et al. (P < 0.05) were not statistically significant, we excluded this study and included the remaining two in the meta-analysis. Heterogeneity analysis revealed I2 = 6.2% and P > 0.05, thus a fixed-effects model was adopted. The results (Figure 5) indicated that high expression of lncRNA TUG1 and low expression of lncRNA MEG3 were associated with a reduced EFS in AML patients (HR = 1.51, 95%CI 1.19–1.90, P = 0.001), with a statistically significant difference.
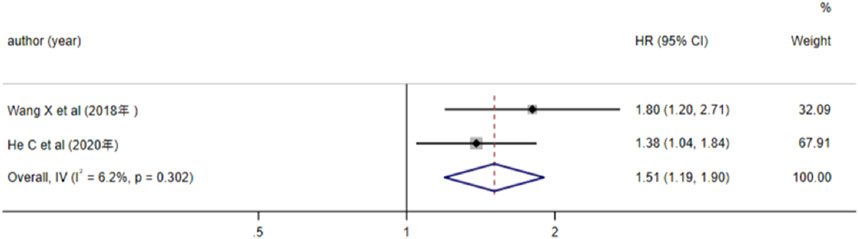
Figure 5. Forest plot of the relationship between lncRNA expression levels and EFS in AML patients.
5 studies investigated the relationship between aberrant lncRNA expression and RFS in AML patients. Heterogeneity testing (I2 = 0.0%, P > 0.05) led to the use of a fixed-effects model for meta-analysis. The results (Figure 6) showed that aberrant expression of lncRNA (high expression of NORAD, ANRIL, SNHG14, HOXA-AS2, etc., and low expression of IRAIN) was associated with a reduced RFS in AML patients (HR = 2.82, 95%CI 2.03–3.91, P < 0.001), with a statistically significant difference.

Figure 6. Forest plot of the relationship between lncRNA expression levels and RFS in AML patients.
3.3.4 The relationship between the high expression of lncRNA HOXA-AS2, CRNDE, ZEB2-AS1, TUG1, and MALAT-1 and the prognosis of AML patientsIt is evident from Table 2 that multiple studies included in the literature have investigated the impact of high expression of lncRNA HOXA-AS2, CRNDE, ZEB2-AS1, TUG1, and MALAT-1 on the prognosis of AML patients. Due to the different result types of the studies on lncRNA MALAT-1, these results could not be combined. Therefore, we conducted a Meta-analysis to examine the relationship between lncRNA ZEB2-AS1 and DFS, and the relationships between the other three lncRNAs (CRNDE, TUG1, and HOXA-AS2) and OS. The results (as shown in Figure 7) indicated that high expression of lncRNA CRNDE and TUG1 was associated with a reduced OS in AML patients, and high expression of lncRNA ZEB2-AS1 was associated with a decreased DFS in AML patients, with statistically significant differences. However, high expression of lncRNA HOXA-AS2 was associated with a reduced OS in AML patients, but this association was not statistically significant.
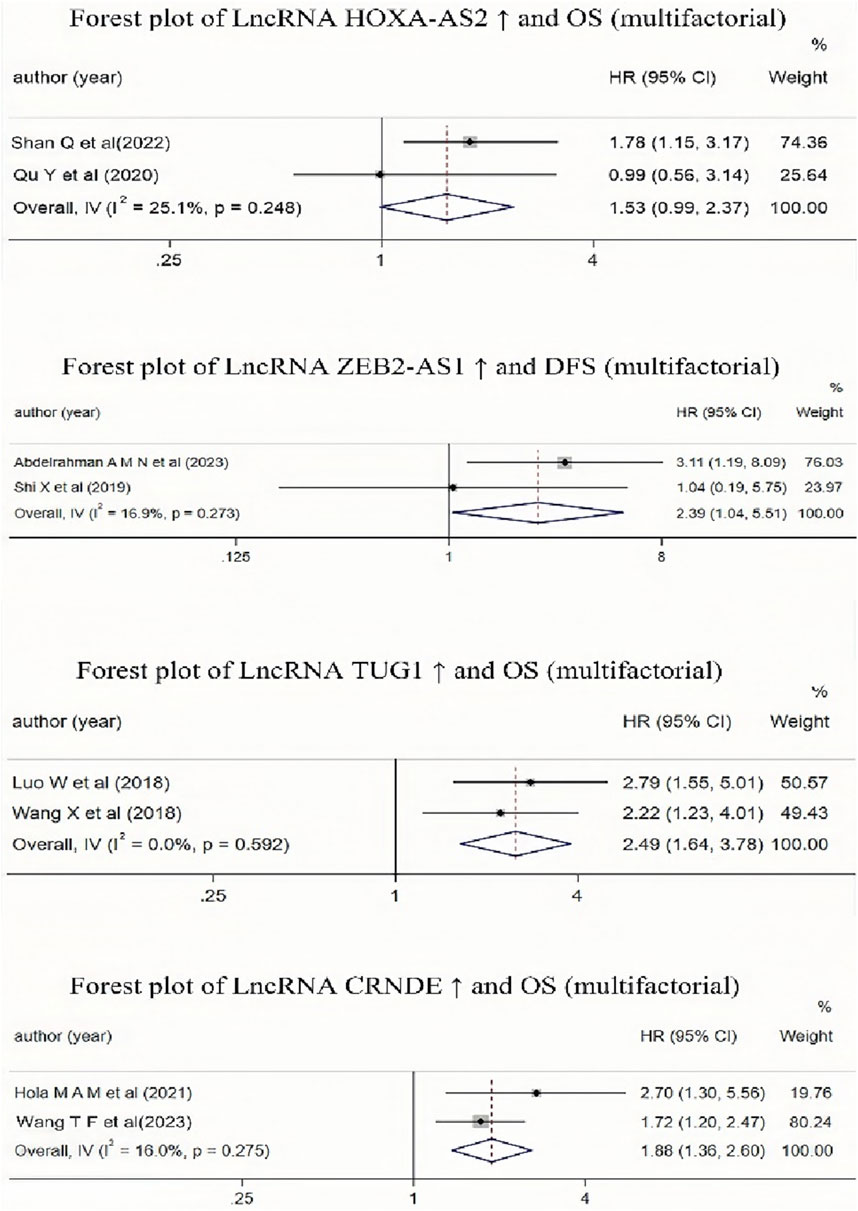
Figure 7. Forest plot collection of different lncRNAs and OS (multifactor) or lncRNAs and DFS(multifactor).
3.3.5 Publication bias and sensitivity analysisWe conducted a publication bias assessment using the included literature with OS (single factor or multiple factors) as the outcome indicator. Begg’s test (P = 0.944) and Egger’s test (P = 0.346) indicated no publication bias in the literature with OS (single factor) as the outcome indicator. However, the literature with OS (multiple factors) as the outcome indicator showed a certain degree of publication bias (Begg’s test: P = 0.020, Egger’s test: P < 0.001).
A sensitivity analysis was performed for the OS of AML patients in the multiple-factor analysis. The results (Figure 8) indicated that the studies by Jia et al. (2018) and He et al. (2020) significantly influenced the heterogeneity of the Meta-analysis. After excluding these studies and re-analyzing the heterogeneity, the heterogeneity analysis result decreased from I2 = 53% to I2 = 9.3%, with P = 0.345, HR = 2.147, 95%CI (1.839 - 2.507), P < 0.001. No significant change was observed in the combined effect size after exclusion, indicating good stability of the results.
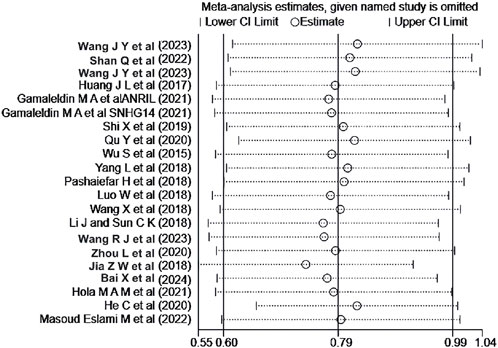
Figure 8. Sensitivity analysis of the relationship between abnormal expression of lncRNA and OS (multifactorial) in AML patients.
4 DiscussionAML is a hematological disorder characterized by high malignancy and rapid disease progression, with complex etiological factors and significant heterogeneity. Currently, the primary treatment modalities for AML include intensive chemotherapy, targeted therapy, and hematopoietic stem cell transplantation, which have significantly improved the 5-year survival rate of AML patients. However, some patients still face challenges such as poor prognosis, high drug resistance, and high recurrence rates. Therefore, effective prognosis monitoring and the identification of therapeutic targets have become key research priorities for many scholars.
Initially, lncRNA was discovered and considered as “noise” in gene transcription due to its non-coding nature for proteins. However, with advancements in life sciences and gene sequencing technologies, researchers have gradually recognized the critical role of lncRNA in the regulation of biological processes. The types of lncRNA are extremely diverse, and their mechanisms of action are highly complex, and our understanding of them remains limited. Currently, researchers believe that lncRNA plays significant roles in epigenetics, cell apoptosis, differentiation, and other processes, and its involvement has been identified in the development of many diseases. Mumbach et al. published an article in “Nature Methods” using HiCHIRP technology to elucidate the mechanism by which lncRNA regulates chromatin structure and gene expression, demonstrating the significant application value of lncRNA in the field of life sciences (Mumbach et al., 2019). Additionally, with the application of third-generation sequencing technology, a technical foundation has been established for the research and application of lncRNA in clinical medicine. Therefore, exploring the role of lncRNA in the occurrence and development of AML is a highly significant approach for identifying new therapeutic targets and effective prognosis monitoring indicators for AML patients. We employed a meta-analysis method to incorporate studies examining the relationship between lncRNA and the prognosis of AML patients, providing evidence-based medical evidence for the clinical application of lncRNA and offering a valuable reference for scholars researching the relationship between lncRNA and AML.
Our research findings indicate that the abnormal expression of lncRNA has a negative impact on the overall survival (OS), disease-free survival (DFS), relapse-free survival (RFS), and event-free survival (EFS) of AML patients, thereby reducing their prognosis. This is consistent with the previous meta-analysis results of Lin Hongli et al., Shi et al., and Priya (Lin et al., 2022; Shi Jie et al., 2020; Priya et al., 2024). With the in-depth research on lncRNA and AML by scholars, we designed a relatively rigorous meta-analysis protocol, incorporating more recently published studies, and analyzing the relationship between newly discovered lncRNA and the prognosis of AML patients. In terms of the results, there is a certain degree of heterogeneity in studies with OS (using multi-factor analysis) as the outcome indicator. Through subgroup analysis and sensitivity analysis, we believe that the source of heterogeneity may be the different baseline characteristics of the included studies, such as disease types, sample collection methods, and the ethnicity of the study subjects. Additionally, some indicators could not be analyzed due to the small number of studies, non-inclusion in the research protocol, or a large time span of the studies, making it impossible to establish a unified standard, such as the types of lncRNA, expression levels and cut-off values, and AML treatment regimens. We consider that the differences in these baseline characteristics may be the main source of heterogeneity in the included literature, and we hope that more comprehensive and high-quality related studies will emerge in the future. Furthermore, we summarized the various factors in the multi-factor analysis included in the studies and found that the factors explored in each study were not the same. Some studies included comprehensive factors, covering multiple laboratory indicators, and even considered factors such as disease risk stratification and gene mutations, resulting in more reliable outcomes. Some studies only included factors such as age, gender, and white blood cell count, leading to potential positive biases, which may affect the persuasiveness of the resulting evidence-based medical evidence.
For the lncRNAs (HOXA-AS2, CRNDE, ZEB2-AS1, TUG1, MALAT-1) mentioned in multiple articles, we conducted a separate meta-analysis. The results showed that the high expression of lncRNAs (CRNDE, TUG1) reduces the overall survival rate of AML patients, and the high expression of lncRNA ZEB2-AS1 reduces the disease-free survival rate of AML patients. The conclusions were statistically significant, and the heterogeneity of the included literature was relatively small. This suggests that lncRNAs CRNDE, ZEB2-AS1, and TUG1 may have significant potential to become prognostic monitoring indicators for AML patients. However, the number of included studies on lncRNAs (CRNDE, ZEB2-AS1, TUG1) is relatively small, and these three lncRNAs still require further validation to become prognostic risk stratification indicators for AML patients.
Herein, we summarize the full names, loci, and selected functions of the 22 lncRNAs mentioned in the included literature in tabular form to provide valuable references for readers’ research on the correlation between lncRNAs and AML (Table 5). Due to the limited space in this article, we present an overview of only the 3 lncRNAs that are frequently mentioned in multiple studies (Figure 9).
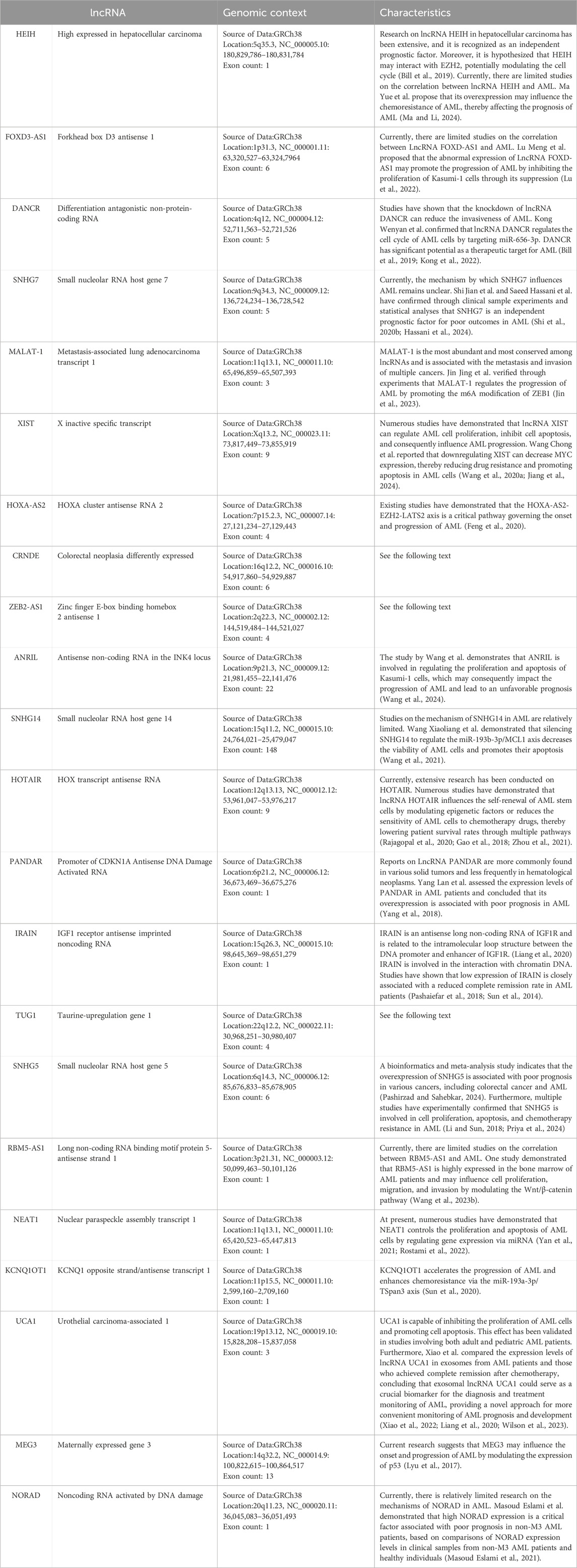
Table 5. Summary Table of Multiple lncRNA Information.
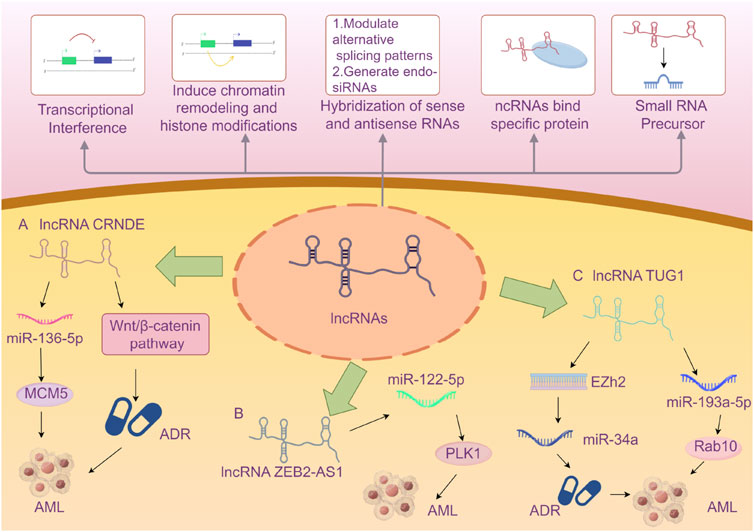
Figure 9. A Brief Introduction to the Functions of lncRNAs (Wilusz et al., 2009) AND The Roles of Three lncRNAs in Acute Myeloid Leukemia. A The lncRNA CRNDE enhances the drug resistance of acute myeloid leukemia (AML) to adriamycin (ADR) by activating the Wnt/β-catenin signaling pathway. CRNDE participates in the CRNDE/miR-136-5p/MCM5 axis to regulate cellular processes and thereby influences the disease progression of AML. B lncRNA ZEB2-AS1 regulates cell proliferation and apoptosis in acute myeloid leukemia via the miR-122-5p/PLK1 axis. C lncRNA TUG1 confers Adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2. And it regulates cell viability and apoptosis by modulating the miR-193a-5p/Rab10 axis in acute myeloid leukemia (AML). Note: Extensive research has explored the influence of the aforementioned three specific lncRNAs on AML disease progression via the microRNA/protein axis. However, no definitive consensus has been established to date. The current review may be subject to bias due to potentially incomplete literature inclusion. Future studies should aim to elucidate the mechanisms by which various lncRNAs influence AML progression through more rigorous and compelling experimental designs.
LncRNA CRNDE (Colorectal Neoplasia Differently Expressed) was initially identified with aberrant expression in colorectal neoplasms and is located on the reverse strand of chromosome 16 (Thierry-Mieg and Thierry-Mieg, 2006). Subsequent in-depth studies have revealed that lncRNA CRNDE exhibits abnormal expression in various solid tumors, such as glioma, hepatocellular carcinoma, and lung cancer, as well as in hematological malignancies. It has been demonstrated that lncRNA CRNDE holds significant value in the diagnosis, treatment, and prognosis of certain tumors (Wang T. et al., 2023; Hola et al., 2021; Ma et al., 2022; Ma et al., 2019). Numerous studies have indicated that lncRNA CRNDE may be involved in the occurrence and development of cancer through its regulatory interactions with microRNAs and proteins (Hor et al., 2023; Xie et al., 2018). Ma et al. demonstrated that knocking out CRNDE in acute promyelocytic leukemia (APL) mice could induce APL cell differentiation, inhibit proliferation, and extend the survival time of APL mice (Ma et al., 2020). Liu et al. proved that CRNDE participates in the CRNDE/miR-136-5p/MCM5 axis to regulate cellular processes and thereby influences the disease progression of AML (Liu et al., 2021). From an evidence-based medicine perspective, our research has shown that the risk of poor prognosis in AML patients with high expression of lncRNA CRNDE is 1.88 times higher than that in patients with low expression. CRNDE may become an important indicator of poor prognosis in AML in the future, but further exploration is still necessary.
LncRNA ZEB2-AS1 (zinc finger E-box binding homeobox 2 antisense 1) is an antisense long non-coding RNA that undergoes antisense transcription during protein coding and is located on the long arm of chromosome 2 at band 2q3. Studies have indicated that lncRNA ZEB2-AS1 shows increased expression in various cancers (Mahboobeh et al., 2020). Guan et al. verified through bioinformatics analysis and mouse experiments that lncRNA ZEB2-AS1 regulates the proliferation and apoptosis of AML cells via the miR-122-5p/PLK1 axis (Guan et al., 2020). The study by Abdelrahman et al., which we included, concluded that lncRNA ZEB2-AS1 can be regarded as a predictor of poor prognosis for AML in Egyptians (Abdelrahman et al., 2023). We combined this study with the one by Shi et al. for meta-analysis and concluded that high expression of lncRNA ZEB2-AS1 reduces the disease-free survival (DFS) of AML patients. ZEB2-AS1 is expected to become an important indicator for prognosis monitoring of AML patients in the future, but more comprehensive studies are still required for validation.
LncRNA TUG1 (taurine-upregulated gene 1) is a 7.1-kb long non-coding RNA located on human chromosome 22. Multiple studies have confirmed that the abnormal expression of lncRNA TUG1 is closely related to the disease progression of various conditions, particularly cardiovascular diseases (Chang and Su, 2024; Liu B. et al., 2024; Liu Y. et al., 2024). It may influence the occurrence and progression of diseases by regulating gene expression or interacting with specific proteins as a competitive endogenous RNA (ceRNA) (Li et al., 2016). Sun et al. reconstructed the lncRNA-miRNA-mRNA network and identified three lncRNAs (XIST, TUG1, GABPB1-AS1) as potential key lncRNAs for cytogenetically normal AML from a bioinformatics perspective, demonstrating that lncRNA TUG1 might be related to the prognosis of AML. This article was the first to propose that lncRNA GABPB1-AS1 might be an important biomarker related to the prognosis of AML (Sun et al., 2022). Other studies have suggested that TUG1 might be associated with doxorubicin resistance in AML, thereby affecting the prognosis of AML (Li et al., 2019). We combined two related studies on TUG1 and concluded that the risk of reduced overall survival (OS) in AML patients with high expression of lncRNA TUG1 is 2.49 times higher than that in patients with low expression. TUG1 may become a new tool for risk stratification of AML in the future, but further research on its mechanism of action is still awaited.
From the existing studies, it is evident that no single lncRNA can directly predict the prognosis of AML or provide unambiguous evidence for risk stratification of AML. Similar to the complex pathogenesis of AML, the regulatory mechanisms of lncRNAs involved in the disease are also intricate, likely influencing the disease through the joint participation of one or more lncRNAs. Therefore, this highlights the need to consider the variations in the expression of multiple lncRNAs in the disease progression when researching the prognostic risk stratification of lncRNAs for AML. Scores should be assigned based on the significance of their roles in the process, thereby establishing a comprehensive scoring system for evaluating the prognosis of AML using lncRNAs. Additionally, the current highly efficient testing techniques can provide substantial assistance for the diagnosis, treatment, and prognosis detection of AML.
Limitations of this study: ① In some studies, hazard ratios (HRs) were not provided directly; instead, we extracted survival data from Kaplan-Meier (KM) curves, which may introduce certain errors. ② The types, expression levels, and cutoff values of lncRNAs included in this study varied, potentially impacting the reliability of the results. ③ The majority of the included studies originated from China, which may affect the generalizability of the findings. ④ The majority of the included studies reported positive results, suggesting the presence of publication bias that could not be entirely eliminated.
In conclusion, the abnormal expression of lncRNAs is associated with poor prognosis in AML, demonstrating considerable potential to serve as prognostic monitoring indicators and tools for risk stratification of AML. Specifically, the evidence from evidence-based medicine indicates a clear correlation between the high expression of lncRNAs (CRNDE, ZEB2-AS1, TUG1) and poor prognosis in AML. Future research can further validate these findings through various approaches, including bioinformatics, basic experiments, and clinical studies. However, whether lncRNAs can become reliable prognostic risk stratification tools for AML remains to be determined. More large-scale, multi-center, high-quality studies are needed to explore the relationships between the expression levels of different lncRNAs and the baseline characteristics of AML patients, such as different subtypes of AML, the effectiveness of different treatment regimens, and patient age.
Author contributionsGL: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. LS: Investigation, Methodology, Supervision, Writing–original draft. PL: Methodology, Project administration, Software, Supervision, Writing–original draft, Writing–review and editing. RQ: Methodology, Project administration, Visualization, Writing–original draft, Writing–review and editing. LW: Investigation, Project administration, Software, Validation, Writing–original draft, Writing–review and editing. AJ: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This paper has received support from two research funds. One is the research fund of the Inner Mongolia Physicians Association applied for by the corresponding author, AJ. The project title is: Exploring the Real-World Study of Romiplostim Versus Recombinant Human Thrombopoietin in the Treatment of ITP, and the fund code is: YSXH2024KYD01. The other is the fund of the Inner Mongolia Autonomous Region Health and Wellness Science and Technology Plan Project applied for by LS. The project title is: Research on Molecular Diagnosis of Acute Myeloid Leukemia Patients Based on Liquid Biopsy Technology, and the fund code is: 202201035.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAbdelrahman, A. M. N., Diab, S. M., Shabaan, H. M. K., Ahmed, M. N. A., and Nabil, R. (2023). The study of long noncoding RNA TUG 1 and ZEB2-AS1 expression in newly diagnosed Egyptian adult acute myeloid leukemia patients. Egypt. J. Med. Hum. Genet. 24, 46. doi:10.1186/s43042-023-00423-z
CrossRef Full Text | Google Scholar
Bai, X., Wu, Y., Li, Z., Chen, X., Wang, M., and Li, J. (2024). Expression of lncRNA UCA1 in patients with acute myeloid leukemia and its clinical significance. Chin. J. Exp. Hematol. 32, 999–1004. doi:10.19746/j.cnki.issn.1009-2137.2024.04.004
PubMed Abstract | CrossRef Full Text | Google Scholar
Bill, M., Papaioannou, D., Karunasiri, M., Kohlschmidt, J., Pepe, F., Walker, C. J., et al. (2019). Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 33, 2169–2182. doi:10.1038/s41375-019-0429-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Chang, C., and Su, Q. (2024). Research progress of lncRNA-TUG1 as a competitive endogenous RNA in cardiovascular diseases. Chin. J. Pathophysiol. 40, 557–563. doi:10.3969/j.issn.1000-4718.2024.03.021
CrossRef Full Text | Google Scholar
Döhner, H., Estey, E., Grimwade, D., Amadori, S., Appelbaum, F. R., Büchner, T., et al. (2017). Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447. doi:10.1182/blood-2016-08-733196
PubMed Abstract | CrossRef Full Text | Google Scholar
Feng, Y., Hu, S., Li, L., Peng, X., and Chen, F. (2020). Long noncoding RNA HOXA-AS2 functions as an oncogene by binding to EZH2 and suppressing LATS2 in acute myeloid leukemia (AML). Cell death and Dis. 11, 1025. doi:10.1038/s41419-020-03193-3
PubMed Abstract | CrossRef Full Text | Google Scholar
Gamaleldin, M. A., Ghallab, O., and Abo Elwafa, R. A. (2021). Prognostic significance of long non coding RNA ANRIL and SNHG14 in acute myeloid leukemia. Asian Pac J. Cancer Prev. 22, 3763–3771. doi:10.31557/APJCP.2021.22.12.3763
PubMed Abstract | CrossRef Full Text | Google Scholar
Gao, S., Zhou, B., Li, H., Huang, X., Wu, Y., Xing, C., et al. (2018). Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp. Hematol. 67, 32–40.e3. doi:10.1016/j.exphem.2018.08.005
PubMed Abstract | CrossRef Full Text | Google Scholar
Guan, J., Liu, P., Wang, A., and Wang, B. (2020). Long non-coding RNA ZEB2-AS1 affects cell proliferation and apoptosis via the miR-122-5p/PLK1 axis in acute myeloid leukemia. Int. J. Mol. Med. 46, 1490–1500. doi:10.3892/ijmm.2020.4683
PubMed Abstract | CrossRef Full Text | Google Scholar
Hassani, S., Rostami, P., Pourtavakol, M., Karamashtiani, A., and Sayyadi, M. (2024). Correlation of SNHG7 and BGL3 expression in patients with de novo acute myeloid leukemia; novel insights into lncRNA effect in PI3K signaling context in AML pathogenesis. Biochem. biophysics Rep. 40, 101850. doi:10.1016/j.bbrep.2024.101850
PubMed Abstract | CrossRef Full Text | Google Scholar
He, C., Wang, X., Luo, J., Ma, Y., and Yang, Z. (2020). Long noncoding RNA maternally expressed gene 3 is downregulated, and its insufficiency correlates with poor-risk stratification, worse treatment response, as well as unfavorable survival data in patients with acute myeloid leukemia. Technol. Cancer Res. Treat. 19, 1533033820945815. doi:10.1177/1533033820945815
PubMed Abstract | CrossRef Full Text | Google Scholar
He, X., Hu, Y., Xu, S., Yang, K., and Ma, C. (2022). Expression levels of serum lncRNA-MALAT-1 and miR-143 in patients with acute myeloid leukemia and their relationship with clinical characteristics and prognosis. Chin. Med. Her. 19, 101–105. doi:10.20047/j.issn1673-7210.2022.35.23
CrossRef Full Text | Google Scholar
Hola, M. A. M., Ali, M. A. M., ElNahass, Y., Salem, T. A. E., and Mohamed, M. R. (2021). Expression and prognostic relevance of long noncoding RNAs CRNDE and AOX2P in adult acute myeloid leukemia. Int. J. Lab. Hematol. 43, 732–742. doi:10.1111/ijlh.13586
PubMed Abstract | CrossRef Full Text | Google Scholar
Huang, J. L., Liu, W., Tian, L. H., Chai, T. T., Liu, Y., Feng, Z., et al. (2017). Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol. Rep. 38, 1353–1362. doi:10.3892/or.2017.5802
PubMed Abstract | CrossRef Full Text | Google Scholar
Jia, Z., Li, Y., Cui, G., Zhao, H., Li, P., and Luo, J. (2018). Expression of LncRNA KCNQ1OT1 in patients with acute myeloid leukemia and its clinical significance. Chin. J. Exp. Hematol. 26
留言 (0)