Plant defenses against microbial pathogens are activated upon detection of infectious agents. A series of pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs) to activate pattern-triggered immunity (PTI) (Wang L. et al., 2016). PTI represents the first line of plant innate immunity, offering broad-spectrum resistance against pathogen invasion. Adapted pathogens deploy effector proteins that undermine PTI and promote pathogen proliferation. In response, plants have evolved nucleotide-binding leucine-rich repeat proteins that recognize specific effector proteins, initiating effector-triggered immunity, a robust immune response along with the emergence of a hypersensitive response (HR) to restrict pathogen propagation. A key indicator of PTI activation is the accumulation of reactive oxygen species (ROS), typically observed minutes after epitope inoculation in plant leaves and several hours following bacterial infiltration (Boller and Felix, 2009; Nguyen et al., 2010). Functional PTI can also be detected approximately 6 h after infiltrating Nicotiana benthamiana leaves with non-virulent bacteria, as evidenced by the suppression of pathogen effectors translocation, pathogen growth, or HR induction (Wei et al., 2013).
Many bacteria produce small cold-shock proteins (CSPs) to counteract the damaging effects of temperature downshifts and protect cells (Charollais, 2004; Keto-Timonen et al., 2016). CSPs are small nucleic acid-binding proteins, approximately 70 amino acids in length, and are highly-conserved across bacteria. Although CSPs are primarily known for their role in the cold-shock response, recent studies suggest that they are also involved in diverse biological processes. CspD from Escherichia coli suppresses DNA replication and its overexpression is lethal to cells (Yamanaka and Inouye, 2001; Uppal et al., 2014). In Clostridium botulinum ATCC3502, CspB and CspC are crucial for survival under NaCl, pH, or ethanol stress, and mutations in cspA and cspC hinder flagella formation and motility (Derman et al., 2015). Additionally, cspA in Brucella melitensis is critical in regulating virulence and metabolism (Wang Z. et al., 2016). Furthermore, CSPs play crucial roles in plant-microbe interactions. A CSP from Staphylococcus aureus has been identified as a PAMP specifically recognized by tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), and potato (Solanum tuberosum) (Felix and Boller, 2003). Although peptide-inoculation studies have shown that CSPs manipulate plant immunity, the mechanisms underlying CSP–plant interaction in vivo remain poorly understood.
A model system involving the interactions between Pseudomonas fluorescens and P. syringae pv. tomato DC3000 (hereafter, Pst DC3000) with N. benthamiana has been widely used to investigate the molecular mechanism of plant-microbe interactions, providing an ideal system to explore the roles of CSPs in vivo. Pst DC3000 is a model pathogen for Arabidopsis, tomato, and N. benthamiana (when the avirulent gene hopQ1-1 is deleted) and is equipped with a functional Type III Secretion System (T3SS) which translocates virulent effectors into plant cells to cause disease. P. fluorescens Pf0-1 is a non-virulent strain that is deficient in a typical T3SS apparatus but can induce PTI in plants. However, Pf0-1 can translocate type III effectors when carrying the functional hrp/hrc (hypersensitive response and pathogenesis/conserved) T3SS gene cluster on the cosmid pHIR11 (Mastropaolo et al., 2012; Wei et al., 2013; Ngou et al., 2021; Jayaraman et al., 2023).
In this study, we aimed to explore the role of CSPs in modulating plant immunity in the interactions between Pseudomonas and N. benthamiana. Our findings demonstrate that CSPs play a role in modulating plant immunity through a genetic strategy, providing deep insights into the PTI elicited by cytoplasmic bacterial proteins.
Materials and methods Strains, plasmids, primers, peptides, and plant materialsThe strains, plasmids, and primers used in this study are listed in Supplementary Tables S1–S3, respectively. All peptides used were synthesized by GenScript and are summarized in Supplementary Table S4. P. syringae and P. fluorescens were cultured in King’s medium B (KB) (King et al., 1954), Agrobacterium tumefaciens was grown in Luria-Bertani (LB) broth at 28°C, and E. coli was grown in LB broth at 37°C. The following antibiotic concentrations were used: ampicillin, 50 mg/mL; rifampicin, 50 mg/mL; spectinomycin 50 mg/mL; and tetracycline 20 mg/mL. N. benthamiana plants were grown in a chamber maintained at 16 h light/8 h dark, 65% humidity, and temperatures of 24°C during the day and 22°C at night.
ROS assayROS measurements were performed as previously described (Wei et al., 2013), with minor modifications. Briefly, fresh Pf0-1 and its derivatives were infiltrated into N. benthamiana leaves. The concentrations of bacterial suspensions are indicated in the figure legends. Leaf disks from the infiltrated area were excised 15 h post-inoculation (hpi) (36 hpi for transient expression) and placed into the wells of a 96-well plate pre-supplied with 100 μL of sterile water. Then, 100 μL of 0.5 mM L-012 (Wako, Japan) in 10 mM morpholinepropanesulfonic acid–KOH buffer (pH 7.4) was added to each well. ROS accumulation was measured by monitoring chemiluminescence using a microplate reader (Tecan, Switzerland).
Agrobacterium-mediated transient expressionTransient expression was performed as previously described (Wei et al., 2013). Briefly, transformed Agrobacterium strains were resuspended in 10 mM MES (pH 5.5) containing 200 μM acetosyringone to the desired optical density and incubated at 28°C for 3 h. The suspension was then infiltrated into N. benthamiana leaves using a 1 mL sterile syringe (without the needle). The inoculated plants were maintained in a greenhouse for subsequent assays.
Challenged-inoculation HR and bacterial growth assaysThe challenged-inoculation assay was performed as previously described (Wei et al., 2015). Briefly, fresh streaks of Pf0-1 and its derivatives, as well as Pst DC3000, were prepared from isolated colonies and grown overnight at 28°C on KB plates. For the HR suppression assay, Pf0-1 and its derivatives were pre-inoculated into N. benthamiana leaves at 2 × 108 CFU/mL. Six hours later, Pst DC3000 was infiltrated into the pre-treated areas at 2 × 107 CFU/mL. Images were captured 2 d after Pst DC3000 infiltration. For the bacterial growth assay, Pf0-1 and its derivatives were inoculated into N. benthamiana leaves at 4 × 107 CFU/mL. Six hours later, the strain Pst DC3000ΔhopQ1–1 was infiltrated into the pre-treated leaves at 5 × 105 CFU/mL. Leaf disks were harvested 4 d post-inoculation (dpi) to assess the bacterial population.
Effector translocation assayThe effector translocation assay was performed as previously described (Wei et al., 2013), with minor modifications. Briefly, bacterial infiltrations were performed using varying inoculum levels, as indicated in the figure legends. Seventy-two hours later, Pf0-1 (pCPP6225 + pCPP3221) was inoculated into N. benthamiana leaves transiently expressing CSPs or FliC derivatives, and leaf disks were collected 6 h after infiltration. For the challenge effector translocation assay, Pf0-1 (pCPP6225 + pCPP3221) or Pf0-1 (pCPP5316) was challenge-inoculated 6 h after pre-inoculation with Pf0-1 and its derivatives, and the leaf disks were harvested 6 hpi. To assess suppression of effector translocation induced by flg22, csp22, and csp15, peptides were infiltrated into N. benthamiana leaves at a concentration of 1 μM each. At 6 hpi, Pst DC3000 (pCPP3221) expressing AvrPto-Cya at 1 × 107 CFU/mL was challenge-inoculated into the plant leaves. Leaf disks were harvested 6 hpi, excised using a 1.0-cm-diameter cork borer, flash-frozen in liquid nitrogen, and ground in 250 μL of 0.1 M HCl. cAMP levels were measured using a Correlate-EIA cAMP immunoassay kit (Enzo, United States), according to the manufacturer’s instructions.
RNA preparation and qRT-PCRThe peptides were infiltrated into N. benthamiana leaves at a concentration of 100 nM. Three leaf disks were collected from the infiltrated area 6 hpi and ground in liquid nitrogen. Total RNA was isolated using the Plant RNA Kit (R6827-02, OMEGA, United States), following the manufacturer’s instructions. cDNA was synthesized using the FastKing RT kit (KR116-02; TIANGEN, China). Real-time PCR reactions were performed using the SYBR® Green Premix Pro Taq HS qPCR Kit (AG11701, Accurate Biology, China) and ROX Reference Dye (4 mM) (AG11710, Accurate Biology, China). The expression levels of WRKY22 and ERF1a were quantified using the qPCR primers listed in Supplementary Table S1. NbEF1α was used as a reference gene.
Accession numbersThe protein sequences used in this study are available under the following accession numbers: AAO55887.1 (CspA for Pst DC3000), ABA73672.1 (P. fluorescens), AAZ34509.1 (P. savastanoi pv. phaseolicola 1448A), UZD98366.1 (P. corrugata B21-055), UUI32708.1 (P. putida ATCC 12633), AAO57601.1 (CspB for Pst DC3000), ABA72944.1 (P. fluorescens), AAZ36544.1 (P. savastanoi pv. phaseolicola 1448A), UZD96690.1 (P. corrugata B21-055), UUI33322.1 (P. putida ATCC 12633), AAO54799.1 (CspC for Pst DC3000), ABA76132.1 (P. fluorescens), AAZ35758.1 (P. savastanoi pv. phaseolicola 1448A), UZD94220.1 (P. corrugata B21-055), and UUI36241.1 (P. putida ATCC 12633).
Statistical analysesStatistical analyses were performed using IBM SPSS Statistics software. Different letters indicate significant differences (p < 0.05 by one-way ANOVA).
Results Three CSPs from Pst DC3000 are highly conserved across strains of PseudomonasTo identify the CSPs in Pst DC3000, we performed a BLASTp analysis using the amino acids of csp15 and csp22—two peptides with PAMP activity that contain the RNP-1 epitope of CSP (Felix and Boller, 2003; Wang L. et al., 2016). Three proteins, CspA (AAO55887.1), CspB (AAO57601.1), and CspC (AAO54799.1), were identified by means of this method. Sequence alignment revealed that all three proteins were highly conserved in Pst DC3000, differing by only one amino acid from csp15 and five amino acids from csp22 (Figure 1A). We also performed multiple alignments of the three CSPs with the CSPs of other plant and environment associated-Pseudomonas strains, including the well-studied strain P. fluorescens Pf0-1. The results showed that these three CSPs were well conserved; CspA shares 94% homology with its homologs, CspB shares 95.43%, and CspC shares 96.86%. Collectively, these findings suggest that these CSPs may exhibit similar functions across diverse Pseudomonas strains (Figure 1B).
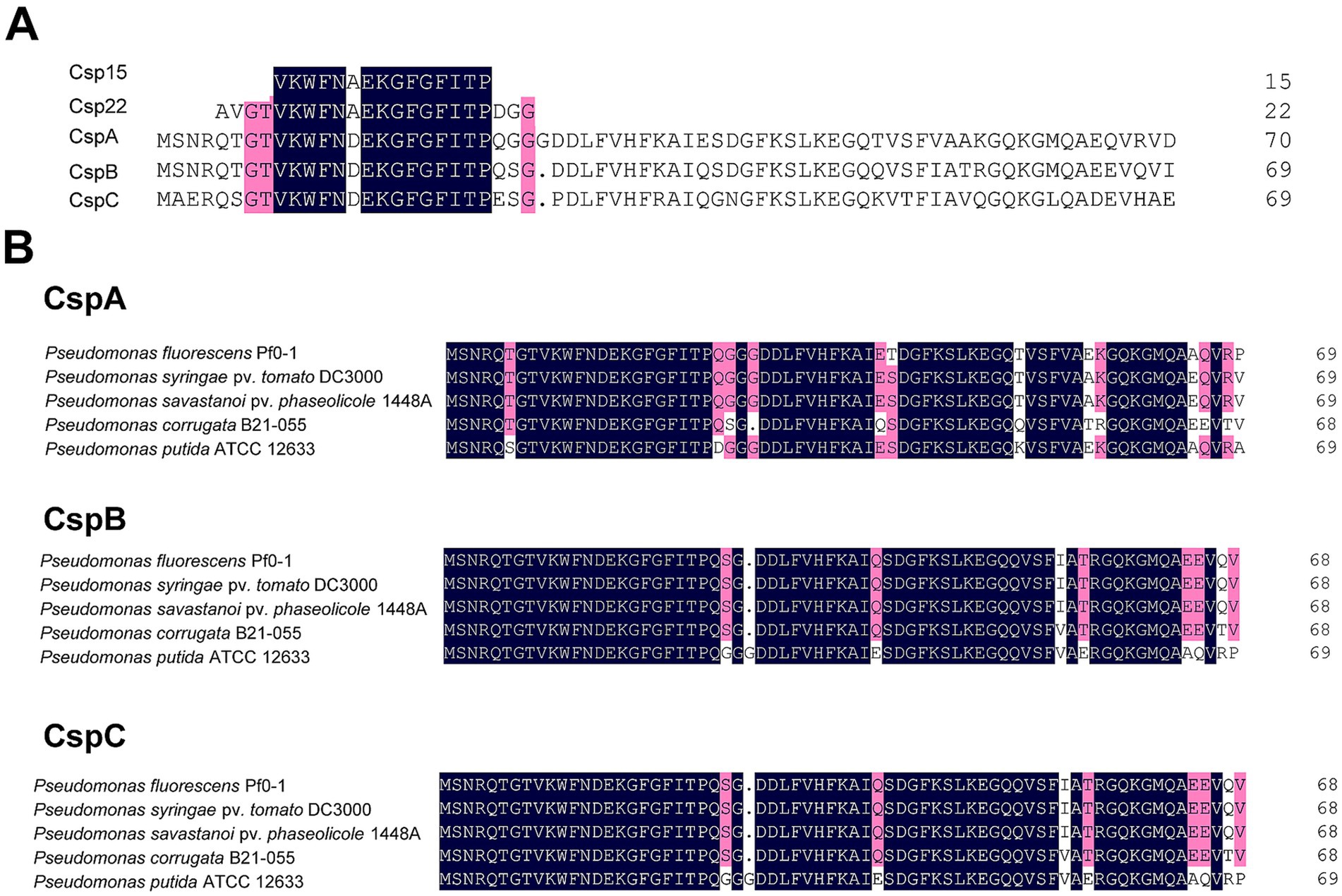
Figure 1. Sequence alignment of CSPs from Pseudomonas strains. (A) Alignment of CspA, CspB, and CspC of Pst DC3000 with csp15 and csp22 peptides. (B) Alignment of CspA, CspB, and CspC in plant-and environment-associated Pseudomonas strains. Amino acids of CSPs were downloaded from NCBI, while the sequences of csp15 and csp22 were obtained from the previous report (Wang L. et al., 2016). Sequence alignment was performed by DNAMAN. The point indicates the absence of amino acids at that position. Dark blue represents identical amino acids, while red indicates similar amino acids sharing homology between 75 and 100%, respectively.
Agrobacterium-mediated transient expression of CSPs from Pst DC3000 suppresses effector translocationTo determine whether the CSPs of Pst DC3000 modulate plant immunity in vivo, we cloned the CDSs of CspA, CspB, and CspC and transiently expressed them in N. benthamiana leaves under the control of the 35S promoter. First, we assessed whether the CSPs stimulated ROS accumulation. It has been well established that FliC was a major PAMP of Pseudomonas recognized by N. benthamiana, we therefore took advantage of FliC and FliC fused with the PR1a signal peptide (SP-FliC) as controls. As previously reported, FliC and SP-FliC induced weak and strong ROS bursts, respectively (Wei et al., 2013). However, CspA, CspB, and CspC did not stimulate ROS accumulation (Figure 2A). In the HR suppression analysis, Agrobacterium tumefaciens harboring pEarleyGateS101 carrying csp genes were inoculated into N. benthamiana leaves at 2 × 108 CFU/mL and then incubated for 48 h before partially overlapping challenge inoculation with a suspension of 4 × 107 CFU/mL of wild-type Pst DC3000. None of the CSPs inhibited the HR induced by Pst DC3000 (Figure 2B). The activation of PTI inhibits the translocation of effector proteins (Anderson et al., 2014). Therefore, we next investigated whether CSPs suppressed effector translocation by employing Pf0-1 harboring plasmids pCPP3221 expressing AvrPto-Cya and pCPP6225 (a derivative of T3SS+ pHIR11 with an unmarked deletion of the genes encoding the effector HopA1 and its chaperone ShcA). N. benthamiana leaves were infiltrated with A. tumefaciens cells and then challenged 72 h later with an overlapping inoculation of Pf0-1 (pCPP6225 + pCPP3221) for the Cya-based translocation assay, in which the translocation of effectors into plant cells can be clearly detected on the basis of the Cya activity as a high production of cAMP (Mukaihara and Tamura, 2009). As shown in Figure 2C, the expression of CspA and CspC, but not CspB, significantly suppressed AvrPto-Cya translocation, as well as SP-FliC, compared to EV and FliC, indicating that CspA and CspC of Pst DC3000 suppress effector translocation when transiently expressed in N. benthamiana leaves. Collectively, these results suggest that the CSPs of Pst DC3000 can suppress effector translocation in N. benthamiana leaves.
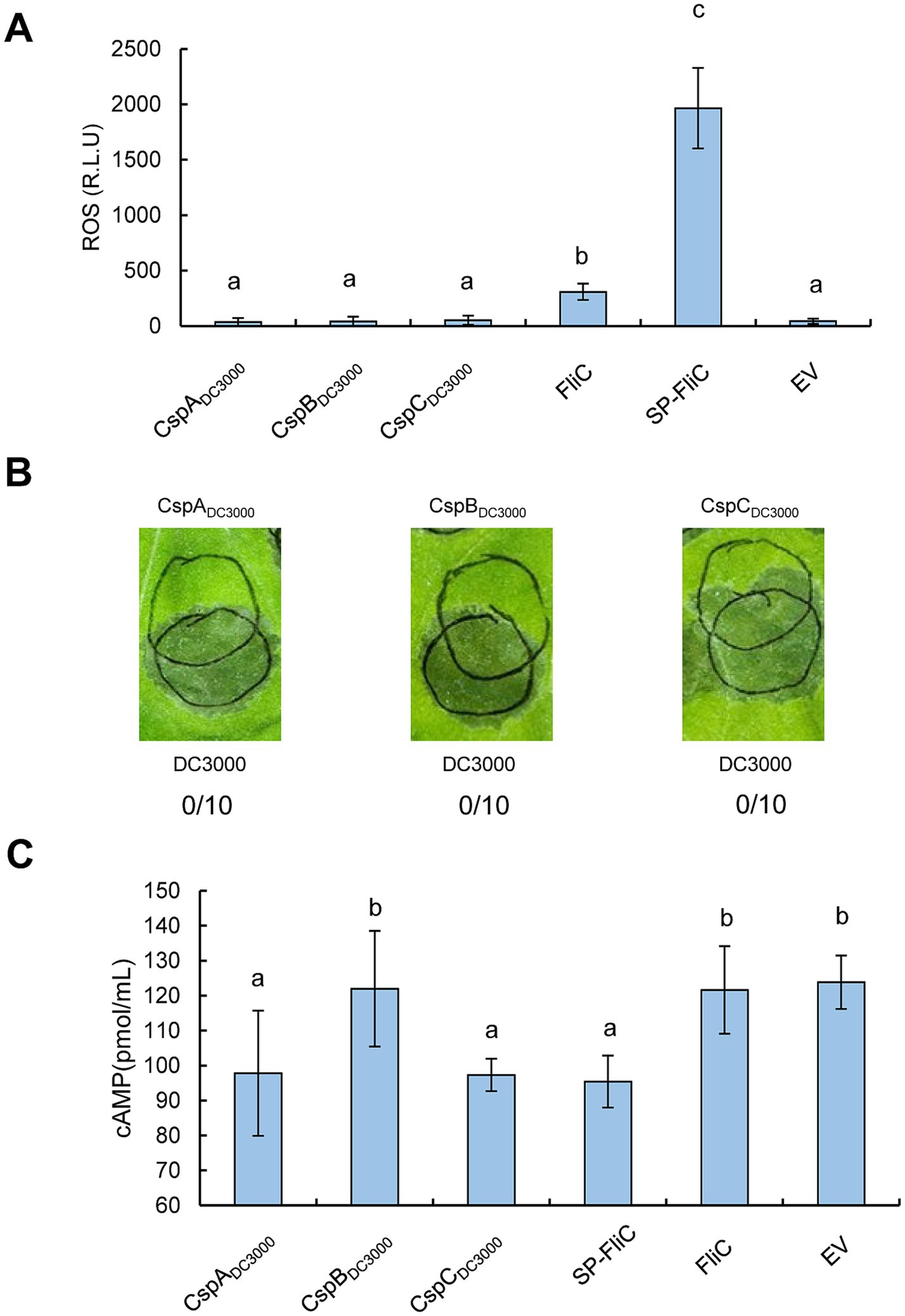
Figure 2. Assessments of PTI induction activities of CSPs from Pst DC3000 by Agrobacterium-mediated transient expression in N. benthamiana leaves. (A) CSPs cannot induce ROS production when transiently expressed in N. benthamiana leaves. CSPs from Pst DC3000 were Agrobacterium-mediated transiently expressed in N. benthamiana leaves at 2 × 108 CFU/mL, leaf disks were collected at 36 hpi to monitor ROS accumulation. (B) CSPs cannot suppress HR when transiently expressed in N. benthamiana leaves. The three CSPs were Agrobacterium-mediated transiently expressed at 2 × 108 CFU/mL prior to an overlapping infiltration of wild-type Pst DC3000 at 4 × 107 CFU/mL. The upper circles indicate transient expression inoculation, while the lower circles indicate infiltration with Pst DC3000. Leaves were photographed 48 h after the challenge inoculation. The number of times each test CSP induced PTI as a fraction of number of times tested are shown below each photograph, respectively. (C) CSPs suppress effector translocation when transiently expressed in N. benthamiana leaves. Agrobacterium cells harboring expression vectors were infiltrated with N. benthamiana leaves at 2 × 108 CFU/mL, and challenged 72 h later with an inoculation of Pf0-1 (pCPP6225 + pCPP3221) at 4 × 107 CFU/mL, leaf disks were harvested at 12 hpi to determine the cAMP level. EV indicates empty vector. Different letters indicate significant differences (p < 0.05 by one-way ANOVA), respectively. All experiments were repeated three times with similar results.
CSPs from Pseudomonas fluorescens Pf0-1 are redundant in suppressing effector translocation independent of FliCTo further probe into the function of CSPs in vivo and to circumvent the effects of effectors, we employed P. fluorescens Pf0-1 in subsequent assays. Mutants lacking individual CSPs (ΔcspA, ΔcspB, or ΔcspC) and the triple mutant ΔcspABC were generated and subjected to ROS burst assay. The ΔfliC mutant served as a positive control, while 10 mM MgCl2 served as a reagent control. As expected, Pf0-1 strongly induced ROS accumulation compared to the mock treatment, whereas ΔfliC induced negligible ROS production (Figure 3A). ROS levels in N. benthamiana leaves treated with ΔcspA, ΔcspB, ΔcspC, or the triple mutant ΔcspABC were comparable to those in leaves treated with Pf0-1, suggesting that deleting the three CSPs could not impair the ROS burst (Figure 3A).
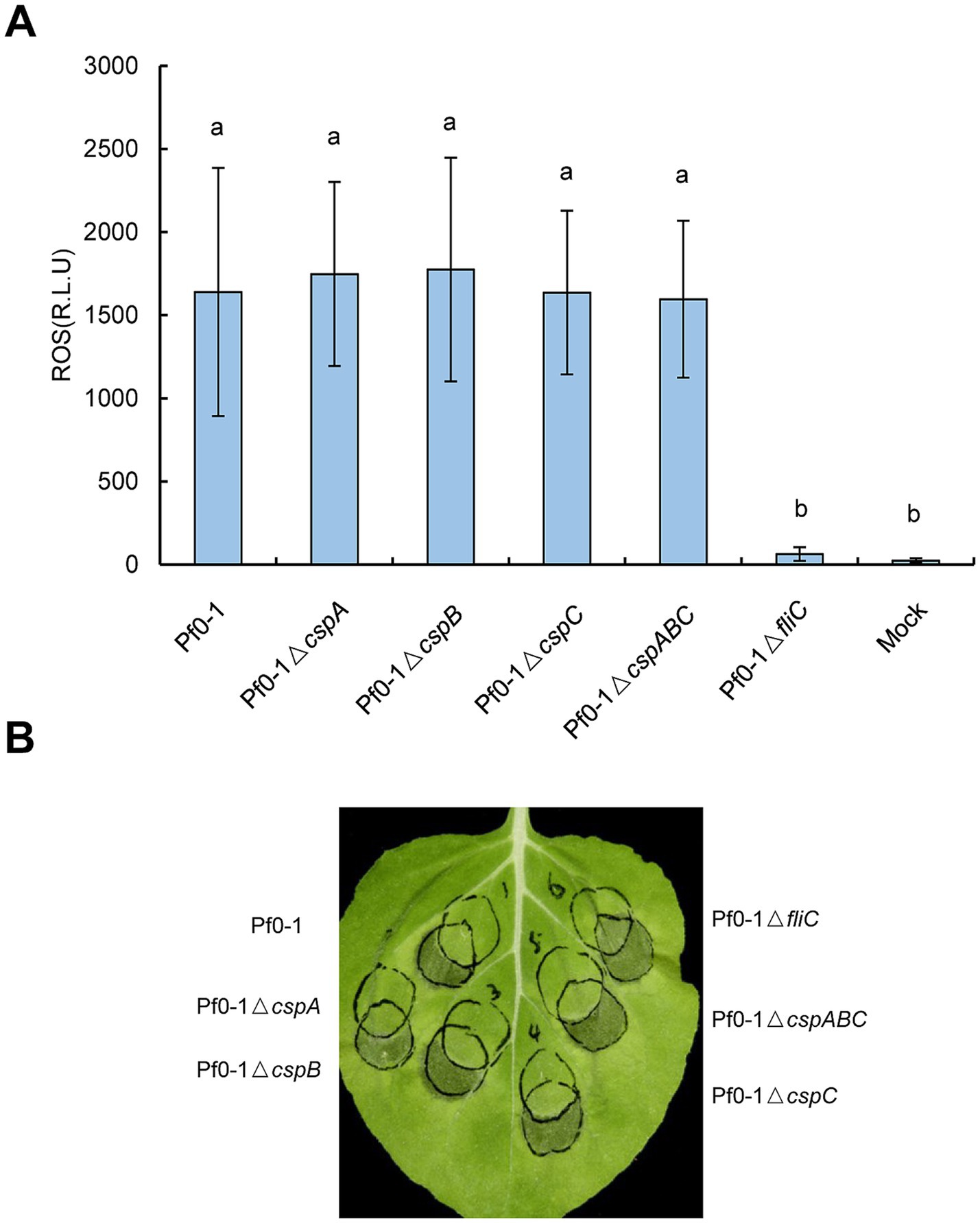
Figure 3. Pseudomonas CSPs have no contributions to the ROS burst or HR inhibition in vivo. (A) Mutations of csp genes have no impact on ROS accumulation. Pf0-1 and its derivatives were inoculated into N. benthamiana leaves at 2 × 108 CFU/mL, leaf disks were collected at 15 hpi to monitor the ROS level. Different letters indicate significant differences (p < 0.05 by one-way ANOVA), respectively. (B) Mutations of csp genes of Pf0-1 did not impair HR suppression ability. Pf0-1 and its derivatives were inoculated into N. benthamiana leaves at 2 × 108 CFU/mL, then 6 h later, Pst DC3000 was overlapped infiltrated with the pre-treated areas at 2 × 107 CFU/mL. For the three treatments on the left side of the leaf, the right or upper circles represent inoculation with Pf0-1 or its derivatives; for the three treatments on the right side of the leaf, the left or upper circles indicate inoculation with Pf0-1 derivatives. The pictures were photographed 48 h after challenge infiltration. All experiments were repeated three times with similar results.
Next, we examined whether the ability of Pf0-1 to suppress HR was associated with CSPs. N. benthamiana plants were inoculated with Pf0-1 and its derivatives at 4 × 107 CFU/mL, incubated for 6 h, and then challenge inoculated with wild-type Pst DC3000 at 2 × 107 CFU/mL. All treatments, including Pf0-1, ΔcspA, ΔcspB, ΔcspC, or the triple mutant ΔcspABC, suppressed the HR induced by Pst DC3000, whereas the control ΔfliC mutant lost this ability, indicating that mutating these three CSPs does not affect the HR suppression ability of Pf0-1 (Figure 3B). Next, we used Pf0-1 (pCPP6225 + pCPP3221) to investigate whether CSPs play a role in suppressing effector translocation. Pf0-1 (pCPP6225 + pCPP3221) was infiltrated into N. benthamiana leaves 6 h after inoculation with Pf0-1 and its derivatives to measure cAMP levels. The results showed that cAMP levels induced by Pf0-1, ΔcspA, ΔcspB, or ΔcspC treatments were analogous, but were significantly lower than the mock treatment. However, ΔcspABC inoculation generated notably higher cAMP levels than Pf0-1, ΔcspA, ΔcspB, or ΔcspC treatments, suggesting that CspA, CspB, and CspC of Pf0-1 were functionally redundant in mitigating the effector injection restriction (Figure 4A).
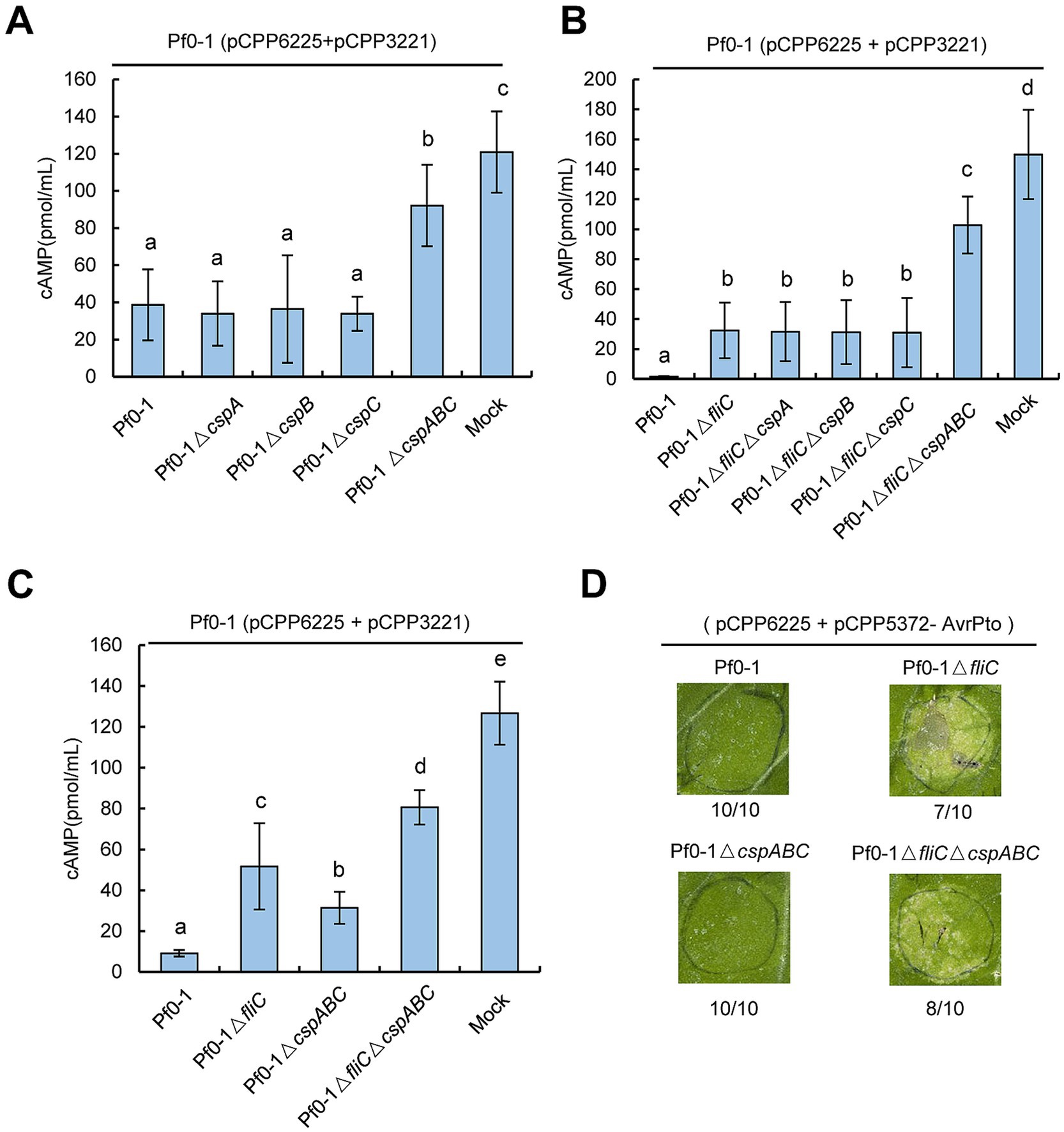
Figure 4. Pseudomonas CSPs share functional redundancy in restricting effector translocation in a FliC-independent manner. (A) Pseudomonas CSPs are redundant in suppressing effector translocation. P. fluorescens strains were infiltrated with N. benthamiana leaves at 4× 107 CFU/mL and challenged 6 h later with an overlapping inoculation of P. fluorescens (pCPP6225 + pCPP3221) expressing AvrPto-Cya at 4 × 107 CFU/mL. At 6 h after the challenge inoculation, leaf disks were collected from areas of overlap between pretreatment and challenge inoculations to determine the cAMP level. (B) Pseudomonas CSPs suppress effector translocation independent of FliC. P. fluorescens strains were infiltrated with N. benthamiana leaves at 2 × 108 CFU/mL, challenge inoculation was done as above. (C) FliC suppresses more AvrPto translocation than CSPs. P. fluorescens strains were infiltrated with N. benthamiana leaves at 2 × 108 CFU/mL, challenge inoculation was done as above. (D) Mutations of fliC but not csp genes generate AvrPto-induced chlorosis. Pf0-1 derivatives carrying pCPP6225 and pCPP5372-AvrPto were infiltrated with N. benthamiana leaves at 5 × 108 CFU/mL. Pictures were photographed at 72 hpi. Relative numbers of chlorosis in inoculated zones are shown. Different letters indicate significant differences (p < 0.05 by one-way ANOVA), respectively. All experiments were repeated three times with similar results.
FliC is a major Pseudomonas PAMP recognized by N. benthamiana. Therefore, we investigated whether the ability of CSPs to suppress effector translocation was related to FliC. We constructed Pf0-1ΔfliC mutants with individual or combined deletions of cspA, cspB, and cspC. Effector translocation assays showed that ΔfliC induced higher cAMP levels than Pf0-1 (Figure 4B). Moreover, cAMP levels induced by Pf0-1ΔfliC, ΔfliCΔcspA, ΔfliCΔcspB, or ΔfliCΔcspC were similar, but significantly lower than those induced by ΔfliCΔcspABC (Figure 4B). These data indicate that CSPs are redundant in suppressing effector translocation independent of FliC.
CSPs-mediated suppression of effector translocation is modest compared to that triggered by FliCTo compare the relative contributions of FliC and CSPs to effector translocation inhibition, we conducted further assays. N. benthamiana leaves were inoculated with Pf0-1 and its derivatives at 2 × 108 CFU/mL and challenged 6 h later with Pf0-1 (pCPP6225 + pCPP3221) at 4 × 107 CFU/mL. Effector translocation assays showed that Pf0-1ΔfliC induced significantly higher cAMP levels than Pf0-1ΔcspABC, indicating that FliC inhibits effector translocation more effectively than CSPs (Figure 4C). Furthermore, we transformed pCPP5372-AvrPto into Pf0-1 derivatives carrying pCPP6225, infiltrated them into N. benthamiana leaves at 5 × 108 CFU/mL, and kept the plants in the chamber for 3 days. The results showed that ΔfliC and ΔfliCΔcspABC, but not ΔcspABC, caused AvrPto-induced chlorosis, suggesting that mutation of FliC allowed more AvrPto translocation than CSPs (Figure 4D). Additionally, ΔfliCΔcspABC induced significantly higher cAMP levels than ΔfliC and ΔcspABC, further supporting the conclusion that CSPs suppress effector translocation independently of FliC (Figure 4C). We further analyzed the effect of these mutations on HopA1 translocation. Pf0-1 was transformed with pCPP5316 carrying the functional T3SS cluster and HopA1-Cya, and challenge-inoculation effector translocation assays were performed. The results showed that ΔfliC induced significantly higher cAMP levels than Pf0-1, but lower than ΔfliCΔcspABC, suggesting that both fliC and cspABC mutations increase HopA1 translocation (Supplementary Figure S1). We also investigated whether mutations in CSPs alleviated PTI-associated inhibition of challenge-inoculated pathogen growth. Pf0-1 and its derivatives were infiltrated into N. benthamiana leaves at 4 × 107 CFU/mL and incubated for 6 h before challenge inoculation with Pst DC3000ΔhopQ1–1 at 5 × 105 CFU/mL. The results showed that at 4 dpi, no significant difference in pathogen population suppression was observed between the CSP-deleted mutants and Pf0-1 or Pf0-1ΔfliC (Supplementary Figure S2). Collectively, these results suggest that the suppression of effector translocation mediated by CSPs is moderate compared to that mediated by FliC.
csp22 and csp15 suppress effector translocation in Nicotiana benthamianaTo further validate the role of CSPs in restricting effector translocation, we synthesized csp22 and csp15 peptides, which represent the RNP-1 epitope of CSP with PAMP activity as well as the conserved flg22 epitope. We aimed to test whether inoculation with these peptides could induce PTI-associated suppression of effector translocation by assessing their ability to inhibit HR and the cAMP levels produced by Cya fusion. We found that HR was nearly eliminated in N. benthamiana leaves pre-infiltrated with flg22 following challenge inoculation with Pst DC3000, whereas it was partially eliminated in leaves pre-infiltrated with csp22 or csp15 (Figure 5A). Moreover, pre-infiltration with flg22, csp22, or csp15 resulted in significantly lower cAMP levels than the mock treatment, indicating that these peptides effectively suppressed effector translocation in N. benthamiana (Figure 5B). Additionally, inoculation with these peptides upregulated the PTI marker gene—WRKY22—and the ethylene-related marker gene—ERF1a —consistent with previous reports (Felix and Boller, 2003; Saur et al., 2016) (Supplementary Figure S3). Collectively, these findings suggest that csp22 and csp15 trigger PTI-associated suppression of effector translocation.
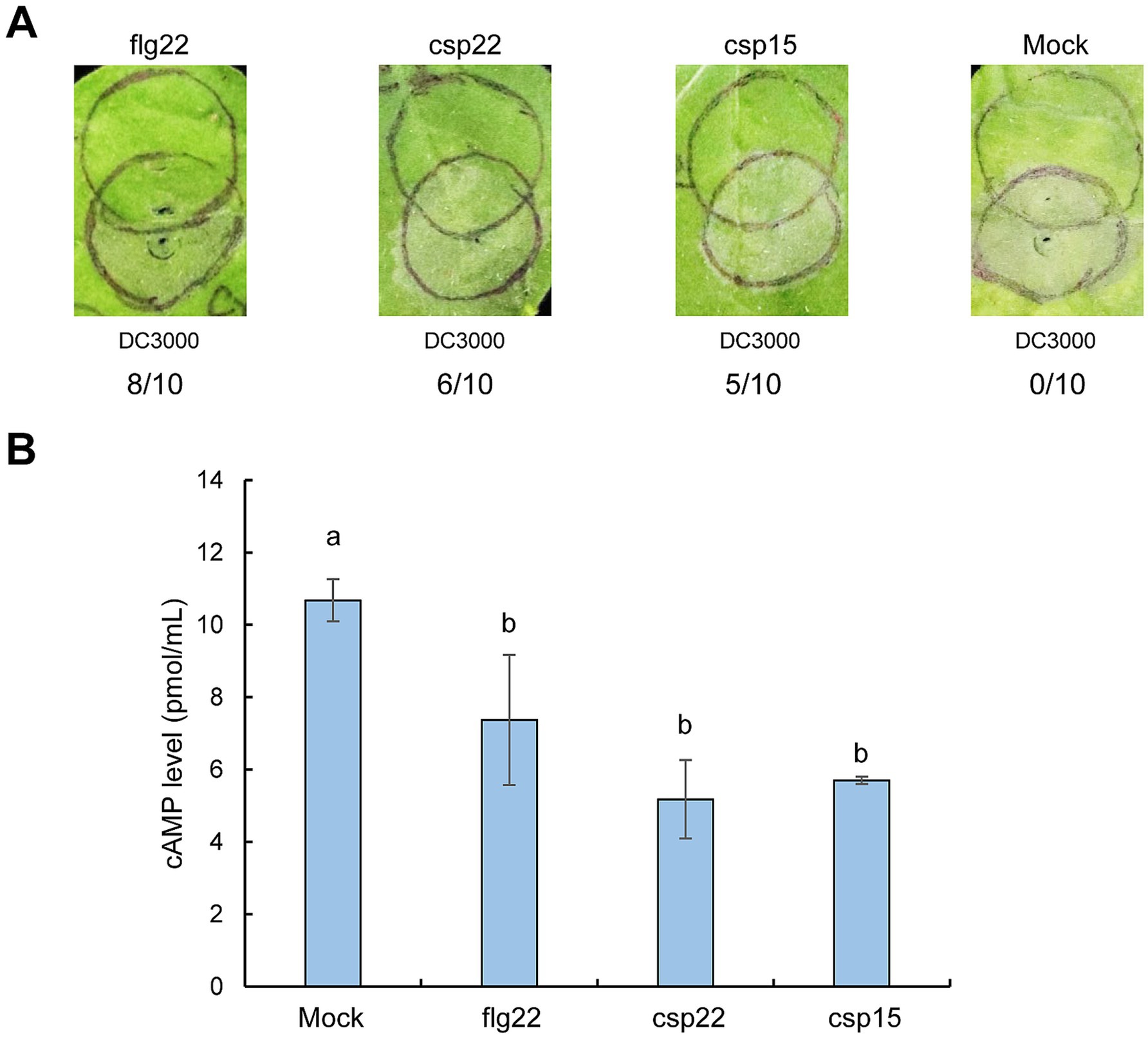
Figure 5. Inoculation of csp22 and csp15 restricts effector translocation in N. benthamiana plants. (A) Inoculation of csp22 and csp15 suppresses HR induced by Pst DC3000. Peptides were infiltrated with N. benthamiana leaves at a concentration of 1 μM. At 6 hpi, Pst DC3000 suspension was challenge-inoculated into plant leaves at 5 × 106 CFU/mL. Pictures were photographed at 72 h post final inoculation. The upper circles were inoculated with peptides, while the lower circles were infiltrated with Pst DC3000. The number of times each test peptide induced PTI as a fraction of number of times tested are shown below each photograph, respectively. (B) Inoculation of csp22 and csp15 suppresses effector translocation. The inoculation of peptides was performed as above and challenge 6 h later with Pst DC3000 (pCPP3221) expressing AvrPto-Cya at 1 × 107 CFU/mL. At 6 h after the challenge inoculation, leaf disks were collected from areas of overlap between pretreatment and challenge inoculations to determine the cAMP level. Different letters indicate significant differences (p < 0.05 by one-way ANOVA), respectively. All experiments were repeated three times with similar results.
DiscussionIn this study, we employed genetic approaches to investigate the role of Pseudomonas CSPs in modulating plant immunity. Multiple alignments revealed that the CSPs were highly conserved in pathogenic and nonvirulent Pseudomonas strains, consistent with a recent finding suggesting that these CSPs shared similar functions (Chen et al., 2024) (Figure 1B). Further genetic assessments indicated that CSPs from Pst DC3000 and Pf0-1 did not contribute to ROS burst or the suppression of HR induced by Pst DC3000 in vivo. However, CSPs were redundant in suppressing bacterial effector translocation in a FliC-independent manner (Figures 2–4). Additionally, inoculation with csp15 and csp22 peptides suppressed effector translocation (Figure 5B). These findings suggest that CSPs from different bacterial species are functionally conserved in suppressing bacterial effector translocation.
Over two decades ago, a highly conserved CSP was identified as a PAMP that is recognized by the immune system of Solanales plants (Felix and Boller, 2003). Subsequent studies identified two PRRs—CORE in tomato and NbCSPR in N. benthamiana—that recognize the csp22 epitope (Saur et al., 2016; Wang et al., 2016). Stable transgenic A. thaliana plants expressing NbCORE or NbCSPR exhibited stronger resistance to Pst DC3000, implying that Pseudomonas CSPs can be recognized by these receptors in N. benthamiana plants in vivo (Saur et al., 2016; Wang et al., 2016). However, genetic analysis in the current study showed that Pseudomonas CSPs did not stimulate ROS production or suppress HR, likely due to the age-dependent low expression of NbCORE and NbCSPR in 4-week-old N. benthamiana plants (Figures 2A,B, 3) (Saur et al., 2016; Wang et al., 2016). Transient expression of CspA and CspC from Pst DC3000 restricted effector injection; a result similar to that observed when FliC was transiently expressed in N. benthamiana leaves (Figure 2C). This finding suggests that some PAMPs, when strongly expressed under the control of the 35S promoter, may leak from the cytoplasm, enabling extracellular perception by PRRs (Wei et al., 2013). We hypothesized that the transient expression of cspB might be too weak to allow the leakage of CspB from the cytosol, accounting for the absence of restriction on effector translocation in our study (Figure 2C). Although Agrobacterium-mediated transient expression has been extensively utilized in exploring the biological function of protein in planta, there are still certain limitations in yield and sustainability of expressed proteins. Furthermore, the efficiency of protein expression is affected by the concentration and timing of the inoculation. Therefore, further investigation into the CSPs-triggered PTI responses in the stable expression system of plants will provide deeper insights into the roles of CSPs in plant-microbe interactions.
Gene family expansion is widespread across bacterial genomes and often results in functionally conserved members (Hahn et al., 2007; Collins et al., 2011). The flg22 epitopes from commensal bacteria of Arabidopsis thaliana induced similar immune responses (Colaianni et al., 2021). Similar phenomenon was observed in our study which demonstrates that conserved Pseudomonas CSPs share functional redundancy in suppressing effector translocation, which is a critical step for bacterial pathogenesis (Figure 4A). Furthermore, we established that CSPs-and FliC-mediated inhibition of effector translocation occured via independent signaling pathways, with FliC suppressing more effector injection than CSPs (Figures 4B,C). This difference in inhibition is likely due to the different localization of FliC and CSPs in bacterial cells; FliC is an extracellular protein more readily exposed to plant membranes to induce immune responses, whereas CSPs are cytoplasmic bacterial proteins, with only a small number present in the secretomes of bacteria (Song et al., 2009; Kühn et al., 2018). This difference in inhibition accounts for FliC being the primary PAMP and the lack of impact from csp gene deletion in Pf0-1 on ROS accumulation or HR suppression (Figure 3). Additionally, deletion of csp from Pf0-1 or Pf0-1ΔfliC did not affect the growth of Pst DC3000ΔhopQ1-1 in the challenge inoculation assay (Supplementary Figure S2). A similar finding, where deletion of fliC did not significantly impact the survival of pathogen Pst DC3000ΔhopQ1-1, was likely due to the high virulence of Pst DC3000ΔhopQ1-1 which masked the minor growth promotion triggered by the defective PTI response (Kvitko et al., 2009). Collectively, our results illustrate significant roles for CSPs in modulating plant immunity, providing deep insights into the mechanisms by which cytoplasmic bacterial proteins-triggered PTI responses.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributionsSC: Investigation, Writing – original draft. J-ZL: Investigation, Methodology, Resources, Writing – original draft. M-RZ: Data curation, Formal analysis, Methodology, Writing – original draft. H-LW: Conceptualization, Writing – original draft, Writing – review & editing. WZ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Key R&D Program of China (2022YFD1901300), the National Natural Science Foundation of China (32472514), the Science and Technology Programs of the Zunyi Tobacco (2021XM03), and the Major Science and Technology Project of China National Tobacco Corporation [110202201005 (JY-05)]. H. -L. Wei and J. -Z. Li were supported by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-CSCB-202401) and the Beijing Innovation Consortium of the Agriculture Research System (BAIC04-2024).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statementThe authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1539906/full#supplementary-material
ReferencesAnderson, J. C., Wan, Y., Kim, Y.-M., Pasa-Tolic, L., Metz, T. O., and Peck, S. C. (2014). Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 111, 6846–6851. doi: 10.1073/pnas.1403248111
PubMed Abstract | Crossref Full Text | Google Scholar
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
PubMed Abstract | Crossref Full Text | Google Scholar
Charollais, J. (2004). CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32, 2751–2759. doi: 10.1093/nar/gkh603
PubMed Abstract | Crossref Full Text | Google Scholar
Chen, C., Buscaill, P., Sanguankiattichai, N., Huang, J., Kaschani, F., Kaiser, M., et al. (2024). Extracellular plant subtilases dampen cold-shock peptide elicitor levels. Nat. Plants 10, 1749–1760. doi: 10.1038/s41477-024-01815-8
PubMed Abstract | Crossref Full Text | Google Scholar
Colaianni, N. R., Parys, K., Lee, H.-S., Conway, J. M., Kim, N. H., Edelbacher, N., et al. (2021). A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 29, 635–649.e9. doi: 10.1016/j.chom.2021.02.006
PubMed Abstract | Crossref Full Text | Google Scholar
Derman, Y., Söderholm, H., Lindström, M., and Korkeala, H. (2015). Role of csp genes in NaCl, pH, and ethanol stress response and motility in Clostridium botulinum ATCC 3502. Food Microbiol. 46, 463–470. doi: 10.1016/j.fm.2014.09.004
PubMed Abstract | Crossref Full Text | Google Scholar
Felix, G., and Boller, T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208. doi: 10.1074/jbc.M209880200
PubMed Abstract | Crossref Full Text | Google Scholar
Jayaraman, J., Yoon, M., Hemara, L. M., Bohne, D., Tahir, J., Chen, R. K. Y., et al. (2023). Contrasting effector profiles between bacterial colonisers of kiwifruit reveal redundant roles converging on PTI-suppression and RIN4. New Phytol. 238, 1605–1619. doi: 10.1111/nph.18848
PubMed Abstract | Crossref Full Text | Google Scholar
Keto-Timonen, R., Hietala, N., Palonen, E., Hakakorpi, A., Lindström, M., and Korkeala, H. (2016). Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 7:1151. doi: 10.3389/fmicb.2016.01151
PubMed Abstract | Crossref Full Text | Google Scholar
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307
PubMed Abstract | Google Scholar
Kühn, M. J., Schmidt, F. K., Farthing, N. E., Rossmann, F. M., Helm, B., Wilson, L. G., et al. (2018). Spatial arrangement of several flagellins within bacterial flagella improves motility in different environments. Nat. Commun. 9:5369. doi: 10.1038/s41467-018-07802-w
PubMed Abstract | Crossref Full Text | Google Scholar
Kvitko, B. H., Park, D. H., Velásquez, A. C., Wei, C.-F., Russell, A. B., Martin, G. B., et al. (2009). Deletions in the repertoire of Pseudomonas syringae pv. Tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. doi: 10.1371/journal.ppat.1000388
PubMed Abstract | Crossref Full Text | Google Scholar
Mastropaolo, M. D., Silby, M. W., Nicoll, J. S., and Levy, S. B. (2012). Novel genes involved in Pseudomonas fluorescens Pf0-1 motility and biofilm formation. Appl. Environ. Microbiol. 78, 4318–4329. doi: 10.1128/AEM.07201-11
PubMed Abstract | Crossref Full Text | Google Scholar
Mukaihara, T., and Tamura, N. (2009). Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrp B-regulated gene products. Microbiology 155, 2235–2244. doi: 10.1099/mic.0.027763-0
PubMed Abstract | Crossref Full Text | Google Scholar
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
PubMed Abstract | Crossref Full Text | Google Scholar
Nguyen, H. P., Chakravarthy, S., Velásquez, A. C., McLane, H. L., Zeng, L., Nakayashiki, H., et al. (2010). Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant-Microbe Interact. 23, 991–999. doi: 10.1094/MPMI-23-8-0991
PubMed Abstract | Crossref Full Text | Google Scholar
Saur, I. M. L., Kadota, Y., Sklenar, J., Holton, N. J., Smakowska, E., Belkhadir, Y., et al. (2016). NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 113, 3389–3394. doi: 10.1073/pnas.1511847113
PubMed Abstract | Crossref Full Text | Google Scholar
Uppal, S., Shetty, D. M., and Jawali, N. (2014). Cyclic AMP receptor protein regulates csp D, a bacterial toxin gene, in Escherichia coli. J. Bacteriol. 196, 1569–1577. doi: 10.1128/JB.01476-13
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, L., Albert, M., Einig, E., Fürst, U., Krust, D., and Felix, G. (2016). The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2:16185. doi: 10.1038/nplants.2016.185
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, Z., Liu, W., Wu, T., Bie, P., and Wu, Q. (2016). RNA-seq reveals the critical role of Csp a in regulating Brucella melitensis metabolism and virulence. Sci. China Life Sci. 59, 417–424. doi: 10.1007/s11427-015-4981-6
PubMed Abstract | Crossref Full Text | Google Scholar
Wei, H.-L., Chakravarthy, S., Mathieu, J., Helmann, T. C., Stodghill, P., Swingle, B., et al. (2015). Pseudomonas syringae pv. Tomato DC3000 type III secretion effector polymutants reveal an interplay between hop AD1 and Avr PtoB. Cell Host Microbe 17, 752–762. doi: 10.1016/j.chom.2015.05.007
留言 (0)