Akabane virus (AKAV) is a culicoides-borne arbovirus that primarily affects cattle and small ruminants (Tohru et al., 2019). AKAV infection can result in congenital defects, abortion, premature birth, stillbirth (Kirkland, 2015), and even encephalitis in pregnant animals (Lee et al., 2002). Historical outbreaks of AKAV-induced animal diseases have caused significant economic losses. For instance, in 2010, AKAV led to encephalomyelitis in over 500 cattle in South Korea (Jeong et al., 2017), and in 2011, an outbreak of AKAV-induced encephalomyelitis occurred Shimane Prefecture, western Japan (Hayama et al., 2016). Between 2013 and 2016, numerous adult and juvenile bamboo rats in Guangxi Province, China, succumbed to AKAV infection and died (Tang et al., 2017). The spread of AKAV has consequently imposed substantial losses and financial burdens on the animal husbandry.
AKAV is crucial vectors by biting midges (Kurogi et al., 1987; Jeong et al., 2017), and its spread is largely influenced by the geographical distribution of these vectors. The virus has been detected in various parts of the temperate and subtropical regions, including Australia (Della-Porta et al., 1977), Asia (Bryant et al., 2005; Hayama et al., 2016; Jeong et al., 2017), the Middle East (Stram et al., 2004), and parts of African (Metselaar and Robin, 1976). AKAV was first isolated from mosquito specimens in Japan (Oya et al., 1961). Recently, the virus was detected in Yunnan Province, China, and has been isolated from local Anopheles vagas, Culex quinquefasciatus (Feng et al., 2015), as well as from sentinel goats, and cattle (Meng et al., 2019, 2020; Gao et al., 2022). Yunnan’s subtropical climate, diverse ecosystems, and extensive agricultural activities provide optimal conditions for the breeding of biting midges, the primary vector of AKAV. Moreover, Yunnan’s borders with several Southeast Asian countries, where cross-border livestock movement is common, further heightens the risk of AKAV introduction and spread. However, despite the crucial role of biting midges in AKAV transmission, the virus has not yet been isolated from biting midges collected on the Chinese mainland.
In this study, two AKAV strains (No. 52 and 55) were isolated from biting midges collected near cattle shelters in Shizong County, Yunnan Province, China, in 2013. The complete genome of both strains was sequenced, and neutralizing antibodies to AKAV were detected in domestic animals. These findings suggest that AKAV is present in natural populations of biting midges in mainland China. Furthermore, this virus transmitted by these biting midges appears to have established a natural cycle involving local domestic animals.
2 Materials and methods2.1 Collection of biting midge samplesBiting midges were collected using light traps (Gongfu Xiaoshua, 12 V, 300 mA, Wuhan, China) placed near cattle, goats, and pig helters in Wulong Township, Shizong County, Yunnan Province, in July 2013. The collection occurred overnight, from 18:00 to 8:00 the following morning (Zhai et al., 2006).
The collected biting midges were killed by freezing at −20°C for 30 minutes and identified based on their morphological characteristics. They were then pooled into groups, with each group containing approximately 50 biting midges, and placed into individua tubes. Each tube was labeled and stored in liquid nitrogen until transported to the laboratory for virus isolation.
2.2 Collection of animal serum samplesCattle and goats over one year of age, with no record of AKAV vaccination, were randomly selected from Shizong, Jiangcheng, and Shuangjiang counties in Yunnan Province. Jugular vein blood samples (5 mL) were collected from each animal using vacuum blood collection tubes. The blood was allowed to coagulate, after which the tubes were centrifuged at 3000 rpm for 10 minutes. The separated serum samples were then frozen at −20°C until further use.
2.3 Cells culture and isolation of virusesBaby hamster kidney (BHK-21)cells, Madin-Darby bovine kidney (MDBK) cells, African green monkey kidney (VERO) cells, and Aedes albopictus cells (C6/36) were stored in the Yunnan tropical subtropical animal virus diseases laboratory. BHK-21, MDBK, and VERO were cultured in Dulbecco’s modified Eagle’s medium (DMEM) at 37°C, while C6/36 was cultured in Roswell Park Memorial Institute medium 1640 at 28°C. All cells were grown in respective media, supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, at a pH of 7.4.
Biting midge pools were removed from liquid nitrogen and homogenized in 1 mL of grinding fluid in sterile Eppendorf tube using a high-throughput tissue grinder (QIAGEN TissueLyser II, Germany) at −20°C for 10 minutes at a frequency of 30 times per second. The tubes were then centrifuged at 8000 rpm for 10 minutes, and 100 μL of supernatants was inoculated into each well containing a monolayer of C6/36, BHK-21, MDBK, or VERO cells in 24-well plates respectively. The cells were observed daily (days 1–7 post-inoculation) for cytopathic effects (CPE). A blind passage was sustained for three generations, and the supernatants from CPE-positive cultures were used for expanded cultivation and stored at –80°C for identification.
2.4 Preliminary identification of virusViral RNA was extracted from the CPE-positive cell culture supernatants using RNAiso Plus (TaKaRa, Dalian, China), following the manufacturer’s instructions.
The virus was identified through reverse transcriptase-polymerase chain reaction (RT-PCR). First-strand cDNA was synthesized using random primers P(d)N6 (TaKaRa, Dalian, China) and PrimeScript1 Reverse Transcriptase (TaKaRa, Dalian, China), as per the manufacturer’s instructions. The cDNA was then amplified via PCR using specific primers for Flavivirus (Kuno, 1998), Alphavirus (Pfeffer et al., 1997), and Simbu serogroup (Camarão et al., 2019). The PCR products were separated by electrophoresis on 1.3% agarose gel containing nucleic acid dye 1× TAE buffer. The results were visualized under ultraviolet light and sequenced using next-generation sequencing.
2.5 Full genome sequencingThe PCR primers were designed based on the AKAV strain sequence DHL10M110 (KY284023, KY284022, KY284021) in GenBank (Table 1). RT-PCR was performed on viral RNA to obtain all AKAV gene fragments. The amplified DNA fragments were purified using a DNA Fragment Purification Kit (TaKaRa, Dalian, China). Then cloned into the pMD19-T cloning vector (TakaRa, Dalian, China) and transformed into the chemically competent Escherichia coli DH5α cells (TakaRa, Dalian, China). The PCR products and the selected transformed colonies were cultured and sequenced by Beijing Tsingke Biotech (Beijing, China).
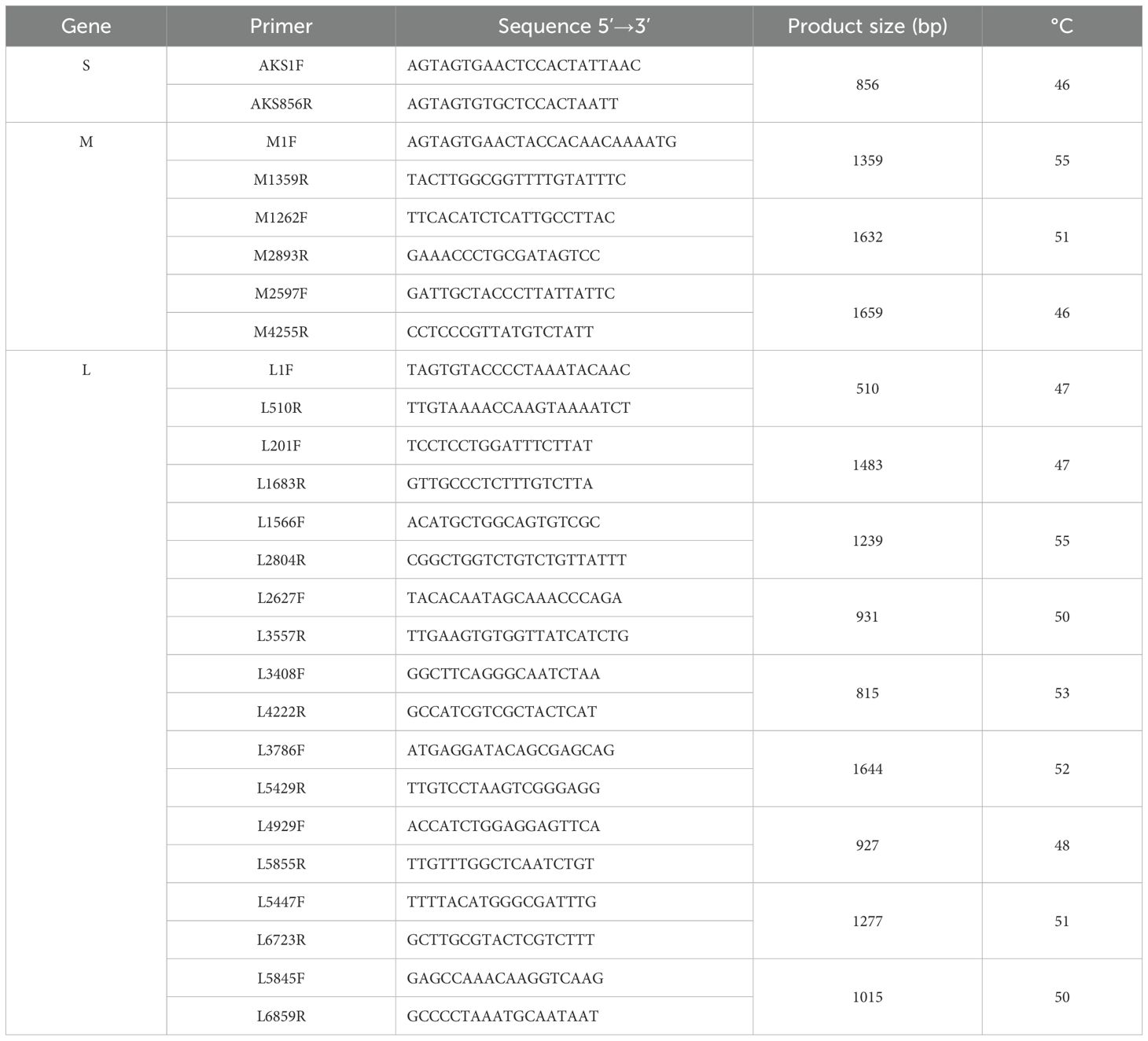
Table 1. The primers used for the amplification of the S, M, L segments of the Akabane virus genome.
The initial sequence assembly and analyses were performed using Seqman software in the DNAStar software package. The open reading frame (ORF) sequences of the S, M, and L segments of AKAV strains isolated from China, Japan, South Korea, Australia, and Kenya, as well as Aino virus strains belonging to the Simbu serogroup, were downloaded from GenBank. Homology and alignment analyses were conducted using MegAlign software in the DNAStar software package. With MEGA 6.1 based on the maximum likelihood assay, phylogenetic and evolutionary analyses were conducted. The bootstrap value was 1,000.
2.6 Viral neutralization antibodyAntibodies in serum samples were detected using a neutralization test. After heat treatment at 56°C for 30 minutes, each serum sample was diluted to 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:256, and 1:512, with two replicates per dilution. Each serum dilution (50 μL) was mixed with an equal volume of the virus supernatant (100 TCID50/50 μL) and inoculated into 96-well plates. Viral supernatants of 100TCID50, 10TCID50, and 1TCID50, along with a blank control, were included as the controls. The plates were shaken for 5 min and then incubated for 1 hour at 37°C under 5% CO2 to allow for neutralization. Following this, 100 μL of BHK-21 cell suspension (1 × 106 cells/mL) was added to each well. The plates were observed daily for 7 days, and the experimental results were evaluated according to previously reported criteria (Meng et al., 2023). A serum sample was considered positive for virus neutralizing antibody if the titer was ≥1:4 (Xu et al., 2007).
3 Results3.1 Sample collection statisticsIn July 2013, a total of 3704 biting midges were collected from Shizong County, comprising 2437 (49 pools), 117 (2 pools), and 1150 (23 pools) from cattle, goat, and pig helters, respectively. In total, 74 pools of biting midges were collected.
Additionally, 466 serum samples were collected from three sampling sites, snamely 147 (cattle 98 and goats 49) from Shizong County, 154 (cattle 101 and goats 53) from Jiangcheng County, and 165 (cattle 86 and goats 79) from Shuangjiang County.
3.2 Virus isolation and identificationTwo pools of biting midges (No. 52, 55) from cattle helters induced CPE in the BHK-21, MDBK, and VERO cells (Figure 1). The BHK-21 cells became spherical and detached after 24 hours, with CPE reaching 50% at 48 hours post-inoculation. The MDBK cells fragmentized and detached after 24 hours, with CPE reaching 50% at 48 hours post-inoculation. The VERO cells exhibited reticulated and detached after 24 hours, with CPE reaching 50% at 48 hours. No significant CPE was observed in the C6/36 cells.
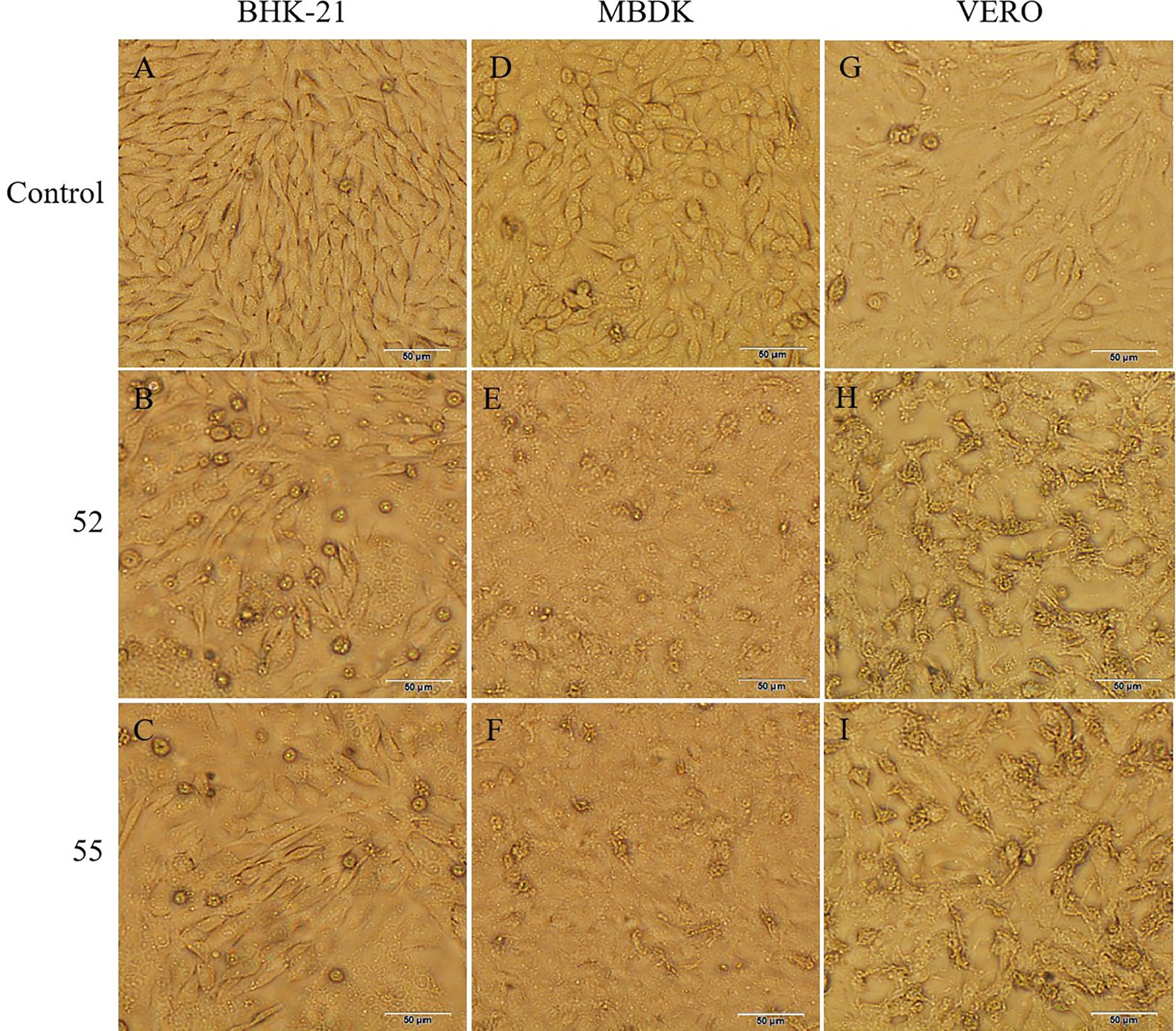
Figure 1. CPE caused by 52 and 55 on BHK-21, MDBK, and VERO cells 48 h postinoculation (100×). (A) Control BHK-21 cells. (B) Cytopathic effect of 52 in BHK-21 cells. (C) Cytopathic effect of 55 in BHK-21 cells. (D) Control MDBK cells. (E) Cytopathic effect of 52 in MDBK cells. (F) Cytopathic effect of 55 in MDBK cells. (G) Control VERO cells. (H) Cytopathic effect of 52 in VERO cells. (I) Cytopathic effect of 55 in VERO cells. Scale bar 50μm.
Positive samples for strains 52 and 55 were amplified using RT-PCR with Simbu serogroup primers, yielding approximately 600 nucleotides for sequencing. BLAST analyzed on NCBI revealed that the obtained sequences had over 97% similarity to the S segments of AKAV, indicating that viruses 52 and 55 are closely related to AKAV.
3.3 Genome organization and characteristics of AKAVThe S, M, and L segments of the two virus isolates were obtained through whole genome sequencing. The S segment was 856-nucleotide long and contained two overlapping open reading frames (ORFs) of 699 and 285 nucleotides, respectively, encoding a 233-amino-acid nucleocapsid (N) protein and a 91-amino-acid nonstructural (NSs) protein, respectively. The M segment contained a 4,206-nucleotide ORF encoding 1,401 amino acids, while the L segment contained a 6756-nucleotide ORF encoding 2251 amino acids. The complete AKAV genome sequences for strains 52 and 55 were submitted to GenBank (Table 2).
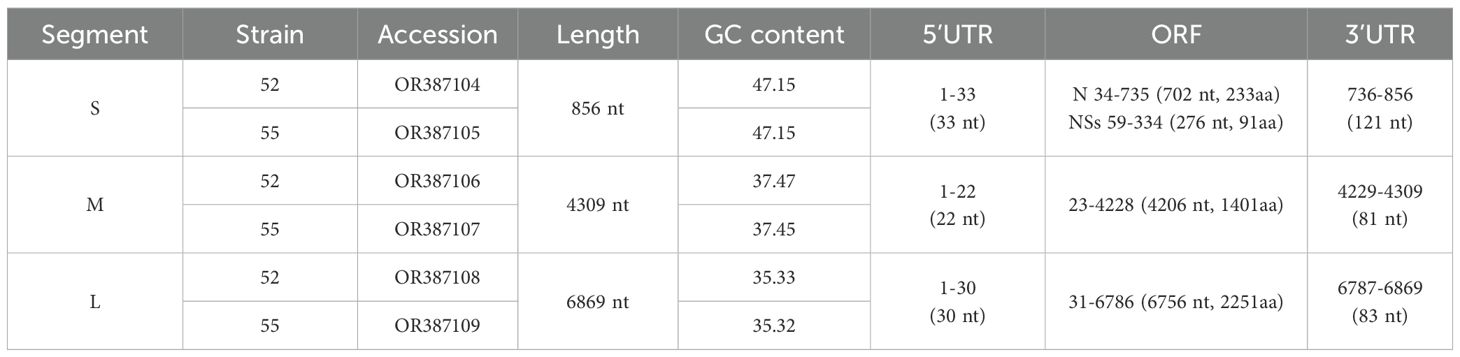
Table 2. Length and number of nucleotide sequences of the complete genome of AKAV strains newly isolated from Yunnan.
The nucleotide (and amino acids) of S, M, and L segments between the 52 and 55 strains were 100% (100% for N and NSs), 99.7% (99.6%), and 99.9% (99.9%), respectively. The nucleotide and amino acid homology of strain 55 was compared with 12 representative AKAV strains from China, Japan, South Korea, Australia, and Kenya (Table 3). The nucleotides (amino acids) of S, M, and L segments between strain 55 and genogroup I exhibited 95.6%–99.4% (98.3%–100% for N protein and 95.6%–97.8% for NSs protein), 91.9%–98.9% (95.4%–99.3%), and 94.8%–99.1%(98.6%–99.6%) identity, respectively. The nucleotides (amino acids) of S, M, and L segments between strain 55 and genogroup II exhibited 95.4%–96.4% (98.7% for N protein and 95.6%–97.8% for NSs protein), 87.5%–88.3% (93.6%–94.4%), and 92.0%–92.7% (97.6%–97.8%) identity, respectively. The nucleotides (amino acids) of S and M segments between strain 55 and genogroup III exhibited 92.6% (97.4% for N protein and 94.5% for NSs protein) and 84.3% (91.9%) identity, respectively. The nucleotides (amino acids) of S and M segments between strain 55 and genogroup IV displayed 83.8% (90.6% for N protein and 73.6% for NSs protein) and 70.5% (74.2%) identity, respectively. The 52 and 55 strains shared a high nucleotide and amino acid homology with genogroup Ia. Among them, the CX-01 strain isolated from Yunnan goats exhibited the highest homology.
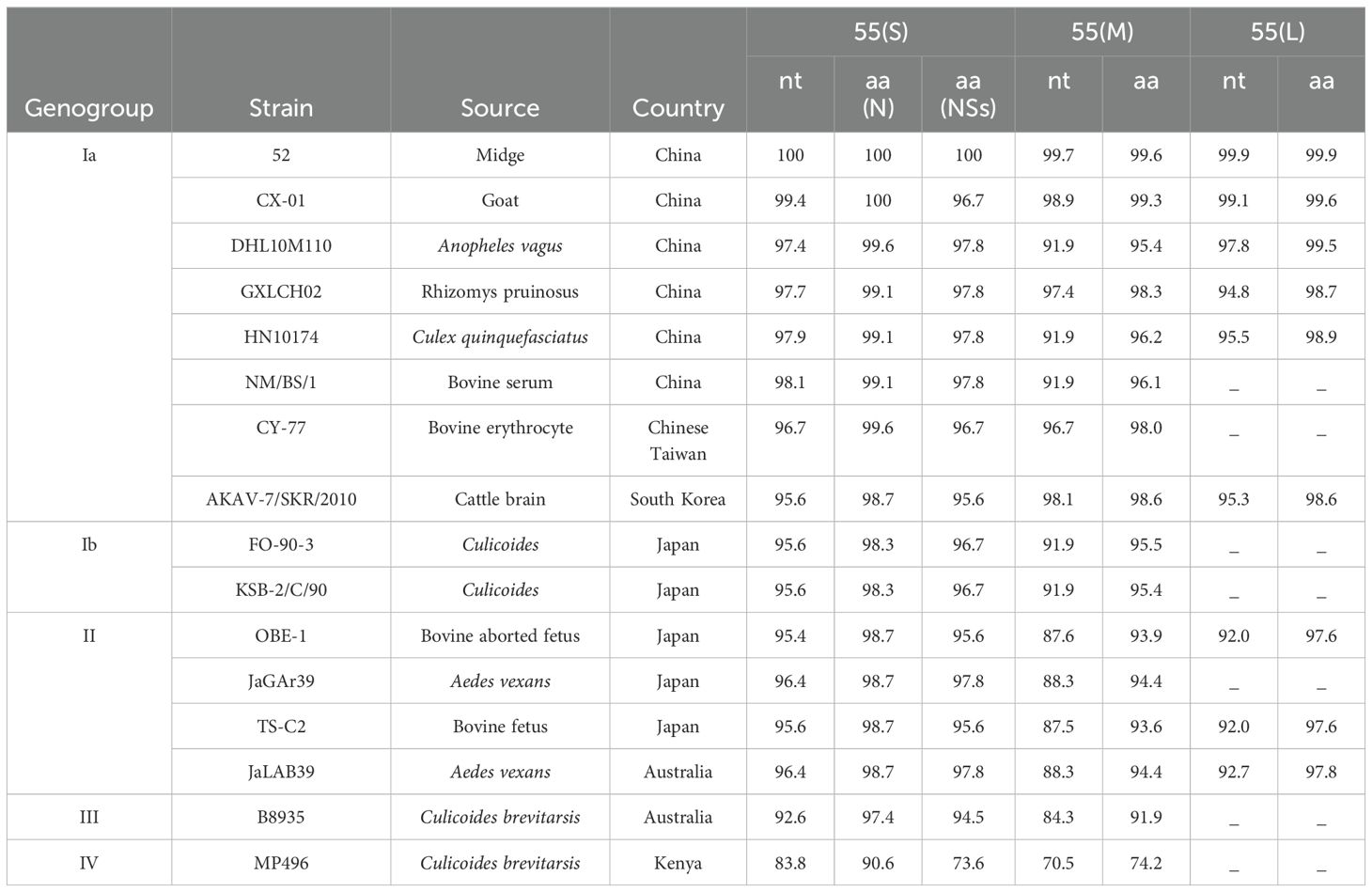
Table 3. Nucleotide and amino acid sequence identity (%) between 52, 55, and representatives of recognized AKAV.
Representative AKAV strains from various sources and countries were selected, encompassing four genogroups Ia, Ib, II, III, and IV. In total, 16 strains were analyzed for amino acid variation. Compared to the Chinese strain GXLCH02, strains 52 and 55 isolated from Yunnan biting midges, exhibited unique amino acid mutations in the NSs protein (position 79) and the L segment (position 763). The four strains isolated from Yunnan shared the same amino acid mutation at position 563 of the L segment. Additionally, eight strains from China displayed a common amino acid mutation at position 206 of the N protein, while seven strains from the Chinese mainland showed the same amino acid mutation at position 76 of the NSs protein. Genotype Ia strains contained unique amino acid variations at four positions (255, 610, 936, and 1279) in the M segments and five position (145, 1106, 1185, 1335 and 1524) in the L segments. Furthermore, genotype I strains exhibited three unique amino acid sites (positions 14, 758, and 1077) in the M segment (Table 4).
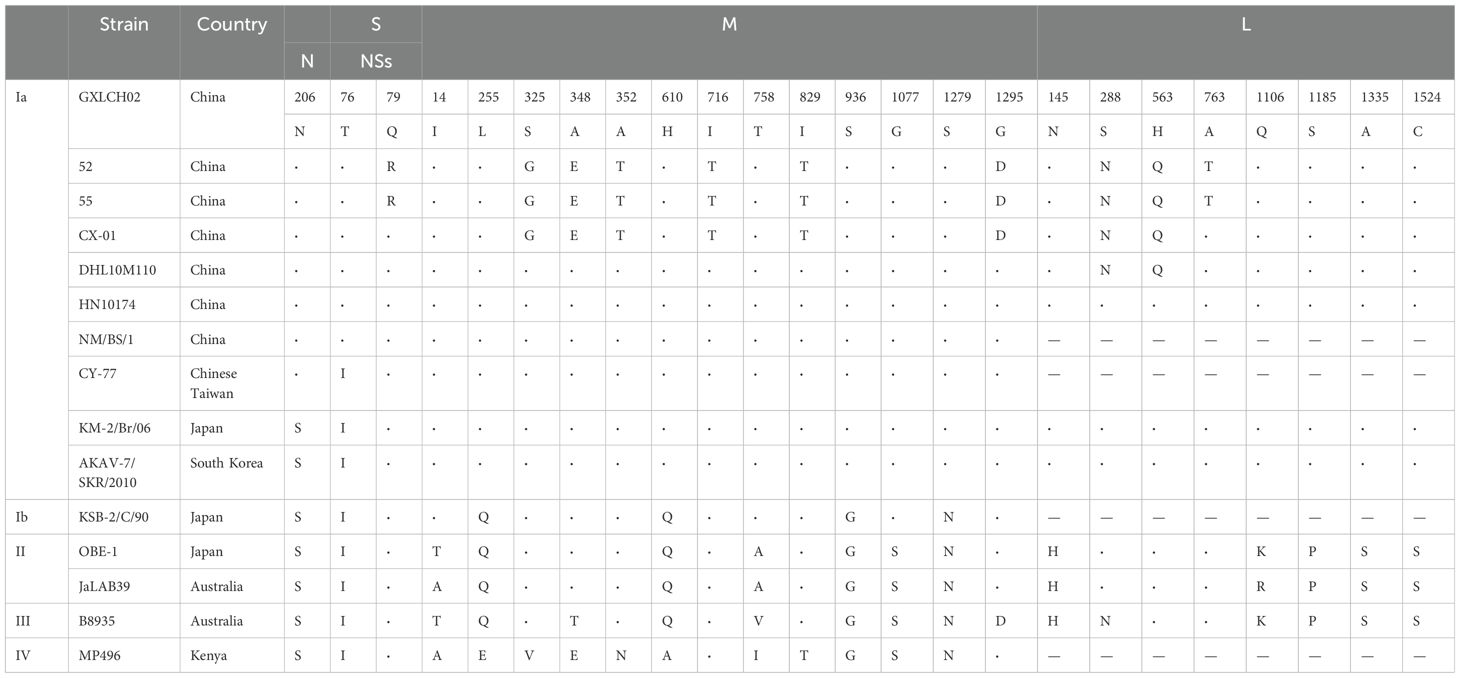
Table 4. Amino acid changes in the deduced sequences of the N, NSs, M, and L proteins of AKAV strains compared with those of 52 and 55.
3.4 Phylogenetic classification of 52 and 55Figure 2 illustrates the phylogenetic trees based on the nucleotide sequences of the S segment N protein (Figure 2A), M segment (Figure 2B), and L segment (Figure 2C) for viruses 52 and 55, alongside representative AKAV strain from various countries, hosts, and vectors, as well as the Aino virus strain from the Simbu serogroup. The analysis revealed that AKAV strains fall into genotypes I–IV, with genotype I further subdivided into two branches: Ia and Ib. Phylogenetic analysis of the N protein indicated that the viruses 52 and 55, isolated from biting midges in the study, belong to the branch as genotype Ia. Notably, AKAV isolates from the Chinese mainland formed a separate branch within the genotype Ia cluster, separate from viruses isolated from Chinese Taiwan, Japan, and South Korea. The phylogenetic tree based on the nucleotide sequences of the M and L segments further confirmed that strains 52 and 55 are classified within genotype Ia, with isolates from China, Japan, and Korea clustering closely in similar evolutionary groups.
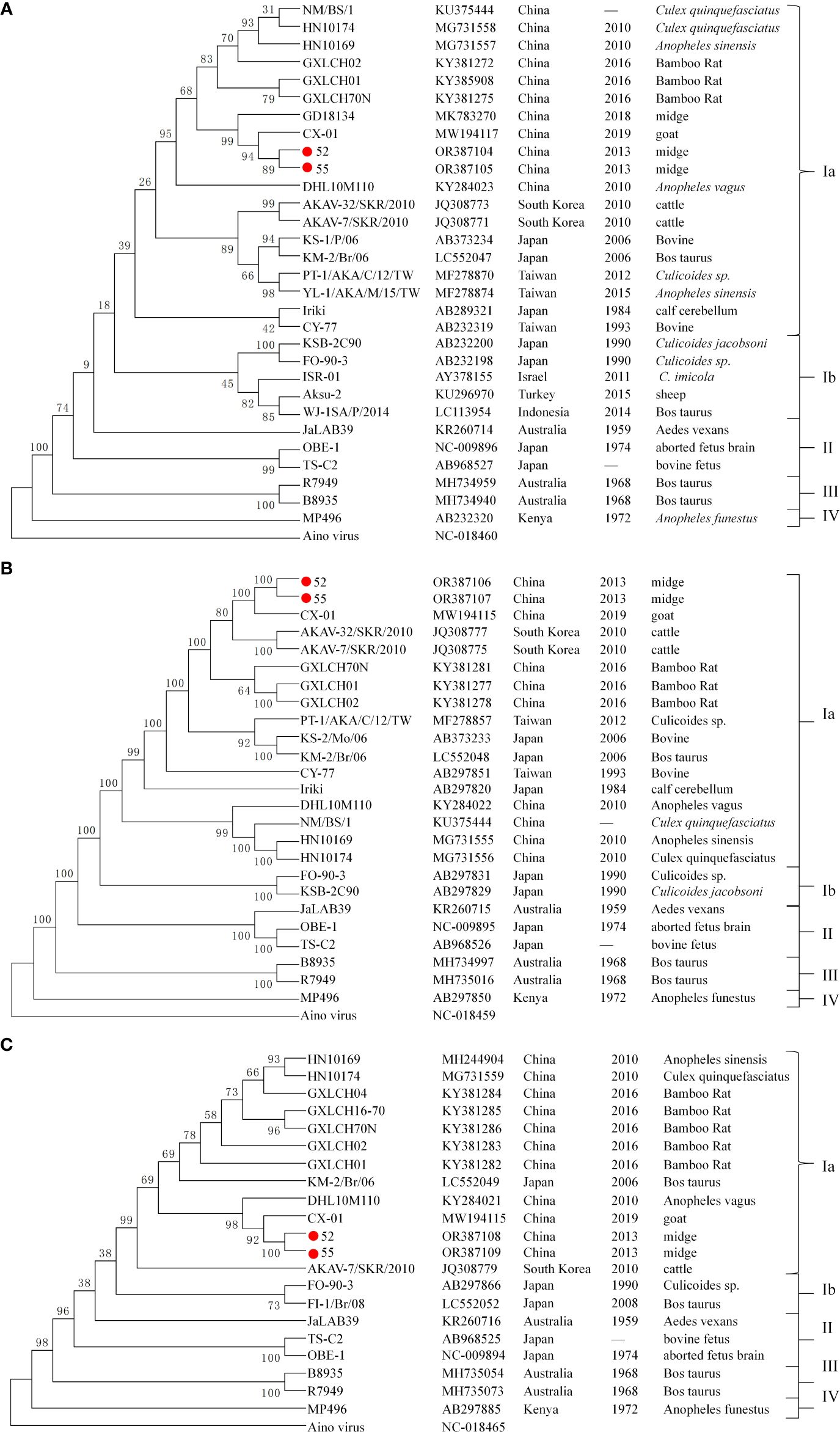
Figure 2. Phylogenetic trees of AKAV based on the ORF nucleotide sequence of S segment N protein (A), M segment (B), and L segment (C). The trees were constructed using the maximum likelihood assay. Sequences obtained in this study are marked with a round red spot ( ). The nucleotide sequences of other AKAV strains used here were obtained from GenBank.
). The nucleotide sequences of other AKAV strains used here were obtained from GenBank.
In total, 466 serum samples were collected from Shizong, Jiangcheng, and Shuangjiang counties and tested for AKAV neutralizing antibodies. The neutralizing antibody titer for AKAV was ≥1:4 in 202 serum samples, resulting in a positive rate of 43.35% (200/466). The detection of seropositive cattle and goats in this study suggests natural exposure to AKAV.
At the three sampling sites, the positive rate of AKAV neutralizing antibody was higher in cattle serum samples compared to goat serum samples. In Shizong county, the positive rates were 41.84% (41/98, 95% confidence interval (CI): 32–52) for cattle and 26.53% (13/49, 95% CI: 14–39)for goats. In Jiangcheng county, the rates were 43.56% (44/101, 95% CI: 34–53) for cattle and 37.74% (20/53, 95% CI: 24–51) for goats. In Shuangjiang county, the rates were 62.79% (54/86, 95% CI: 52–73) for cattle and 37.94% (30/79, 95% CI: 27–48) for goats (Figure 3A).
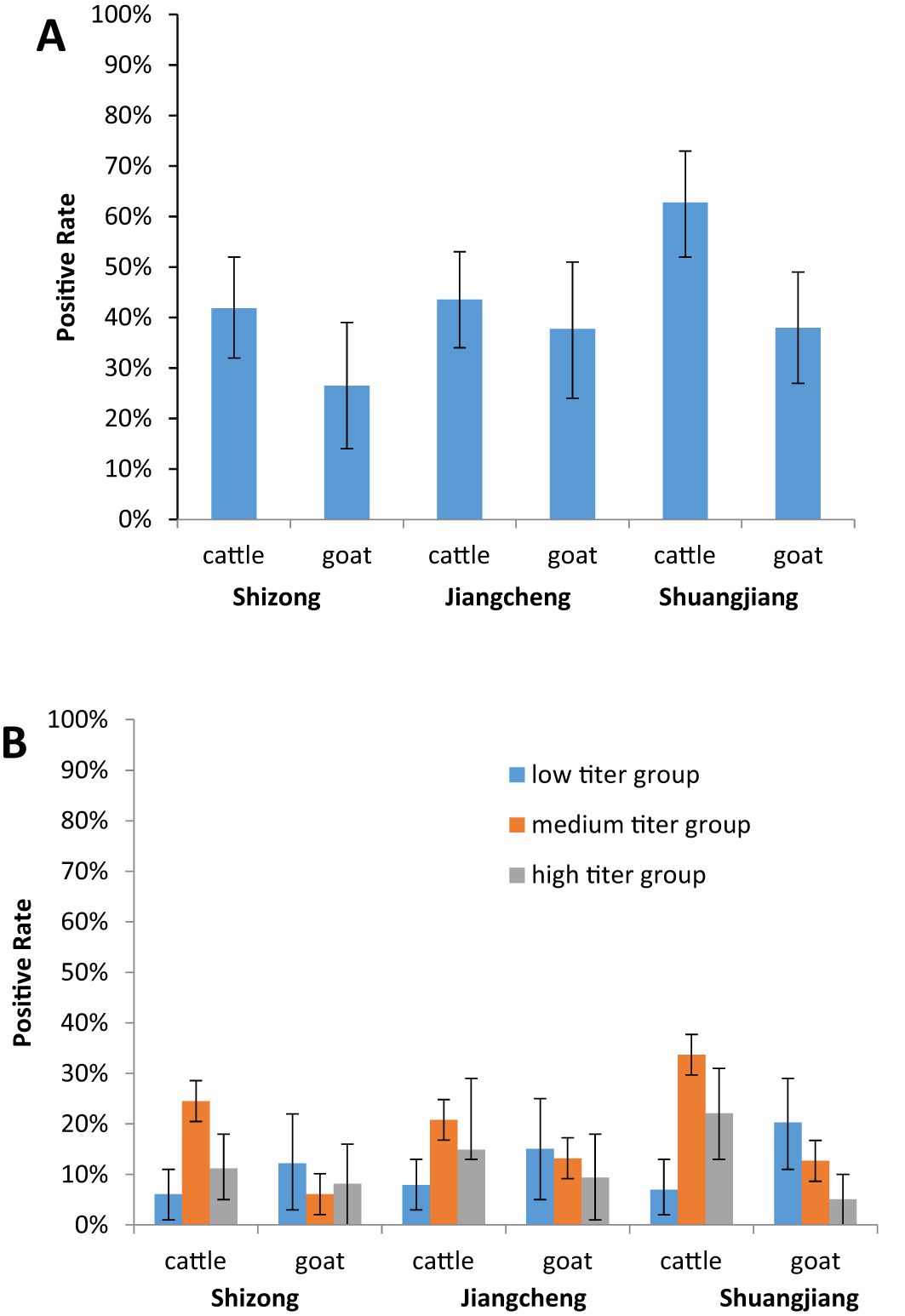
Figure 3. Serology of AKAV in two domestic animals (cattle, and goats) in different cities in Yunnan, China. (A) The positive rates of the AKAV antibody in the cattle and goats serum samples between the three sampling sites. (B) To evaluate the differences in AKAV neutralizing antibody titers in the cattle and goats serum samples between the three sampling sites.
To assess differences in AKAV neutralizing antibody titers between cattle and goat serum samples across the three sampling sites, the positive serum neutralizing antibody titers were divided into the low titer group (1:4–1:16), medium titer group (1:32–1:128), and high titer group (above 1:256). Most neutralizing antibody titers in cattle serum samples were concentrated in the medium titer group, with positive rates of 58.54% (24/41), 47.73% (21/44), and 53.7% (29/54) at the three sites, respectively. Conversely, the majority of neutralizing antibody titers in goat serum samples were found in the low titer group, with positive rates of 46.15% (6/13), 40% (8/20), and 53.33% (6/30) across the sites (Figure 3B).
4 DiscussionWe isolated two AKAV strains (52 and 55) from biting midges samples collected in Shizong County, Yunnan Province. These strains were shown to infect mammalian kidney cells, resulting in CPE. The complete genome sequences of the two strains revealed presence of three gene segments (S, M, and L). with nucleotide sequences closely resembling those of AKAV. The sequences comparisons indicated highly similar topologies, with only three amino acid variations in the M segment being different. When compared to the whole genome sequences of representative AKAV strains from both domestic and international sources, strains 52 and 55clustered within the same evolutionary branch as AKAV and belonged to the Ia branch of genotype I. The nucleotide and amino acid homology of strains 52 and 55 with other AKAV strains reported in China exceeded 90%. Overall, sequence analysis confirmed, that strains 52 and 55 were indeed AKAV.
In China, seroprevalence studies of AKAV have been conducted in regions such as Guangdong, Sichuan, and Inner Mongolia, with varied results reflecting regional differences in climate, livestock management practices, and vector distribution. For instance, studies from Guangdong reported relatively high seropositivity in cattle populations, likely due to the subtropical climate favoring vector activity year-round (Wang et al., 2019). Conversely, studies in more temperate regions like Inner Mongolia have shown lower AKAV prevalence, correlating with the shorter active season for biting midges vectors (Li et al., 2023). To investigate natural AKAV infection in local domestic animals in Yunnan, three sampling sites were selected to collect 466 serum samples from cattle and goats over one year of age, all without AKAV vaccination records. AKAV antibody detection revealed that 202 samples tested positive, resulting in a positive rate of 43.35%. Notably, the positive rate was higher for cattle (48.77%) than for goats (34.81%), which confirmed that, to a certain extent, cattle were more susceptible to infection than goats under natural conditions (Tang et al., 2019). The three sampling sites, Jiangcheng County, bordering Laos and Vietnam; Shuangjiang County, near Myanmar; and Shizong County, across the river from Guangxi Province. Represent the south, west, and east regions of Yunnan Province, respectively. The serological survey results reflect AKAV prevalence in Yunnan, with moderate to high seroprevalence rates that align more closely with results from subtropical regions like Guangdong, highlighting the influence of climate and ecological factors on AKAV transmission.
AKAV nucleic acid was previously been detected in midges collected from Sichuan and Hainan provinces in China, however, virus isolates were not obtained from these midges (Song et al., 2018; Fan et al., 2020). This study marks the first isolation AKAV from midges in China, with high titer of neutralizing antibodies against the AKAV virus detected in cattle and goat serum samples. These findings confirm that midges are vectors of AKAV, These findings confirm that midges are exists in natural midges in China and forms a natural cycle between midges and local domestic animals in Yunnan Province. The comparison of our findings with previous national and international studies highlights the important role of the midges in AKAV transmission patterns. Importantly, climate change is expected to alter the distribution of biting midges, potentially expanding AKAV’s range into previously unaffected regions. Warmer temperatures and changes in rainfall patterns could lead to longer breeding seasons and increased vector activity in temperate zones, raising concerns about AKAV outbreaks in regions traditionally considered low risk. This has already been observed in Europe, where other vector-borne diseases, such as Bluetongue virus (BTV), transmitted by biting midges, have expanded northward in recent decades due to milder winters and prolonged summers (Mullens et al., 2015; Peter and Estelle, 2015). Given these global changes, the findings from Yunnan have broader implications. Yunnan, with its climate and high livestock density, may serve as a model for understanding how AKAV could spread in other subtropical and temperate regions under changing environmental conditions. Enhanced surveillance and vector control strategies will be essential, not only in Yunnan but also globally, to mitigate the risk of AKAV transmission as climate change continues to reshape vector dynamics.
Data availability statementThe datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR387104; https://www.ncbi.nlm.nih.gov/genbank/, OR387105; https://www.ncbi.nlm.nih.gov/genbank/, OR387106; https://www.ncbi.nlm.nih.gov/genbank/, OR387107; https://www.ncbi.nlm.nih.gov/genbank/, OR387108; https://www.ncbi.nlm.nih.gov/genbank/, OR387109.
Ethics statementThe animal studies were approved by The Ethical Review Committee of Yunnan Animal Science and Veterinary Institute. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributionsJM: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft, Writing – review & editing. YH: Investigation, Methodology, Writing – original draft. NL: Data curation, Software, Writing – original draft. ZY: Data curation, Formal analysis, Writing – original draft. SF: Resources, Writing – original draft. DW: Resources, Writing – original draft. AX: Supervision, Writing – original draft. JW: Project administration, Validation, Writing – review & editing. GL: Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2022YFC2601603), Basic Research Projects of Yunnan Province (202301AT070028, 202201AS070062), National Natural Science Foundation of China (32260896), the central government to guide local scientific and technological development (202207AB110006), Special Fund for Agro-scientific Research in the Public Interest (201303035).
AcknowledgmentsWe are grateful to researcher Huachun Li for his financial support in the process of sample collection. We acknowledge the cooperation and enthusiasm of the Animal Disease Prevention and Control centers in Shizong prefecture.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesBryant, J. E., Crabtree, M. B., Nam, V. S., Yen, N. T., Duc, H. M., Miller, B. R. (2005). Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am. J. Trop. Med. Hyg. 73, 470–473. doi: 10.4269/ajtmh.2005.73.470
PubMed Abstract | Crossref Full Text | Google Scholar
Camarão, A. A. R., Swanepoel, R., Boinas, F., Quan, M. (2019). Development and analytical validation of a group-specific RT-qPCR assay for the detection of the Simbu serogroup orthobunyaviruses. J. Virol. Methods 271, 113685. doi: 10.1016/j.jviromet.2019.113685
PubMed Abstract | Crossref Full Text | Google Scholar
Della-Porta, A. J., O’Halloran, M. L., Parsonson, I. M., Snowdon, W. A., Murray, M. D., Hartley, W. J., et al. (1977). Akabane disease: isolation of the virus from naturally infected ovine foetuses. Aust. Vet. J. 53, 51–52. doi: 10.1111/j.1751-0813.1977.tb15825.x
PubMed Abstract | Crossref Full Text | Google Scholar
Fan, N., Sun, D. W., Cheng, R., Fu, S. H., Zeng, L. H., Wu, Q., et al. (2020). Isolation and identification of Arbovirus in Hainan province 2017-2018. Chin. J. Epidemiol. 41, 236–243. doi: 10.3760/cma.j.issn.0254-6450.2020.02.018
PubMed Abstract | Crossref Full Text | Google Scholar
Feng, Y., He, B., Fu, S. H., Yang, W. H., Zhang, Y. Z., Tu, C. C., et al. (2015). First isolation of akabane virus from mosquitoes in yunnan province, China. Chin. J. Virol. 31, 51–57. doi: 10.13242/j.cnki.bingduxuebao.002626
Crossref Full Text | Google Scholar
Gao, H. F., Wang, J. P., Yang, Z. X., Xie, J. R., He, Y. W., Hong, Q. H., et al. (2022). Genetic and pathogenic characterisation of a virulent akabane virus isolated from goats in Yunnan, China. J. Vet. Res. 66, 35–42. doi: 10.2478/jvetres-2022-0007
PubMed Abstract | Crossref Full Text | Google Scholar
Hayama, Y., Moriguchi, S., Yanase, T., Ishikura, Y., Abe, S., Higashi, T., et al. (2016). Spatial epidemiological analysis of bovine encephalomyelitis outbreaks caused by Akabane virus infection in western Japan in 2011. Trop. Anim. Health Prod. 48, 843–847. doi: 10.1007/s11250-016-1014-9
PubMed Abstract | Crossref Full Text | Google Scholar
Jeong, H., Oem, J.-K., Yang, M.-S., Yang, D., Kim, M.-S., Lee, K.-H., et al. (2017). Experimental infection of goats with a newly isolated strain of akabane virus that causes encephalomyelitis. J. Comp. Pathol. 157, 220–229. doi: 10.1016/j.jcpa.2017.05.003
PubMed Abstract | Crossref Full Text | Google Scholar
Kurogi, H., Akiba, K., Inaba, Y., Matumoto, M. (1987). Isolation of Akabane virus from the biting midge Culicoides oxystoma in Japan. Vet. Microbiol. 15, 243–248. doi: 10.1016/0378-1135(87)90078-2
PubMed Abstract | Crossref Full Text | Google Scholar
Lee, J. K., Park, J. S., Choi, J. H., Park, B. K., Lee, B. C., Hwang, W. S., et al. (2002). Encephalomyelitis associated with akabane virus infection in adult cows. Vet. Pathol. 39, 269–273. doi: 10.1354/vp.39-2-269
PubMed Abstract | Crossref Full Text | Google Scholar
Li, C., Chi, T. Y., Li, B. W., Pan, J. H., Wang, K. C., Tang, J. L., et al. (2023). Retrospective monitoring and analysis of bovine akabane disease in China from 2020 to 2022. China Animal Health Inspection. 40, 12–16. doi: : 10.3969/j.issn.1005-944X.2023.12.003
Crossref Full Text | Google Scholar
Meng, J. X., Li, N., He, Y. W., Kou, M. L., Wang, J. L., Ding, H., et al. (2020). Akabane virus isolated from Yunnan sentinel goat in 2015. Chin. J. Exp. Clin. Virol. 34, 411–414. doi: 10.3760/cma.j.cn112866-20200417-00134
Crossref Full Text | Google Scholar
Meng, J. X., Li, N., Xiao, L., He, Y. W., Kou, M. L., Song, J. L., et al. (2019). Sequence analysis of S gene of two strain of Akabane virus isolated from Yunnan Province. Chin. J. Vet. Sci. 39, 1703–1709. doi: 10.16303/j.cnki.1005-4545.2019.09.08
Crossref Full Text | Google Scholar
Meng, J. X., Wang, F., He, Y. W., Li, N., Yang, Z. X., Yao, J., et al. (2023). In vivo and in vitro characterization of a new Oya virus isolate from Culicoides spp. and its seroprevalence in domestic animals in Yunnan, China. PloS Negl. Trop. Dis. 17, e0011374. doi: 10.1371/journal.pntd.0011374
PubMed Abstract | Crossref Full Text | Google Scholar
Oya, A., Okuno, T., Ogata, T., Obayashii, K., Matsuyama, T. (1961). Akabane, a new arbor virus isolated in Japan. Jpn. J. Med. Sci. Biol. 14, 101–108. doi: 10.7883/yoken1952.14.101
PubMed Abstract | Crossref Full Text | Google Scholar
Pfeffer, M., Proebster, B., Kinney, R. M., Kaaden, O. R. (1997). Genus-specific detection of alphaviruses by a semi-nested reverse transcription-polymerase chain reaction. Am. J. Trop. Med. Hyg. 57, 709–718. doi: 10.4269/ajtmh.1997.57.709
PubMed Abstract | Crossref Full Text | Google Scholar
Song, S., Fu, S. H., Zhou, X. X., Zhang, J. K., Li, W., Liu, L. J., et al. (2018). Mosquitoes, midges and related arboviruses in southeast Sichuan province. Chin. J. Epidemiol. 39, 1381–1386. doi: 10.3760/cma.j.issn.0254-6450.2018.10.017
PubMed Abstract | Crossref Full Text | Google Scholar
Stram, Y., Brenner, J., Braverman, Y., Banet-Noach, C., Kuznetzova, L., Ginni, M. (2004). Akabane virus in Israel: a new virus lineage. Virus Res. 104, 93–97. doi: 10.1016/j.virusres.2004.03.004
PubMed Abstract | Crossref Full Text | Google Scholar
Tang, H. B., Chen, F. L., Rao, G. B., Bai, A. B., Jiang, J. J., Du, Y. C., et al. (2017). Characterization of Akabane virus from domestic bamboo rat, Southern China. Vet. Microbiol. 207, 280–285. doi: 10.1016/j.vetmic.2017.06.018
PubMed Abstract | Crossref Full Text | Google Scholar
Tang, H. B., Ren, P. F., Qin, S. Y., Lin, J., Bai, A. B., Qin, S. M., et al. (2019). Isolation, genetic analysis of the first Akabane virus from goat in China. J. Vet. Med. Sci. 81, 1445–1449. doi: 10.1292/jvms.18-0602
PubMed Abstract | Crossref Full Text | Google Scholar
Tohru, Y., Tomoko, K., Yoko, H., Hiroaki, S., Makoto, Y., Shogo, T. (2019). Oral susceptibility of Japanese culicoides (Diptera: ceratopogonidae) species to akabane virus. J. Med. Entomol. 56, 533–539. doi: 10.1093/jme/tjy201
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, J. D., Blasdell, K. R., Yin, H., Walker, P. J. (2019). A large-scale serological survey of Akabane virus infection in cattle, yak, sheep and goats in China. Vet. Microbiol. 207, 228–266. doi: 10.1016/j.vetmic.2018.11.007
PubMed Abstract | Crossref Full Text | Google Scholar
Xu, R. S., Liu, Z. Y., Dong, Z. Z., Yang, Y. J., Ma, B. H., Wu, S. H., et al. (2007). Protocol of quarantine for Akabane. SN/T 1128-2007. Industrial Code of the People’s Republic of China Entry-Exit Inspection and Quarantine, General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China.
Zhai, S. Y., Huang, G., Dong, J. Z., Wang, X. M. (2006). The fauna of blood-sucking Dipterous in China. Acta Parasitol. Et. Med. Entomol. Sin. 13, 178caca.
留言 (0)