Motor evoked potentials (MEPs) are crucial in transcranial magnetic stimulation (TMS) studies. MEPs induced by TMS were first introduced by Barker et al. in 1985, when magnetic pulses were applied to a participant’s motor cortex and induced a muscle contraction in the participant’s hand. This demonstrated that MEPs can be used as a non-invasive assessment of corticospinal excitability (Barker et al., 1985; Rossini et al., 2015). Enabled by technological advancements in TMS, MEPs have been studied across various stimulation paradigms, either strengthening or challenging previous hypotheses and exploring novel mechanisms in many areas of neuroscience (Rossini et al., 2015; Siebner et al., 2022).
Each MEP feature provides unique insights into the underlying neurophysiological mechanisms (Rossini et al., 2015; Siebner et al., 2022). Peak-to-peak amplitude (Amp) and onset latency (Lat) are the two most commonly used MEP features. Amp represents the size of the MEP; a higher Amp indicates higher cortical excitation, as demonstrated by increasing stimulation intensity or navigating closer to the representative area of the targeted muscle at the primary motor cortex (Pellegrini et al., 2018). Lat and the time point of the first major peak (T1T) are both timing features of MEPs, but they reflect different neuronal mechanisms. Lat is the duration required for an activation potential to travel from the stimulated area to the distal muscle. Therefore, it has been utilized in studying central motor conduction mechanisms (Rusu et al., 2014). The T1T, or peak latency, is the time point at which Amp is the highest. It is almost consistent despite changes in SI and might reflect the precision of timed motor performance (Nguyen et al., 2023).
Less commonly studied features, such as MEP polyphasia and duration (Dur), offer valuable information about the corticospinal tract’s status. MEP polyphasia is characterized by the number of turns and phases (NT and NP, respectively) present in its waveforms. High MEP polyphasia indicates a hyper-excitable motor neuron system in multiple sclerosis (Perretti et al., 2004; Neva et al., 2016; Snow et al., 2019). Abnormal Dur has been detected in several movement disorders and motor neuron diseases, such as prolonged Dur in multiple sclerosis (Snow et al., 2019; Šoda et al., 2023) and shortened Dur in acute stroke (Brum et al., 2016). In addition to these intrinsic features, combined features can be derived from these original features, such as the area under the curve (AUC) and thickness, defined as the ratio of AUC to Amp (Sollmann et al., 2021).
Despite the development of several toolboxes for MEP feature extraction, many are tailored to specific studies, such as recruitment curve analysis or online feature extraction, and often provide incomplete sets of MEP features. To address these limitations, this study introduces MEPFeatX, a robust package designed to automatically extract and visualize MEP features, covering essential information about MEP size, critical time points, and other morphological characteristics, including duration and polyphasia. Figure 1 illustrates an example of an MEP with its features extracted and visualized using MEPFeatX. This package was validated with MEPs recorded across various stimulation paradigms and is available at the following GitHub repository: https://github.com/NeuromodulationUEF/MEPFeatX. By clearly outlining the significance of MEP features and their applications, we aim to enhance users’ understanding of their practical utility in diverse contexts, thereby reinforcing the contribution of our work to the field of TMS research.
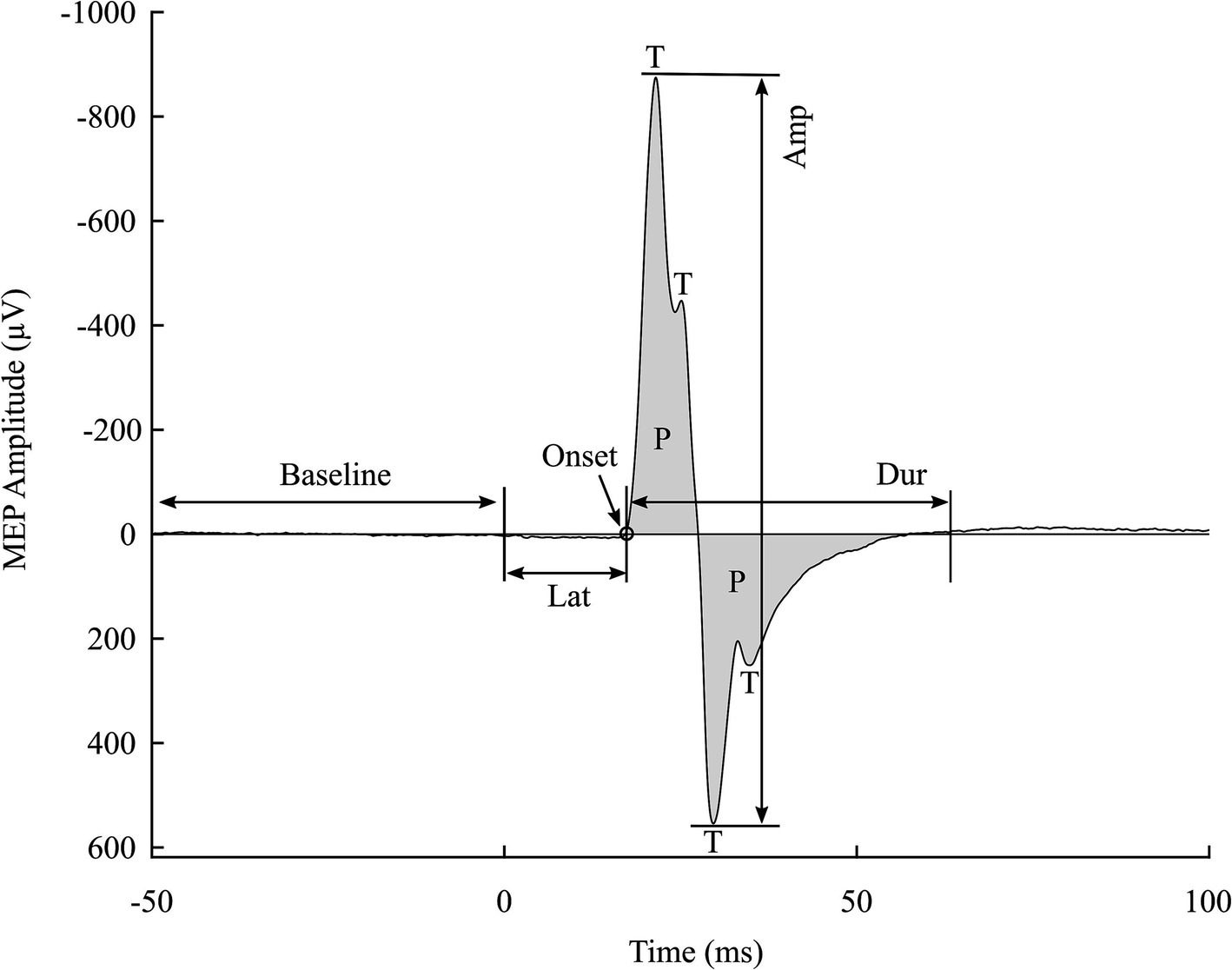
Figure 1. Visualization of an individual MEP with some of its features denoted. Amp, amplitude; Dur, duration; Lat, latency; P, phase; and T, turn. The two major peaks are denoted by T1 and T2, each providing a feature of timing and amplitude (T1T, T1A, T2T, and T2A). In addition to the number of turns (NT) and number of phases (NP), the area under the curve (AUC) is the sum of all phases’ area. Thickness is defined as a ratio of the AUC to Amp.
2 Existing tools for MEP feature extractionThe usual data pipeline in MEP studies includes data acquisition, preprocessing of raw electromyography (EMG) recordings, feature extraction from preprocessed MEPs, data analysis and visualization, and reporting. Data acquisition outputs the recorded EMG data and metadata of the experiment. Extracting the data from these formats requires a tailored process, depending on the devices/software used. Preprocessing includes filtering, artifact removal, and segmentation of data into trials. Next, features of interest are extracted from each trial, analyzed using appropriate statistical methods, and visualized to facilitate the interpretation of the results. Reports are generated afterward.
Several toolboxes have been developed over the years for various steps in this data pipeline, such as MAVIN, CortExTool, Motometrics, VETA, and MEP-ART. A summary of these toolboxes is provided in Table 1.
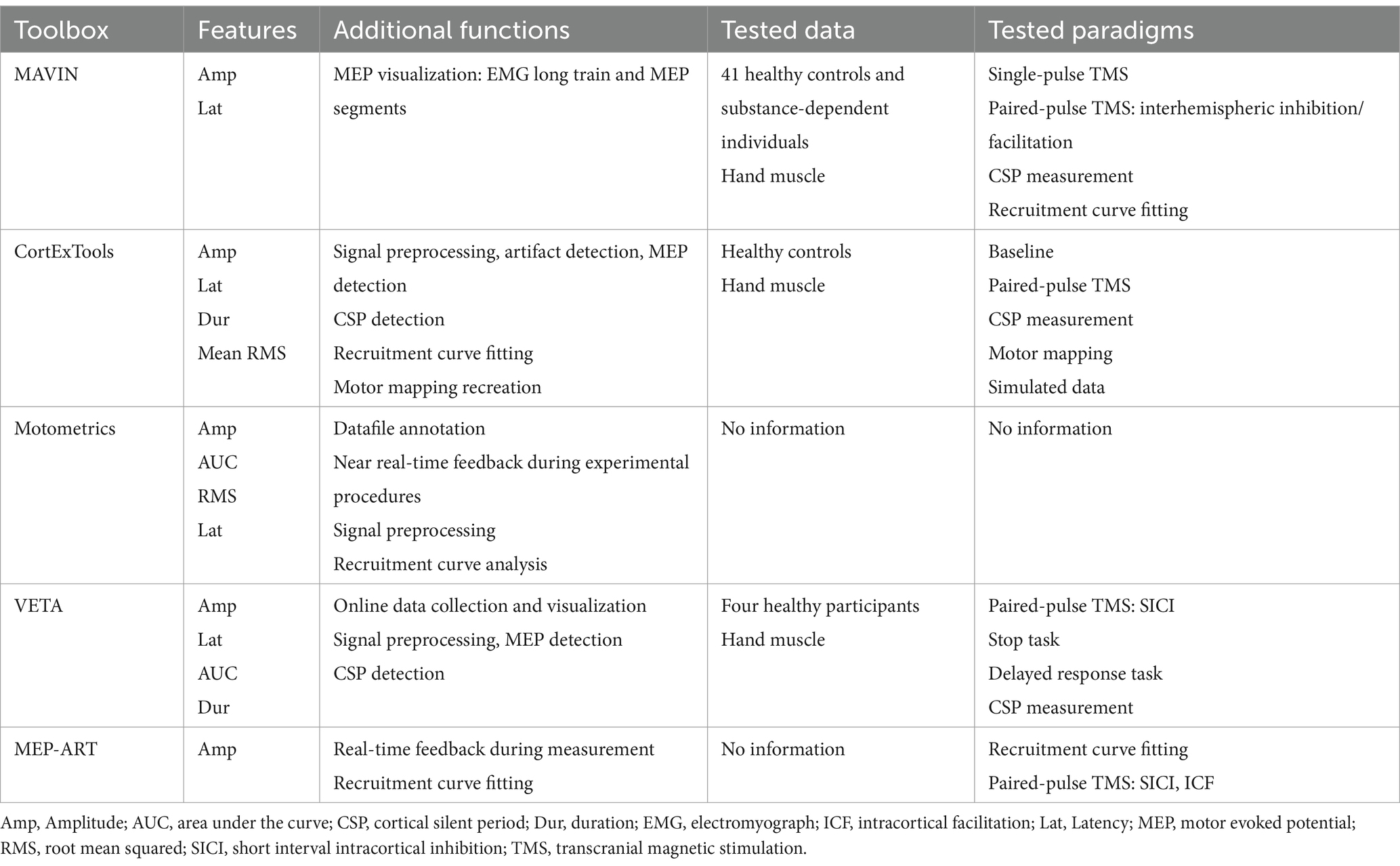
Table 1. Existing toolboxes for MEP feature extraction.
MAVIN (Mullins and Hanlon, 2016) is an open-source tool for offline MEP visualization and analysis and can be utilized in studies involving basic/paired associative stimulation, as well as the cortical silent period (CSP) and recruitment curves. Motometrics (Ratnadurai Giridharan et al., 2019) focuses specifically on recruitment curve analysis.
VETA (Jackson and Greenhouse, 2019) provides real-time data acquisition and feature extraction. MEP-ART (Skelly et al., 2020), is a real-time feedback and MEP analysis tool used to investigate the reliability of recorded data. It is compatible with the Magstim 200 (and BiStim) system (Magstim Inc., Eden PrairieMN). VETA, on the other hand, focuses on detecting MEPs from EMG recordings and then analyzes and visualizes the detected MEPs. CortExTool (Harquel et al., 2016) is a comprehensive MATLAB toolbox designed for MEP analysis. It includes features for EMG preprocessing, automatic MEP detection, the extraction of various MEP features, and the analysis of I/O curves and cortical silent periods.
MAVIN and Motometrics were inaccessible. In addition, we were unable to run MEP-ART, CortExTool, and VETA, primarily due to incompatible data formats.
3 Methods 3.1 DesignIn comparison to the existing toolboxes mentioned in Section II, MEPFeatX focuses on feature extraction. In total, it outputs a set of 11 MEP features: Amp, Lat, T1T, Dur, AUC, NT, NP, amplitude of the first major turn (T1A), time point of the second major turn (T2T), amplitude of the second major turn (T2A), and thickness.
The first version of the package was developed to explore MEP patterns in single-pulse TMS performed at the cortical representation area for the hand and to estimate the minimum number of trials required to reliably represent the entire dataset using bootstrapped principal component regression (Nguyen et al., 2019). The current version of this package was further improved to offer greater flexibility, allowing it to work on MEPs in various conditions, such as across multiple age groups, muscles, and stimulation paradigms (Table 2) (Nguyen et al., 2019, 2023). The use cases can be found on the MEPFeatX Wiki, under the “Use cases” page.
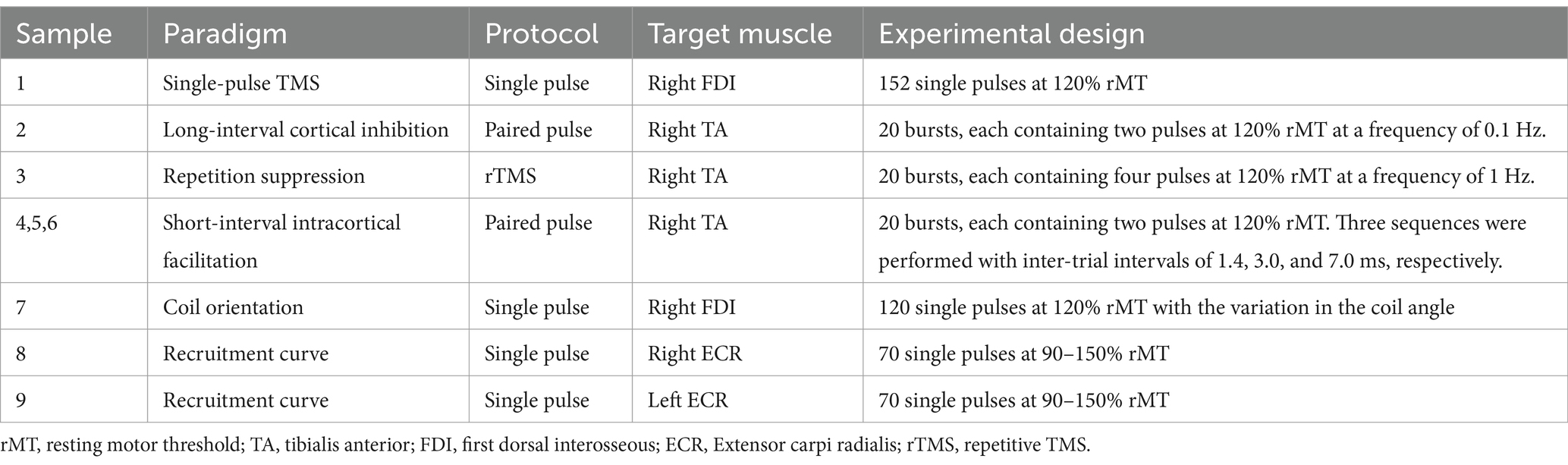
Table 2. Metadata table for the sample datasets.
In addition to feature extraction, the package also provides a workflow for feature extraction, particularly for MEPs, including a combination of metadata and features as a comprehensive table that can be utilized for exploratory data analysis, logs of the extraction process, and reliable evaluation of feature quality through plots. Documentation on MEPFeatX use cases was created to enhance the package’s usability and ensure the reproducibility of its functions in any MEP analysis pipeline (Romano and Moore, 2020). Furthermore, the user can select several stimulation paradigms, such as long-interval intracortical inhibition (LICI), repetition suppression (RS) at 1 Hz, and short-interval intracortical facilitation (SICF). After the feature table is created in MATLAB, the user can opt to use R notebooks for analyzing the table. Several R scripts are provided to analyze the exported feature table through descriptive analysis and/or causal analysis to identify the decisive factors in each study, such as stimulation protocols and subject demographics (Figure 2).
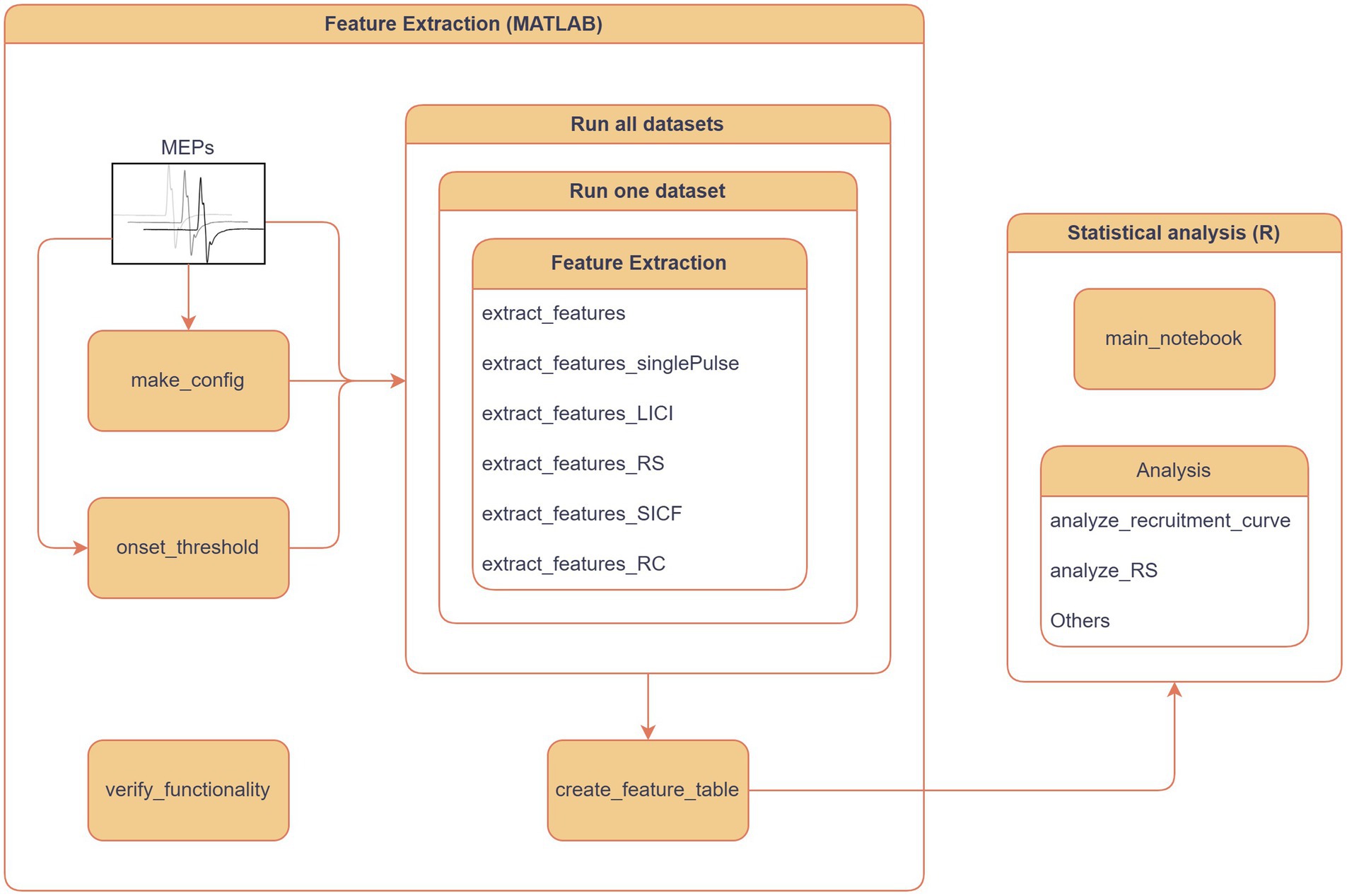
Figure 2. MEPFeatX workflow. The primary component of MEPFeatX is feature extraction, programmed in MATLAB, designed to extract features from MEP responses across various stimulation paradigms. Users can select from several paradigms, including long-interval intracortical inhibition (LICI), repetition suppression (RS) at 1 Hz, and short-interval intracortical facilitation (SICF). Once the feature table is generated in MATLAB, users can utilize R notebooks for further analysis, including descriptive statistics and linear mixed-effects modeling.
MEPFeatX offers a more streamlined approach focused solely on feature extraction compared to the existing toolboxes. It accepts MATLAB.mat files as input, eliminating data format mismatches. In addition, MEPFeatX is fully and freely available to the community, with version control for both code and documentation managed through Git. Updates and bug fixes for the package can be requested via GitHub or directly from the authors. Version control is implemented for the package scripts and analysis. MEPFeatX outputs are automatically organized into specified categories and saved to folders named by the date the analysis is performed.
MEPFeatX does not include a graphical user interface (GUI) as a GUI requires a specific version of the MATLAB runtime. Instead, the package workflow is programmed similarly to the R Markdown Notebook (Allaire et al., 2024) and Python Jupyter Notebook (Kluyver et al., 2016), with ample comments for easy integration into any analysis pipeline.
3.2 Package componentsGiven that MEPs are already segmented from the recorded EMG, the core functionality of MEPFeatX is the feature extraction algorithm, which simply follows the definition of each feature. First, it detects the two most prominent peaks (positive and negative) appearing in a signal segment from the TMS pulse delivery time to 150 ms afterward. Amp, T1T, T1A, and T2T are extracted from the timing and magnitude of these two peaks.
We used the standard deviation of the 50 ms segment prior to the delivery of the TMS pulse as the background activity. The MEP onset is the time point when the signal first exceeds the background activity within the time window from the pulse delivery time to the T1T. Lat is the time interval between the pulse delivery time and the MEP onset. The endpoint of the MEP is defined as the point when the signal returns to the level of the background activity. It is determined as the first sample of a 10 ms signal segment that is statistically similar to the background activity, meaning this 10 ms segment has a standard deviation comparable to that of the background activity. Dur is the time interval between the MEP onset and its endpoint. Next, the NT is counted as the significant peaks occurring during the MEP Dur, while the NP is counted by the zero-crossing points between the MEP onset and endpoint. Combined features, such as AUC and thickness, are calculated afterward. The AUC is the area under the rectified signal during the Dur. Thickness is the ratio of the AUC to Amp. The visualization of MEPs and their features is shown in Figure 1.
3.3 PrerequisitesFor the current version, the package runs on preprocessed MEP datasets stored in MAT files, and the input dataset must have rows as samples and columns as a stack of trials.
A ready-made configuration file contains control parameters for running MEPFeatX, such as specifying data directories, creating new folders for storing outputs, and defining the sampling frequency. All these parameters must be checked carefully for the extraction to run. Details on the data format and configuration file are provided in the MEPFeatX GitHub Wiki.
3.4 WorkflowA complete workflow is provided with the package to help the user create a new analysis (Figure 2). This workflow demonstrates feature extraction on all datasets or a single dataset stored in the data folder and saves the output to the analysis folder. Therefore, the user can access the data at different stages of the analysis, such as figures, features, or statistical analysis results.
Logs of the current analysis are also recorded and saved to the analysis folder. Errors, such as missed MEPs where the extraction failed, are reported in a table-format file for review. Details on the deployment of MEPFeatX are provided in the MEPFeatX GitHub Wiki.
4 Results 4.1 Package verification and validationMEPFeatX functions were utilized in our previous studies (Nguyen et al., 2019, 2023). The two studies used single-pulse TMS in healthy participants across four age groups: children, preadolescents, adolescents, and adults. MEPs were recorded from upper extremity muscles, such as those in the forearm and hand.
MEPFeatX was validated using a comprehensive dataset of participants from several studies on MEP analysis, two of which have been published (Nguyen et al., 2019, 2023). In the 2019 study (Nguyen et al., 2019), the studied dataset contained data samples from nine healthy adult participants. Each sample included 120 MEPs at a stimulation intensity of 120% rMT. In the 2023 study (Nguyen et al., 2023), 38 healthy participants were categorized into four age groups: children, preadolescents, adolescents, and adults. A total of 70 stimuli were performed with the stimulation intensity (SI) randomly ranging from sub-threshold to supra-threshold. MEPs were recorded from the flexor carpi radialis (FCR), extensor carpi radialis (ECR), abductor digiti minimi (ADM), and first dorsal interosseous (FDI) muscles. Stimulation was performed on both hemispheres, and EMG was recorded from the contralateral muscles. The MEPFeatX output was manually verified in these studies. Thus, this demonstrated the capabilities of MEPFeatX in extracting MEP features across various age groups, muscle groups, and stimulation protocols.
To verify the output of MEPFeatX in lower extremity muscles and other paradigms, we extracted MEP features from 7,799 MEPs recorded from the tibialis anterior (TA) muscle across six paradigms: single-pulse TMS, repetition suppression (RS) at 1 Hz, long-interval intracortical inhibition (LICI) at 100 ms, and short-interval intracortical facilitation (SICF) with three inter-pulse intervals of 1.4 ms, 3.0 ms, and 7.0 ms. Data were collected from 23 healthy controls. The studies involving human participants were reviewed and approved by the Research Ethics Committee of the Hospital District of Northern Savo. The participants provided written informed consent to participate in this study.
Bland–Altman analysis was used to compare the Amp extracted by MEPFeatX with the Amp extracted in real-time during the data acquisition (Figure 3). The Pearson correlation coefficient between the Amp extracted by the two algorithms was r = 0.998 (p < 0.0001), with a coefficient of reproducibility of 36 μV. The online-extracted latency was unreliable and was not compared.
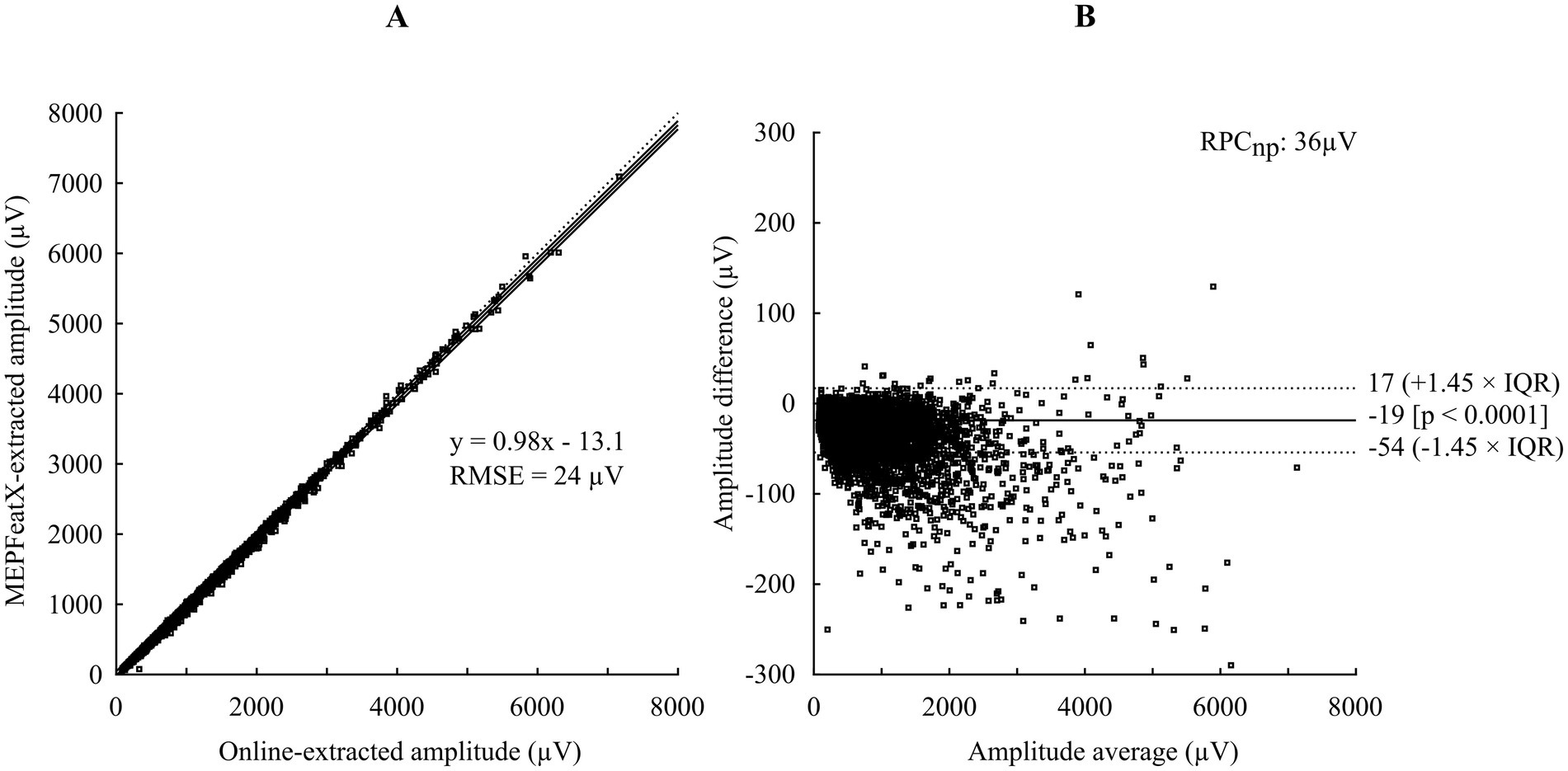
Figure 3. A comparison of online-extracted amplitude and MEPFeatX-extracted amplitude using Bland–Altmann analysis. (A) A regression plot between the amplitude extracted by the online algorithm and MEPFeatX. (B) A plot of pair-wise differences of the amplitude extracted by the two methods. The amplitude extracted by MEPFeatX was lower than the online-extracted amplitude by 19 μV, and the bias became larger with higher amplitude. This bias was due to the effect of the low-passed filter during the preprocessing in MEPFeatX, which reduced high-frequency noises. RMSE, root mean squared error; IQR, inter-quartile range; RPCnp, non-parametric coefficient of reproducibility.
It is recommended to validate the package’s functionality before its first use in a new system. The package includes outputs from anonymized sample datasets and the validation function. The features extracted from the sample datasets and their plots are used as a reference to validate the package’s output before the first use.
4.2 Sample datasets and analysis templatesThe sample datasets and their analysis templates are provided to help users implement MEPFeatX in their own studies (Table 2). Each sample dataset contains raw responses and preprocessed MEPs, both of which have been segmented into 200 ms segments, from 50 ms prior to TMS administration to 150 ms after the pulse. The provided templates help users manipulate and visualize MEP features in specific paradigms. For a detailed explanation, refer to the MEPFeatX GitHub Wiki.
5 DiscussionThis study aimed to enhance the analysis of MEPs by introducing the MEPFeatX package. Our findings indicate that MEPFeatX significantly improves the automatic and comprehensive feature extraction of MEPs across various stimulation paradigms.
When comparing MEPFeatX to existing tools, we found that MAVIN and Motometrics were not accessible for our analysis. In addition, MEP-ART, CortExTool, and VETA could not be executed on our data due to input format mismatches. In contrast, MEPFeatX not only provides a user-friendly interface but also includes visualizations that reliably evaluate analyzed MEPs and their features. Furthermore, the package offers workflows and templates for several use cases, which enhances reproducibility and integrity within MEP analysis pipelines. In addition, MEPFeatX is designed to efficiently handle datasets of varying sizes. The MATLAB codes are segmented into small functions that support parallel computing. This feature allows it to effectively run large datasets.
Despite these advantages, our study has limitations. The accessibility issues with other tools highlight the need for data pre-processing. Thus, it might not work with EMG signals that contain significant interference. Future research should focus on exploring the application of MEPFeatX across additional paradigms to further validate its effectiveness in diverse settings.
Although the output of MEPFeatX was visually observed by two experts in the field, we did not record any objective metrics. Thus, future development and validation of MEPFeatX should follow a structured procedure that includes human evaluation with proven metrics, such as the level of agreement among evaluators.
In conclusion, MEPFeatX emerges as a superior tool for MEP analysis, allowing researchers to focus on exploratory analysis and derive meaningful insights from exported features. We encourage further investigations to expand its applicability and address existing limitations.
Data availability statementThe data analyzed in this study is subject to the following licenses/restrictions: the data that support the findings of this study are available from the corresponding author, DN, upon reasonable request. Requests to access these datasets should be directed to Dao T. A. Nguyen, bmd1eWVuLmRhby5ia0BnbWFpbC5jb20=.
Ethics statementThe studies involving humans were approved by Research Ethics Committee of the Hospital District of Northern Savo. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributionsDN: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. LS: Funding acquisition, Investigation, Writing – review & editing. EK: Conceptualization, Data curation, Methodology, Resources, Validation, Writing – review & editing. PK: Funding acquisition, Project administration, Supervision, Writing – review & editing. SR: Conceptualization, Methodology, Validation, Writing – review & editing. PJ: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie under Grant 713645 and the Academy of Finland (Helsinki, Finland) Grant 322423.
AcknowledgmentsThe authors gratefully acknowledge the invaluable contributions of Dr. Jusa Reijonen, Ph.D., Ms. Meri Julkunen, and Ms. Anna-Leena Voutilainen for providing the sample data.
Conflict of interestPJ has received consulting pay from Nexstim Oyj (Helsinki, Finland) and has a shared patent with Nexstim unrelated to this work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAllaire, J., Xie, Y., Dervieux, C., McPherson, J., Luraschi, J., Ushey, K., et al. (2024). rmarkdown: dynamic documents for R. Available at: https://github.com/rstudio/rmarkdown (Accessed November 19, 2022).
Barker, A. T., Jalinous, R., and Freeston, I. L. (1985). Non-invasive magnetic stimulation of human motor cortex. The Lancet (British edition) 325, 1106–1107. doi: 10.1016/S0140-6736(85)92413-4
PubMed Abstract | Crossref Full Text | Google Scholar
Brum, M., Cabib, C., and Valls-Solé, J. (2016). Clinical value of the assessment of changes in MEP duration with voluntary contraction. Front. Neurosci. 9:505. doi: 10.3389/fnins.2015.00505
PubMed Abstract | Crossref Full Text | Google Scholar
Harquel, S., Beynel, L., Guyader, N., Marendaz, C., David, O., and Chauvin, A. (2016). CortExTool: a toolbox for processing motor cortical excitability measurements by transcranial magnetic stimulation. Available at: https://hal.science/hal-01390016 (accessed May 3, 2023).
Jackson, N., and Greenhouse, I. (2019). VETA: an open-source Matlab-based toolbox for the collection and analysis of electromyography combined with transcranial magnetic stimulation. Front. Neurosci. 13:975. doi: 10.3389/fnins.2019.00975
PubMed Abstract | Crossref Full Text | Google Scholar
Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., et al. (2016). “Jupyter notebooks – a publishing format for reproducible computational workflows” in Positioning and power in academic publishing: Players, agents and agendas. eds. F. Loizides and B. Schmidt (Amsterdam: IOS Press), 87–90.
Mullins, C. R., and Hanlon, C. A. (2016). MAVIN: an open-source tool for interactive analysis and visualization of EMG data. Brain Stimul. 9, 305–306. doi: 10.1016/j.brs.2015.11.009
PubMed Abstract | Crossref Full Text | Google Scholar
Neva, J. L., Lakhani, B., Brown, K. E., Wadden, K. P., Mang, C. S., Ledwell, N. H. M., et al. (2016). Multiple measures of corticospinal excitability are associated with clinical features of multiple sclerosis. Behav. Brain Res. 297, 187–195. doi: 10.1016/j.bbr.2015.10.015
PubMed Abstract | Crossref Full Text | Google Scholar
Nguyen, D. T. A., Julkunen, P., Säisänen, L., Määttä, S., Rissanen, S. M., Lintu, N., et al. (2023). Developmental models of motor-evoked potential features by transcranial magnetic stimulation across age groups from childhood to adulthood. Sci. Rep. 13, 10604–10611. doi: 10.1038/s41598-023-37775-w
PubMed Abstract | Crossref Full Text | Google Scholar
Nguyen, D. T. A., Rissanen, S. M., Julkunen, P., Kallioniemi, E., and Karjalainen, P. A. (2019). Principal component regression on motor evoked potential in single-pulse transcranial magnetic stimulation. IEEE Trans. Neural Syst. Rehabil. Eng. 27, 1521–1528. doi: 10.1109/TNSRE.2019.2923724
PubMed Abstract | Crossref Full Text | Google Scholar
Pellegrini, M., Zoghi, M., and Jaberzadeh, S. (2018). The effect of transcranial magnetic stimulation test intensity on the amplitude, variability and reliability of motor evoked potentials. Brain Res. 1700, 190–198. doi: 10.1016/J.BRAINRES.2018.09.002
PubMed Abstract | Crossref Full Text | Google Scholar
Perretti, A., Balbi, P., Orefice, G., Trojano, L., Marcantonio, L., Brescia-Morra, V., et al. (2004). Post-exercise facilitation and depression of motor evoked potentials to transcranial magnetic stimulation: a study in multiple sclerosis. Clin. Neurophysiol. 115, 2128–2133. doi: 10.1016/j.clinph.2004.03.028
PubMed Abstract | Crossref Full Text | Google Scholar
Ratnadurai Giridharan, S., Gupta, D., Pal, A., Mishra, A. M., Hill, N. J., and Carmel, J. B. (2019). Motometrics: a toolbox for annotation and efficient analysis of motor evoked potentials. Front. Neuroinform. 13:8. doi: 10.3389/fninf.2019.00008
PubMed Abstract | Crossref Full Text | Google Scholar
Rossini, P. M., Burke, D., Chen, R., Cohen, L. G., Daskalakis, Z., Di Iorio, R., et al. (2015). Non-invasive electrical and magnetic stimulation of the brain, spinal cord, roots and peripheral nerves: basic principles and procedures for routine clinical and research application. An updated report from an I.F.C.N. Committee. Clin. Neurophysiol. 126, 1071–1107. doi: 10.1016/j.clinph.2015.02.001
PubMed Abstract | Crossref Full Text | Google Scholar
Siebner, H. R., Funke, K., Aberra, A. S., Antal, A., Bestmann, S., Chen, R., et al. (2022). Transcranial magnetic stimulation of the brain: what is stimulated? – a consensus and critical position paper. Clin. Neurophysiol. 140, 59–97. doi: 10.1016/J.CLINPH.2022.04.022
PubMed Abstract | Crossref Full Text | Google Scholar
Skelly, M., Salameh, A., McCabe, J., and Pundik, S. (2020). MEP-ART: a system for real-time feedback and analysis of transcranial magnetic stimulation motor evoked potentials. Brain Stimul. 13, 1614–1616. doi: 10.1016/j.brs.2020.09.012
PubMed Abstract | Crossref Full Text | Google Scholar
Snow, N. J., Wadden, K. P., Chaves, A. R., and Ploughman, M. (2019). Transcranial magnetic stimulation as a potential biomarker in multiple sclerosis: a systematic review with recommendations for future research. Neural Plast. 2019, 1–22. doi: 10.1155/2019/6430596
PubMed Abstract | Crossref Full Text | Google Scholar
Sollmann, N., Krieg, S. M., Säisänen, L., and Julkunen, P. (2021). Mapping of motor function with neuronavigated transcranial magnetic stimulation: a review on clinical application in brain tumors and methods for ensuring feasible accuracy. Brain Sci. 11:897. doi: 10.3390/brainsci11070897
留言 (0)