Brazil has about 1.59 million heads of buffalo (FAO, 2024) with herds for milk, meat, and dual-purpose production under genetic evaluation (ABCB, 2024). Studies on the population structure of some breeds raised in the country have been conducted and reported inbreeding coefficients of 3.5% (Nascimento et al., 2021) for the Murrah breed, 2.02% for the Mediterranean breed (Malhado et al., 2013), and 4.22% for the Jaffarabadi breed (Ferraz et al., 2015). Although inbreeding coefficients are low, inbreeding depression for weight, milk production, and reproductive traits has been reported in the Murrah and Mediterranean breeds (Santana et al., 2011; Malhado et al., 2013). Nascimento et al. (2021) reported a genomic inbreeding coefficient (in 1 Mb homozygosity regions) of 7.2% for the Murrah breed, and these values are increasing over generations, suggesting monitoring of matings.
Maintaining genetic variability is essential for achieving gains through selection. Inbreeding measures intra-locus genetic variability, but there is the inter-loci genetic variability given by the recombination event. The recombination occurs at specific sites and is guided by the PRDM9 (PR/SET Domain 9) protein (Myers et al., 2010). Each PRDM9 allele promotes a different protein that performs crossing-over in a particular chromosomal region (Neale, 2010). The PRDM9 is a three functional domain protein, with the terminal one being a zinc finger (ZF) of cysteine2 histidine2 (C2H2) type (Parvanov et al., 2010). The tandem array of C2H2 zinc fingers and the single amino-acid change within the zinc fingers are responsible for creating the alleles. Each PRDM9 allele breaks the double stand in a specific spot and promotes the recombination at that site (Parvanov et al., 2010; Myers et al., 2010; Ahlawat et al., 2017). Therefore, increasing the frequency of less recurrent alleles of this gene in the population helps to increase/maintain inter-loci genetic variability (Gonen et al., 2017).
In ruminants livestock (sheep, goat, buffalo, yaks and cattle), some PRDM9 alleles have been characterized (Ahlawat et al., 2016; Ahlawat et al., 2017). In cattle, a concrete involvement in recombination events was confirmed (Shen et al., 2018; Zhou et al., 2018), even affecting fertility (Li et al., 2020; Seroussi et al., 2019). Moreover, the gene was reported to be under selection (Wang et al., 2019) and potentially useful to manage diverse in inbred population (Rocha et al., 2023).
Ahlawat et al. (2017) characterized the alleles of the PRDM9 gene in buffaloes, totaling 14 different alleles. The alleles of this gene are characterized by the number and type of zinc finger (ZF) domains they possess. Thus, the present study aimed to conduct a preliminary investigation of the alleles occurring in populations of three buffalo breeds (Murrah, Jaffarabadi, and Mediterranean) and their potential use for maintaining and promoting genetic variability.
Materials and methodsThe project was approved by the Animal Use Ethics Committee of the Escola de Medicina Veterinária e Zootecnia of the Universidade Federal da Bahia (81/2018). Hair follicles from of a total of 85 buffaloes (females and males) of Murrah (n = 27), Jaffarabadi (n = 41), and Mediterranean (n = 17) breeds from six commercial farms were collected. The farms were located at São Sebastião do Passé-BA, Nova Andradina-MS, Registro-SP, Tietê-SP, Sarapuí-SP and Paracuru-CE, all in Brazil) in order to be as diverse and representative as possible. The DNA extraction was done using the NucleoSpin® Tissue DNA extraction kit (Macherey-Nagel). The initial number of samples per breed was 50 animals, but the amplification of this region is complicated since it is a high polymorphic region with ZF domain repetitions.
For the amplification of the DNA sequence corresponding to the PRDM9 gene region, the primers (F: ACCTAGATGATTAGTGGGGCG and R: GCTGCAGTAATTCTCCTGTGAC) described by Ahlawat et al. (2017) were used. PCR reactions were carried out with a total volume of 25 µL containing: 50–110 ng of genomic DNA, 0.8 µmol of each primer, 9 µL of Promega Taq Mix, and 12.4 µL of deionized water. The PCR reaction was subjected to the Veriti 96 Well Thermal Cycler (Applied Biosystems) using the following conditions: an initial denaturation step at 95°C for 4 min, followed by 34 cycles of 95°C for 45°s, annealing at 60°C for 45°s, extension at 72°C for 45°s, and a final extension of 8 min at 72°C. The amplification products were verified on 2% agarose gel stained with Sybr Gold. The amplified fragments were visualized under ultraviolet light and photographed with the L-PIX Transilluminator Molecular Imaging (Loccus Biotecnologia). Allelic and genotypic frequencies and Hardy-Weinberg equilibrium were calculated using Excel software. Genotypic frequencies were compared across breeds, in pairs, by chi-squared (GraphPad -https://www.graphpad.com/quickcalcs/chisquared1/). The PCR products of homozygous individuals were purified with 20% polyethylene glycol and sequenced using the BigDye v3.1 sequencing kit (Applied Biosystems, Foster City, CA, United States) on a 3500xl Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions, using both primers to determine the ZF domain sequences. The obtained sequences were analyzed in CodonCode Aligner software and later deposited in GenBank (PP830922-26).
Results and discussionFor the three studied breeds, the three described alleles B, C and D, and six genotypes were found, except for the CC genotype for the Murrah breed (Table 1; Figure 1). The samples were run twice in agarose gel to confirm the correct genotype. It was found that the Jaffarabadi population was out of Hardy-Weinberg equilibrium, while the others were in equilibrium (data not presented). After sequencing the homozygous, the buffalo 1 and buffalo 2 alleles (allele B), buffalo 3 (allele C), buffalo 12, and buffalo 15 (allele D) were observed.
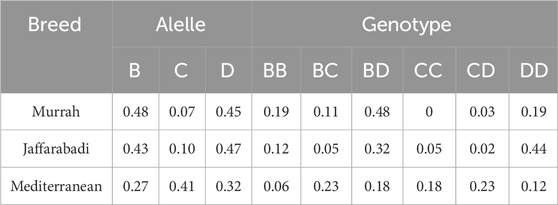
Table 1. Allelic and genotypic frequencies for the PRDM9 gene in buffaloes.
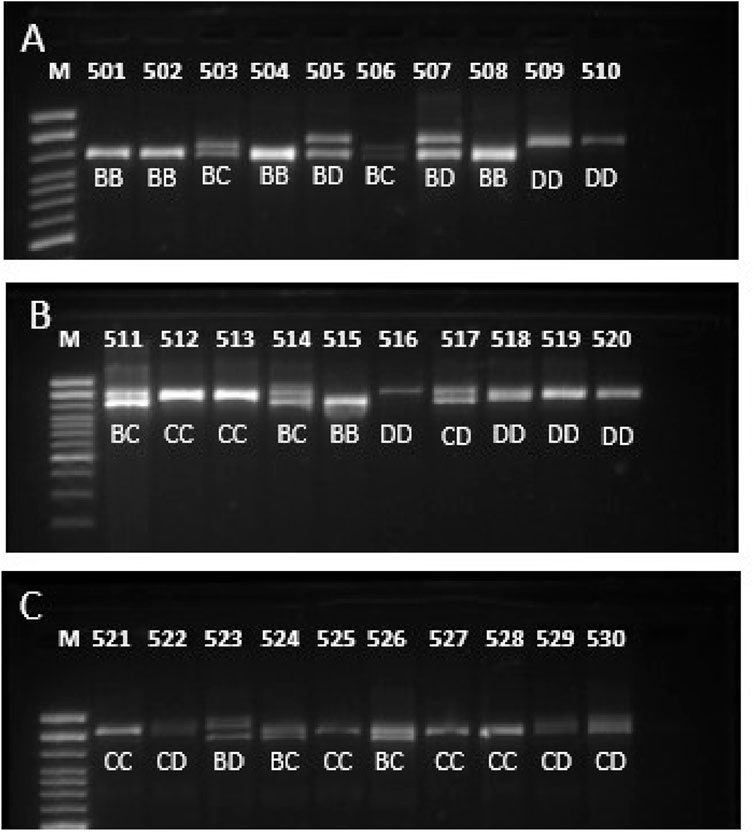
Figure 1. Gel image of PCR product of ZF domain of PRDM9 in three buffalo breeds (A) Murrah, (B) Jaffarabadi and (C) Mediterranean showing some genotypes identified.
The allelic and genotypic frequencies varied between breeds, as reported by Ahlawat et al. (2017), being these the first frequencies estimates of the PRDM9 for the Mediterranean breed. The genotypic frequencies were significantly different across breeds (data not shown). The differences might be related to breed formation and selection strategies. The presented frequencies used a larger number of animals than Ahlawat et al. (2017), who used five to 12 animals for allelic characterization. For Jaffarabadi, the highest genotype (DD) and allele (D) frequencies are the same between studies, but all the six genotypes were herein reported while Ahlawat et al. (2017) reported three. For Murrah, the highest allele (B × D) and genotype (BD × CD) frequencies varied between studies. It might be due to sample size and population characteristics.
The distribution of PRDM9 genotypes in the populations allows defining mating strategies to increase the frequency of some alleles, and genotypes in heterozygosity, aiming to increase the crossing-over rates and inter-loci variability. For Jaffarabadi breed, choosing to mate CC genotype animals with other homozygous tends to increase the frequency of allele C and heterozygous genotypes, promoting crossing-over in different regions and increasing the variety of haplotypes in the population. For Murrah breed, besides mating among homozygous, mating animals with allele C in heterozygosity in their genotype is also interesting for the same purposes. In the Mediterranean breed, genotypic frequencies are equally distributed, indicating a better distribution of recombination in different hotspots. A similar approach was already carried out in the Sindhi cattle (Rocha et al., 2023), which has low intrapopulation variability with good mating strategy options using PRDM9 genotypes. The mating decisions may help to increase genetic variability and increase productivity as consequence.
The HW equilibrium calculation showed that the Jaffarabadi population is out of equilibrium. The higher frequency of homozygous in the observed frequencies compared to the expected ones is an indication of inbreeding, already evidenced in a previous study (Ferraz et al., 2015), being the breed with the highest inbreeding coefficient among those studied. It possibly happens because this breed has the smaller number of animals and natural service is the most common reproductive technique, being more frequent to mate related animals.
The definition of PRDM9 alleles is primarily given by the number of zinc finger domains. Allele B has seven ZF repetitions, allele C has eight repetitions, and allele D is characterized by nine ZF repetitions, initially characterized in agarose gel. However, the ZFs that constitute the alleles can vary, with the final characterization being done by sequencing, using numbers to identify them (buffalo 1-B1, buffalo 2-B2, etc.). All homozygous animals were sequenced, and four alleles (buffalo 1-B1, buffalo 2-B2, buffalo 3-C3, buffalo 12-D12) (Genbank: PP830922-25) out of the 14 described by Ahlawat et al. (2017) and a new one named buffalo 15-D15 (Genbank: PP8309226) were found. The ZF sequences of the new allele was B1-B9-B2-B10-B3-B3-B3-B5 following the initial description by Ahlawat et al. (2017). This allele differs from the others due to the unique sequence combination of ZFs not yet reported. It is important since it provides more options for recombination sites and strategic matings. Of the 41 homozygous animals identified in agarose gel (11 BB, five CC, and 25 DD), it was possible to identify the genotype of five Murrah (two B1B1, two B2B2, and one B1B2), seven Jaffarabadi (three B2B2, one D12D12, and two D15D15), and four Mediterranean (one B1B1, two C3C3, and one D12D12). A sequence example was given in Figure 2. It is important to notice that, for PRDM9 gene and Sanger sequencing, it is possible to identify the alleles of animals with the same ZF number (BB, CC and DD) and with the same sequence (B1B1, B2B2, C3C3, D12D12 and D15D15) or with slight identifiable codon changes (B1B2). It happens because this gene is extremely polymorphic and, with more than one SNP, it is impossible to define the linkage phase to establish which allele it is. This is an interesting result because there is a large number of heterozygous in the populations concerning the ZF sequences, which is related to higher recombination rates.
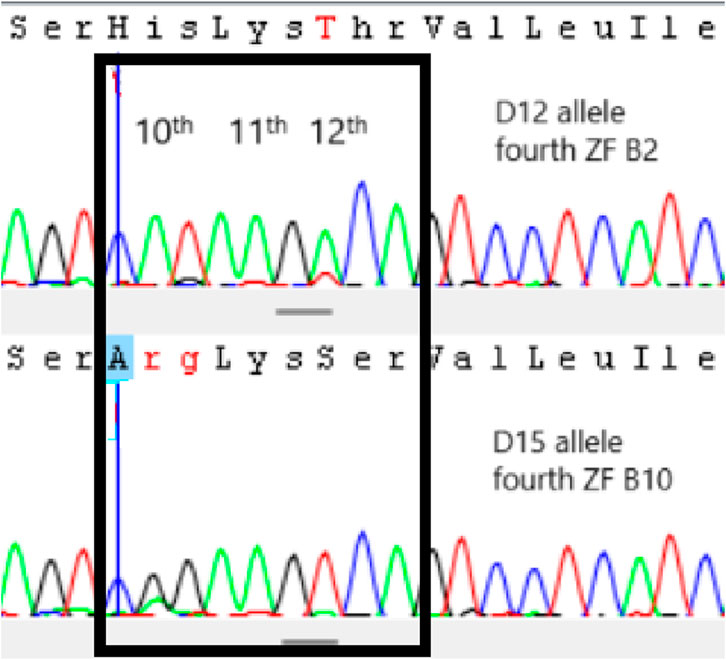
Figure 2. Sequence example of PRDM9 polymorphisms. The black box indicates the codons of the 10th, 11th and 12th amino acids of fourth zinc finger (ZF) of allele D12 (ZF–B2) and D15 (ZF–B10). B2 and B10 ZFs differ from each other in 10th and 12th amino acids, changing a His (CAT) to an Arg (CGG) and a Thr (ACA) to a Ser (TCA), respectively.
PRDM9 genotypes are another tool for maintaining variability that should be used to support other tools. It is important to remember that the way to increase intra-loci variability include mating non-related animals to try introduce new alleles. The methodology presented here can help maintain variability by increasing recombination rates, that is, circulating haplotypes, contributing to inter-loci variability with the possibility of increasing genetic gains (Gonen et al., 2017). Future studies with a larger number of animals and application of next-generation sequencing technology (for precise heterozygous identification) would permit a better scenario mating strategy. Moreover, the effectiveness of mating strategies in increasing variability over generations is a trial that should be planned and was not conducted yet.
ConclusionSeveral circulating alleles of the PRDM9 gene were identified in the three studied buffalo breeds, indicating that they can help as an additional tool in increasing/maintaining genetic variability by choosing sire and dams with different PRDM9 genotypes. It is also a strategy possible to be applied in any other livestock whose conditions were similar.
Data availability statementThe data presented in the study are deposited in the Genbank website repository, with the accession numbers PP830922-26.
Ethics statementThe animal studies were approved by the Animal Use Ethics Committee of the Escola de Medicina Veterinária e Zootecnia of the Universidade Federal da Bahia. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributionsLS: Formal Analysis, Writing–original draft, Writing–review and editing. JA: Formal Analysis, Investigation, Writing–review and editing. FF: Formal Analysis, Writing–review and editing. VR: Writing–review and editing. HT: Writing–review and editing. RC: Writing–review and editing. GdC: Conceptualization, Supervision, Writing–review and editing.
FundingThe author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAhlawat, S., De, S., Sharma, P., Sharma, R., Arora, R., Kataria, R. S., et al. (2017). Evolutionary dynamics of meiotic recombination hotspots regulator PRDM9 in bovids. Mol. Genet. Genomics 292, 117–131. doi:10.1007/s00438-016-1260-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Ahlawat, S., Sharma, P., Sharma, R., Arora, R., Verma, N. K., Brahma, B., et al. (2016). Evidence of positive selection and concerted evolution in the rapidly evolving PRDM9 zinc finger domain in goats and sheep. Anim. Genet. 47, 740–751. doi:10.1111/age.12487
PubMed Abstract | CrossRef Full Text | Google Scholar
Ferraz, P. C., Malhado, C. H. M., Ramos, A. A., Carneiro, P. L. S., Carrillo, J. A., and Malhado, A. C. M. (2015). Population structure and genetic variability of a closed Jaffarabadi buffalo herd from Brazil. Buffalo Bull. 34, 197–207.
Gonen, S., Battagin, M., Johnston, S. E., Gorjanc, G., and Hickey, J. M. (2017). The potential of shifting recombination hotspots to increase genetic gain in livestock breeding. Genet. Sel. Evol. 49, 55–12. doi:10.1186/s12711-017-0330-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, Y. C., Wang, G. W., Xu, S. R., Zhang, X. N., and Yang, Q. E. (2020). The expression of histone methyltransferases and distribution of selected histone methylations in testes of yak and cattle-yak hybrid. Theriogenology 144, 164–173. doi:10.1016/j.theriogenology.2020.01.001
PubMed Abstract | CrossRef Full Text | Google Scholar
Malhado, C. H. M., Malhado, A. C. M., Carneiro, P. L. S., Ramos, A. A., Carrillo, J. A., and Pala, A. (2013). Inbreeding depression on production and reproduction traits of buffaloes from Brazil. Anim. Sci. J. 84, 289–295. doi:10.1111/asj.12006
PubMed Abstract | CrossRef Full Text | Google Scholar
Myers, S., Bowden, R., Tumian, A., Bontrop, R. E., Freeman, C., MacFie, T. S., et al. (2010). Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879. doi:10.1126/science.1182363
PubMed Abstract | CrossRef Full Text | Google Scholar
Nascimento, A. V., Cardoso, D. F., Santos, D. J. A., Romero, A. R. S., Scalez, D. C. B., Borquis, R. R. A., et al. (2021). Inbreeding coefficients and runs of homozygosity islands in Brazilian water buffalo. J. Dairy Sci. 104, 1917–1927. doi:10.3168/jds.2020-18397
PubMed Abstract | CrossRef Full Text | Google Scholar
Rocha, V. C. P., Alves, J. S., Costa, R. B., and de Camargo, G. M. F. (2023). Variability in the PRDM9 gene in Sindhi cattle. Mol. Biol. Rep. 50, 8839–8842. doi:10.1007/s11033-023-08778-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Santana, M. L., Aspilcueta-Borquis, R. R., Bignardi, A. B., Albuquerque, L. G., and Tonhati, H. (2011). Population structure and effects of inbreeding on milk yield and quality of Murrah buffaloes. J. Dairy Sci. 94, 5204–5211. doi:10.3168/jds.2011-4377
PubMed Abstract | CrossRef Full Text | Google Scholar
Seroussi, E., Shirak, A., Gershoni, M., Ezra, E., Santos, D. J. A., Ma, L., et al. (2019). Bos taurus–indicus hybridization correlates with intralocus sexual-conflict effects of PRDM9 on male and female fertility in Holstein cattle. BMC Genet. 20, 71–11. doi:10.1186/s12863-019-0773-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Shen, B., Jiang, J., Seroussi, E., Liu, G. E., and Ma, L. (2018). Characterization of recombination features and the genetic basis in multiple cattle breeds. BMC Genomics 19, 304–310. doi:10.1186/s12864-018-4705-y
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, Z., Ma, H., Xu, L., Zhu, B., Liu, Y., Bordbar, F., et al. (2019). Genome-Wide scan identifies selection signatures in Chinese wagyu cattle using a high-density SNPArray. Anim 9, 296. doi:10.3390/ani9060296
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhou, Y., Shen, B., Jiang, J., Padhi, A., Park, K. E., Oswalt, A., et al. (2018). Construction of PRDM9 allele-specific recombination maps in cattle using large-scale pedigree analysis and genome-wide single sperm genomics. DNA Res. 25, 183–194. doi:10.1093/dnares/dsx048
留言 (0)