Acne vulgaris is a multifactorial chronic inflammatory disease of the pilosebaceous follicle, which includes the hair shafts and sebaceous glands. It is the most common dermatological condition worldwide, with an estimated 650 million people (Moradi Tuchayi et al., 2015). Young people constitute the most compromised population (85%) (Melnik, 2018; Bernhardt and Myntti, 2016).
Disease severity is closely associated with the inflammatory response, mainly to Cutibacterium acnes, a prominent member of the skin microbiota. The skin microbiota is largely composed of Actinobacteria (Corynebacterineae and Propionibacterineae), Proteobacteria, Firmicutes (Staphylococcaceae), and Bacteroidetes (Mayslich et al., 2021). Certain phylotypes of C. acnes have been identified as opportunistic pathogens capable of causing invasive infections and forming biofilms (Mayslich et al., 2021; Achermann et al., 2014; Keshari et al., 2019). Additionally, bacterial interactions, such as those between Staphylococcus epidermidis and C. acnes, may be influenced by changes in host characteristics. These alterations can lead to the selection of pathogenic C. acnes strains that produce virulence factors, thereby increasing their inflammatory potential (Mayslich et al., 2021). Furthermore, shifts in the populations of C. acnes and S. epidermidis on the skin could promote colonization by Staphylococcus aureus, which is associated with acne vulgaris as well as other dermatological conditions, such as atopic dermatitis (Fournière et al., 2020).
Given the complexity of microbial interactions in acne, antibiotics are commonly used for its management. Nevertheless, they do not neutralize secretory toxins but instead exert selection pressure on non-target bacteria at a systematic level (Keshari et al., 2019). This has led to the emergence of resistance to erythromycin, clindamycin, and tetracycline in C. acnes and S. aureus strains, increasing the likelihood of treatment failure (Alkhawaja et al., 2020; Sermswan et al., 2023). This dynamic is particularly concerning in the context of bacterial resistance, as seen in Staphylococcus aureus. In 2019, methicillin-resistant S. aureus (MRSA) was responsible for an estimated 4.95 million deaths globally (Dadgostar, 2019). In addition, other treatment options for acne, such as isotretinoin, also produce side effects, including psychiatric events and inflammatory bowel disease (Costa et al., 2018).
Historically, natural products extracted from plants, animals, and microorganisms have been a prolific source of bioactive compounds. Nonetheless, these treatment options have been applied specifically to help stop the skin aging processes and pigmentation or improve the general appearance (Duarte et al., 2022; Bungau et al., 2023). Actinobacteria, in particular, are responsible for producing about 70% of currently used antibiotics (Dholakiya et al., 2017). Of the approximately 500.000 natural compounds are derived from biological sources, and 70.000 are of microbial origin, of which 29% are obtained from actinomycetes (Siro et al., 2022). Marine microbial communities have been reported as the most diverse source of biologically active compounds (Siddharth, 2019), and unique bioactive natural products have been reported from marine actinobacterial strains, such as marinopyrroles, heronapyrroles, ansalactam ammosamides, salinosporamide A6, and flavonoids, which were initially reported only in plants (Panche et al., 2016; Cheng et al., 2013). Furthermore, marine actinobacteria have been less explored than their terrestrial counterparts, providing an interesting field of study and a source of new bioactive compounds.
Three-quarters of all newly discovered bioactive microbial products in marine environments are produced by bacteria associated with marine invertebrates, and approximately 30% of these compounds originate from marine sponges (Rajasabapathy et al., 2020). Corals rank second after sponges in terms of productivity, with 5,800 compounds derived from corals accounting for nearly 20% of all natural marine products (Siro et al., 2022).
In this study, we explored extracts obtained from marine actinobacteria isolated from sponges and octocorals in the Colombian Caribbean. These extracts were obtained from 13 strains and were subjected to antibacterial analysis against S. epidermidis, S. aureus, and C. acnes. The promising extracts were further evaluated for cytotoxicity against human keratinocytes and fibroblast cell lines. Chemical analysis of the extracts was performed using a metabolomic approach. This analysis indicated that the compounds probably related to antibacterial activity belonged to the alkaloid, terpenoid, naphthalene, and stilbene families, among others, which have previously been reported to have significant antimicrobial potential. Furthermore, molecular identification of the strains producing these promising extracts revealed that most belong to a genera classified as rare actinobacteria, such as Nocardia sp., Micrococcus sp., Rhodococcus sp., and Kocuria sp.
These findings highlight the potential of marine actinobacteria as a promising source of bioactive compounds with significant antimicrobial properties, creating opportunities for the development of new therapeutic alternatives for treating acne vulgaris.
2 Materials and methods 2.1 Actinobacteria strainsMarine actinobacteria were obtained from the Microbial Collection of the Bioprospecting Research Group at Universidad de La Sabana, Colombia. These isolates were originally obtained from sponge and octocorals collected by scuba diving from the Colombian Caribbean, Bahía de Taganga, Punta Venado (11°16′23.9″ N, 74°12′24.9″ W), Bahía de Santa Marta, Punta Betín (11°15′02.1″ N, 74°13′16.0″ W), Magdalena, Colombia, at depths of 13 and 9 m, respectively (Sánchez-Suárez et al., 2021). The bacteria used in the present study were of Colombian origin and were obtained according to Amendment No. 5 to ARG Master Agreement No. 117 of May 26, 2015, granted by the Ministry of Environment and Sustainable Development, Colombia. Supplementary Table S1 describes the actinobacterial strains and their isolation sources.
Staphylococcus epidermidis (ATCC 14990), C. acnes (ATCC 6919), and methicillin-resistant S. aureus (ATCC BAA44) were acquired from the American Type Culture Collection (ATCC).
2.2 Actinobacteria culture and extracts obtentionActinomycete isolates were inoculated in 100 mL of liquid Glucose, Yeast, and Malt extract broth (GYM) or Zobell Marine broth (Zobell), according to the original isolation medium (Sánchez-Suárez et al., 2021). Briefly, the culture started using a 55.81 mm2 plug from a 7-day-old lawn growth plate as a seed in 3 mL of broth. Each culture was incubated at 30°C with agitation at 200 rpm for 7 days. From this culture, 1 mL was used to inoculate 9 mL of GYM or Zobell broth, which was incubated for 7 days at 30°C with agitation at 200 rpm. Finally, the previous culture product (10 mL) was used to inoculate 90 mL of GYM or Zobell broth into 250 mL flasks on a rotary shaker (200 rpm) for 7 days. Then, 25 mL of each culture were placed in 50 mL plastic tubes, then freeze-dried (FreeZone Laboratory Freeze Dryer 2.5 liters, Labconco USA, Kansas), with a pressure of 0.22 millibar and a temperature of −55°C. The freeze-dried culture was transferred to a 50 mL Erlenmeyer flask, mixed with 10 mL of ethyl acetate (EtOAc), and agitated (150 rpm) for 24 h. The extraction was repeated three times using fresh EtOAc. The organic layers were evaporated under vacuum (Heidolph evaporator). Then, a stock concentration of 50 mg/mL was prepared for each actinobacteria extract, which was then used to perform antibacterial assays at the reported concentrations.
2.3 Cutibacterium acnes culture conditionsCutibacterium acnes ATCC 6919 was activated according to the manufacturer instructions. Briefly, two media were used for its activation: ATTC Medium 2107: Modified Reinforced Clostridial (MRC) and ATCC Medium 260: trypticase soy agar/broth with defibrinated sheep blood under aseptic conditions and incubated in an anaerobic atmosphere at 37°C for 48–72 h. Anaerobic conditions were achieved using test tubes with a gas cannula system connected to anaerobic gas. Loose screw caps were placed in the test tubes in an activated anaerobic gas pack jar.
2.4 Antibacterial activity assaysThe antibacterial activity of the extracts was estimated from the growth curve through the optical density using a Synergy H1 Multimode Reader (BioTek, Winooski, VT, USA). The antibacterial activity was determined using a microbroth susceptibility assay in 96-well plates, each containing 100 μL of extracts at different concentrations (initially 0.3, 0.03, and 0.003 mg/mL), following protocols standardized by the Bioprospecting Group of the Universidad de La Sabana, with slight modifications to the yield of the extracts (Sarmiento-Tovar et al., 2024). Extracts that did not show activity at the initial concentrations were further assessed at the highest concentration prepared (1.25 mg/mL), based on the yield of the extracts, with 100 μL of bacterial inoculum (S. epidermidis, MRSA, and C. acnes) at a cell density of 1.5x108 UFC/mL. The absorbance was maintained within the range of 0.08–0.1 at 600 nm (Bauermeister et al., 2019). The optical density (OD) was measured at 600 nm at 30 min intervals for up to 18–20 h for S. epidermidis and MRSA. For C. acnes, the incubation time was 72–150 h the time at which the bacteria reached a stationary phase. The bacteria in Tryptic Soy Broth (TSB) and modified reinforced clostridial broth (MRC) were used as negative controls. The commercial antibiotics vancomycin and erythromycin, which were used at the same concentrations as the extracts, were used as the positive controls. Percentage inhibition was calculated using the following formula (Equation 1) (Rajivgandhi et al., 2016):
%ofinhibition:UntreatedbacteriaOD600nm−TreatedbacteriaOD600nmUntreatedbacteriaOD600nm×100 (1) 2.5 Determination of maximum specific growth rate and maximum ODSpecific growth rates were determined using ComBase. This database contained statistical models that fit the nature of the data. The inhibition percentage was calculated using the following formula (Equation 2):
%inhibition=UntreatedbacteriaGr1−TreatedbacteriaGr1UntreatedbacteriaGr1×1001growth rate.
2.6 Minimal inhibitory concentration of bioactive extractsThe minimal inhibitory concentration (MIC) of extracts was determined using the broth microdilution method, according to the protocol described by Balagurunathan et al. (2020). Actinobacteria extracts and vancomycin and erythromycin (positive controls) were prepared by the serial 2-fold dilution method from the highest concentration observed in the antibacterial activity screening, 1.25 mg/mL (1.25–0.312 mg/mL) in TSB medium for S. epidermidis and S. aureus, and MRC broth for C. acnes. Freshly grown colonies of bacterial cultures were suspended in 10 mL fresh medium. The bacterial suspension was adjusted to a density of 5 × 105 CFU/mL (Wang et al., 2017; Akhter et al., 2018) and added to 96-well plates. Each well contains 100 μL of the bacterial suspension and 100 μL of the extract. The negative controls included 100 μL of bacterial suspension and 100 μL of culture broth. As the extracts were dissolved in isopropyl alcohol (the maximum final concentration of isopropyl alcohol was 0.3%), a control with this solvent was also included. Each experiment was performed in duplicates. Plates were incubated for 20 h at 37°C for S. aureus and S. epidermidis and incubated for 150 h at 37°C under anaerobic conditions for C. acnes because of its naturally slower growth rate. MIC was determined by observing the lowest concentration of the extract that inhibited bacterial growth.
2.7 Cytotoxicity assayCell viability assays were performed to evaluate the cytotoxicity of the extracts against HDFa (Primary Dermal Fibroblast; Normal, Human, Adult ATCC® PCS-201-012™) and HACAT (spontaneously immortalized human keratinocyte cell line derived from a distant periphery of malignant melanoma) following the methodology described by van de Loosdrecht et al. (1991), with some modifications. Briefly, the cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin, using (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) at 0.5 mg/mL an incubated 4 h. Cells (104 cells/well) were seeded in triplicate in 96-well plates and cultured overnight at 37°C and 5% CO2. Bioactive extracts were added to 96-well plates at final concentrations of 0.3, 0.03, and 0.003 mg/mL and 1.25, 0.625, and 0.3125 mg/mL in DMEM supplemented with 10% fetal bovine serum. Dimethyl sulfoxide (DMSO, between 1 and 10% v/v) was used as the negative control. After 24 h of incubation, MTT was removed, and DMSO was added to each well to dissolve the formazan crystals. The amount of formazan was determined by measuring absorbance at 570 nm (Mosmann, 1983). Cell viability was calculated using the following formula (Equation 3) (Sánchez-Suárez et al., 2021):
%viability=ODSamplesODControlgroup×100 (3) 2.8 Sequencing of 16S rRNA gene and phylogenetic analysisGenomic DNA was extracted using a Quick-DNA Fungal/Bacterial Microprep kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer’s instructions. The 16S rRNA gene was amplified using the universal primers 27F (forward primer: 5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (reverse primer: 5′-TACGGYTACCTTGTTACGACTT-3′) under the following cycling conditions: denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 7 min using Thermal Cycler (Bio-Rad). The amplified products were evaluated by electrophoresis of DNA samples on agarose gel. The agarose gel used in this study was 1% agarose gel in a buffer solution of Tris Acetate EDTA (TAE) (Anggelina et al., 2021).
The 16S rRNA sequences were run on the Basic Local Alignment Search Tool (BLAST) using the NCBI BLAST search tool, and the nearest neighbors were identified. MEGA version X (Molecular Evolutionary Genetics Analysis, version 10), using the Tamura 3-parameter model, was used to construct a phylogenetic tree using the neighbor-joining method with a bootstrap test (1,000 replicates).
2.9 HPLC-QTOF-MS metabolomic analysis of extractsMetabolomic analysis of the antibacterial active extracts and non-active extracts was performed using an Agilent Technologies 1,260 Liquid Chromatography system coupled with a Q-TOF 6545 time-of-flight quadrupole mass analyzer with electrospray ionization (ESI). The separation process was performed on a C18 column (InfinityLab Poroshell 120 EC-C18 100 × 3.0 mm, 2.7 μm). Two μL of the extracts were injected at 30°C and a gradient elution composed of: 0.1% (v/v) formic acid in Milli-Q water (Phase A) and 0.1% (v/v) formic acid in acetonitrile (Phase B) with a constant flow rate of 0.4 mL/min. The elution gradient program was: 0–15 min; 2–30% B, 15–17 min; 30–98% B, 17–21 min; 98% B and 21–26 min; 2%B.
Mass spectrometric detection was performed in the positive ESI mode in the full scan mode, and MS/MS was performed from 100 to 1.800 m/z. The following reference masses were used for mass correction during analysis: m/z = 121.0509 (C5H4N4) and m/z = 922.0098 (C18H18O6N3P3F24). The ESI source parameters were as follows: capillary voltage (3,000 V); drying gas (8 L/min); gas temperature (325°C); nebulizer pressure (50 psi); sheath gas temperature (350°C); and sheath gas glow (11 L/min). Q-ToF parameters included fragmentor voltage (175 V), skimmer voltage (65 V), and octapole radiofrequency peak-to-peak voltage (OCT RF Vpp) (750 V). Quality control (QC) samples were prepared according to the technique by pooling equal volumes of all groups of samples. QC pool samples were injected every 10 samples to evaluate stability and reproducibility throughout the analysis.
Data processing was performed using Agilent MassHunter Profinder software (version 10.0), which carried out deconvolution, alignment, and integration. These procedures were executed using the recursive feature extraction (RFE) algorithm. Datasets were filtered to remove features with a coefficient of variation (CV) > 20% in QC samples, retaining only those present in at least 80% of each sample group.
For metabolite identification, the CEU MASS MEDIATOR (accessible at https://ceumass.eps.uspceu.es/ as of August 15, 2023) (Gil-De-La-Fuente et al., 2019) was used to identify statistically significant m/z values. This tool integrates various platforms, including Metlin, Kegg, HDMB, and LipidMaps, with a tolerance of 10 ppm to annotate more abundant molecular features. Additionally, we utilized the StreptomeDB v3.0 (Moumbock et al., 2021) and the Natural Products Atlas v2.0 (Van Santen et al., 2022) databases.
To validate the identity of the metabolites, MS/MS analyses were conducted using MS-DIAL 4.8 (http://prime.psc.riken.jp/compms/msdial/main.html, accessed on August 22, 2023). These analyses involved in silico mass spectral fragmentation using CFM-ID 4.0 (available at https://cfmid.wishartlab.com/ as of August 28, 2023), and manual MS/MS spectral interpretation using the Agilent MassHunter Qualitative Analysis program (version 10.0, USA) (Camargo et al., 2023). The identified metabolites were reported according to the confidence level of compound annotation described by Blaženovi (2018).
2.10 Statistical data analysisData are presented as mean ± standard deviation (SD), and differences were examined using two-way analysis of variance (ANOVA) with a significance level of 5%. ANOVA assumptions of normality, homogeneity of variance, and independence of the data were checked. Dunnett’s test was performed to compare treatments at the 95% confidence level.
For metabolomic analysis, univariate statistical analysis (UVA) and multivariate statistical analysis (MVA) were performed to assess statistically significant differences among the metabolomic profiles of the groups using Metaboanalyst 5.0 and SIMCA 16.0 (Umetrics, Umea, Sweden), respectively. Initially, MVA based on principal component analysis (PCA) was applied to evaluate the quality of the acquired data. This ensured that the quality control (QC) samples were appropriately clustered in these models to guarantee the stability of the analytical system. Subsequently, orthogonal partial least squares discriminant analysis (OPLS-DA) models were constructed to maximize and examine the differences between the active and inactive extracts in the study groups and select the metabolites responsible for group separation.
Pareto scaling was applied for transformation before statistical analysis. In UVA, the p-value with a Benjamini-Hochberg false discovery rate correction (FDR) was determined (Quintero et al., 2022). Significant features were those with an adjusted p-value < 0.05, and variance important in projection (VIP) >1 with a jackknife confidence interval (JK). To determine the predictability and validity of the OPLS-DA model and to avoid overfitting and false positives, the models were subjected to cross-validation using K-fold and permutation tests (n = 100).
3 Results 3.1 Antibacterial activity of actinobacterial extractsThe antibacterial activity of ethyl acetate extracts from 13 actinomycete isolates was evaluated and established as a decrease in bacterial viability compared with that of untreated bacteria. Six extracts demonstrated activity against Staphylococcus epidermidis, MRSA, and C. acnes. The minimum inhibitory concentrations (MICs) were determined based on the concentrations tested. The Z9.216 extract exhibited MIC of 0.3 mg/mL against S. epidermidis and 0.003 mg/mL against both S. aureus and C. acnes. Extracts Z6.29 and Z9.11 had MIC values of 0.312 mg/mL for S. epidermidis, although MICs for S. aureus and C. acnes could not be determined within the tested concentration range due to the limited yield of the extracts. Table 1 shows the bacterial growth inhibition percentages of the extracts that were active against the three bacteria. These were selected for molecular identification by 16S rRNA gene sequencing. Biological profiling was carried out by metabolomic analyses of the end products using High-Performance Liquid Chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry (HPLC-QToF-MS) and mass spectrometry (MS/MS) analyses.
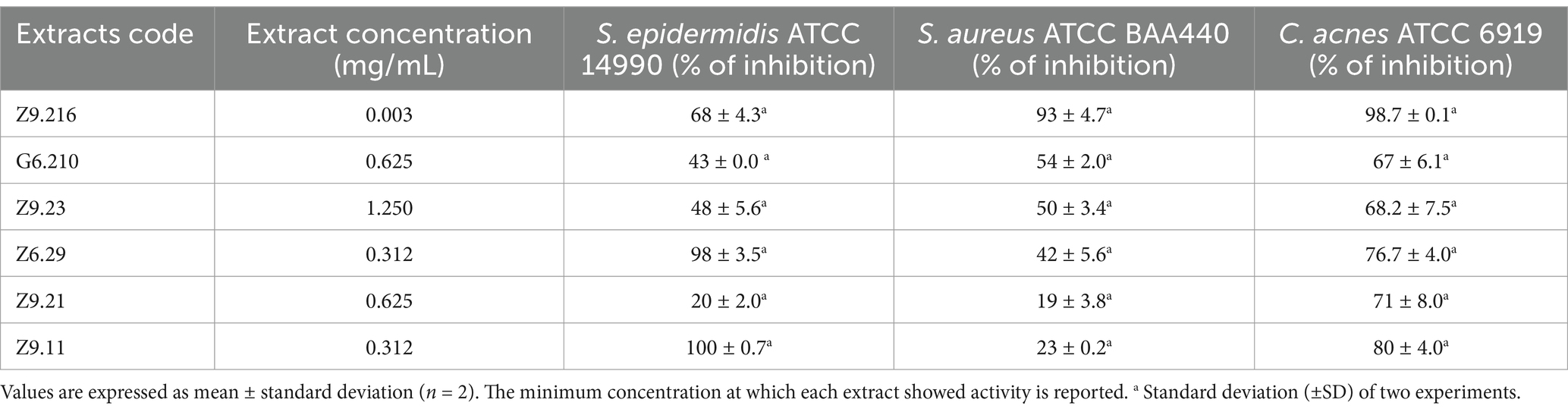
Table 1. Percentage of inhibition of bacterial growth by the most promising ethyl acetate extracts from marine actinobacteria against Staphylococcus epidermidis ATCC 14990, methicillin-resistant Staphylococcus aureus ATCC BA440, and Cutibacterium acnes ATCC 6919.
The most promising isolates were selected because they exhibited more than 50% growth inhibition against some of the bacteria evaluated, namely Z9.216, G6.210, Z9.23, Z6.29, Z9.21, and Z9.11. The extract with the highest percentage of growth inhibition, Z9.216, was selected for further studies. Figure 1 presents the bacterial growth curves of MRSA, S. epidermidis, and C. acnes in the presence of Z9.216 extract.
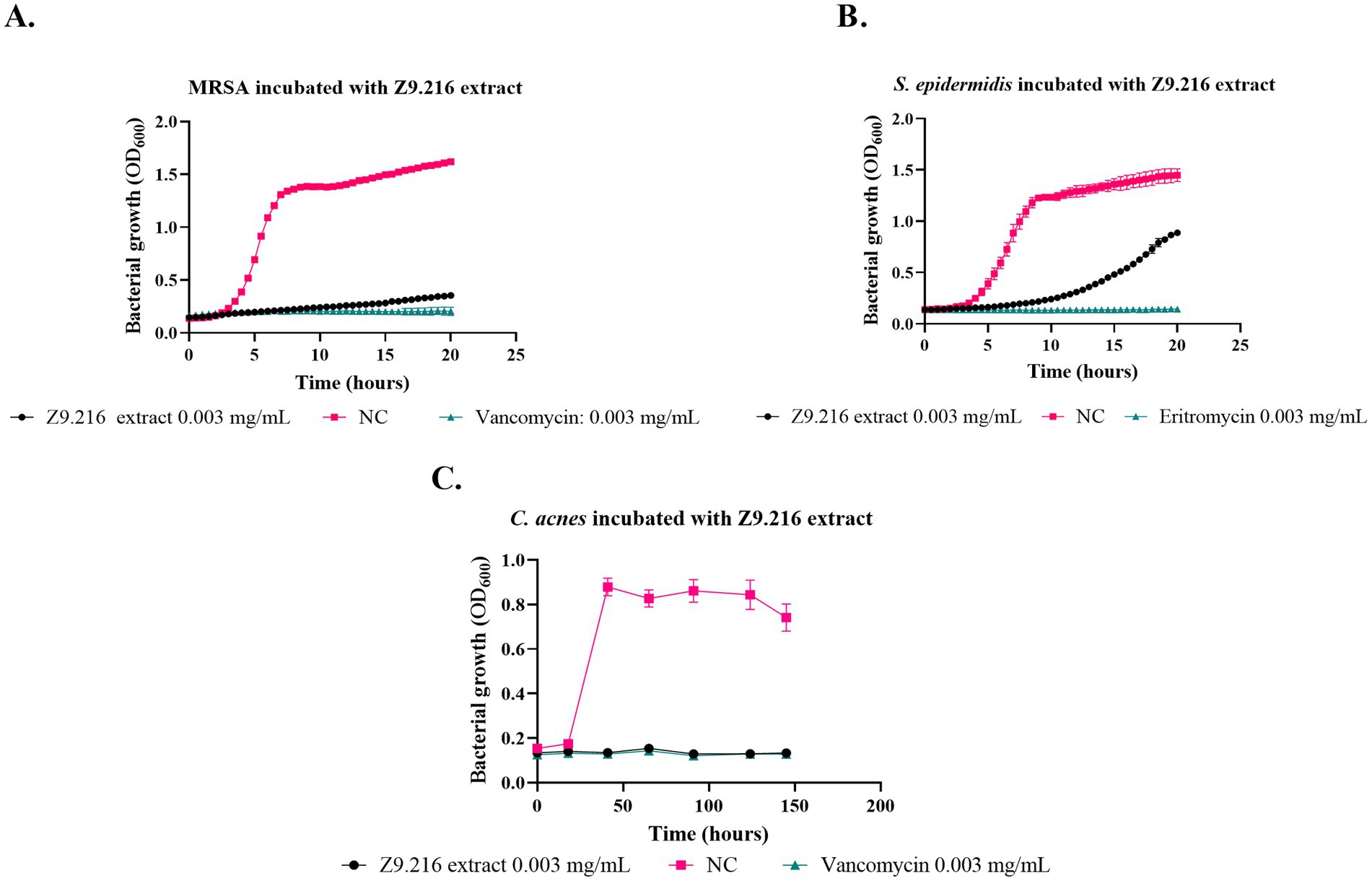
Figure 1. Effect of Z9.216 extract (0.003 mg/mL) on the growth of S. epidermidis ATCC 14990, methicillin-resistant S. aureus (MRSA) ATCC BA440, and C. acnes ATCC 6919. (A) Growth curve of S. aureus ATCC BA440; (B) Growth curve of S. epidermidis ATCC 14990; (C) Growth curve of C. acnes ATCC 6919. NC: Negative control, untreated bacteria.
3.2 Determination of maximum specific growth rateThe growth rate of bacteria treated with various extracts was assessed using the Baranyi and Roberts predictive primary model, because it had the best R2 parameters (>97%) and the lowest squared error (<0.2). The maximum growth rate (measured in units of hours (h−1)), represented by the μ parameter, indicating the exponential growth phase of the bacteria (Dalgaard et al., 1994; Lindqvist and Barmark, 2014), was determined based on this model. Analysis of the data revealed that the bacterial growth rate in the presence of the most promising extract, Z9.216, at concentrations of 0.3, 0.03, and 0.003 mg/mL closely resembled that of the antibiotic vancomycin against MRSA and erythromycin against S. epidermidis and C. acnes. Interestingly, bacteria tested in the presence of extracts G6.210, Z9.23, and Z6.29 at 1.25 mg/mL exhibited growth rates surpassing those of the antibiotics but falling short of the growth rate observed in untreated bacteria. In comparison, the performances of extracts Z9.21 and Z9.11 were found to be similar. Figure 2 illustrates the growth rates of the studied bacteria in the presence of the most promising extract, Z9.216.
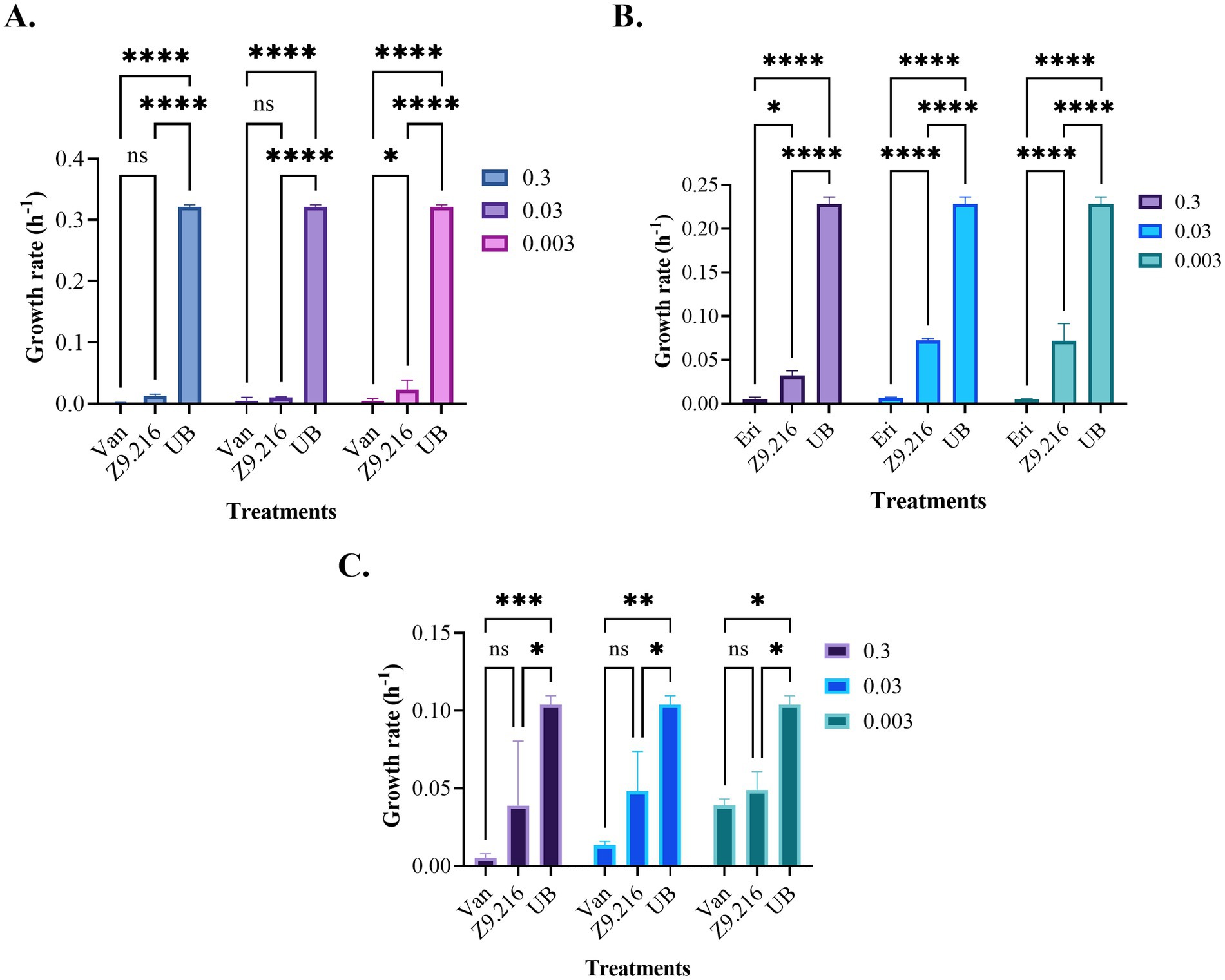
Figure 2. Growth rates of the bacteria studied in the presence of the Z9.216 extract (0.3–0.003 mg/mL). (A) Growth rate of methicillin-resistant S. aureus (MRSA) ATCC BA440. (B) Growth rate of S. epidermidis ATCC 14990. (C) Growth rate of C. acnes ATCC 6919. Values are expressed as mean ± standard error of the mean (SEM) and are compared with the positive control group. Significance levels are indicated as follows. ****p-value < 0.0001; ***p-value < 0.0007; **p-value < 0.0015; *p-value < 0.05.
3.3 In vitro safety evaluation of the actinobacterial extractsThe most promising extract, Z9.216, maintained cell viability above 90% in both the HDFa and HACAT cell lines. In contrast, the bioactive extracts G6.210, Z9.23, and Z9.11, exhibited the highest cytotoxic activity against the HACAT cell line, reducing viability by 44, 36, and 46%, respectively, at the highest concentrations evaluated. In the HDFa cell line, the extracts Z6.29 and Z9.21 demonstrated the highest cytotoxicity, decreasing cell viability by 37 and 47%, respectively, at the highest concentrations tested. For the remaining actinobacterial extracts, cell viability remained above 70% (Figure 3).
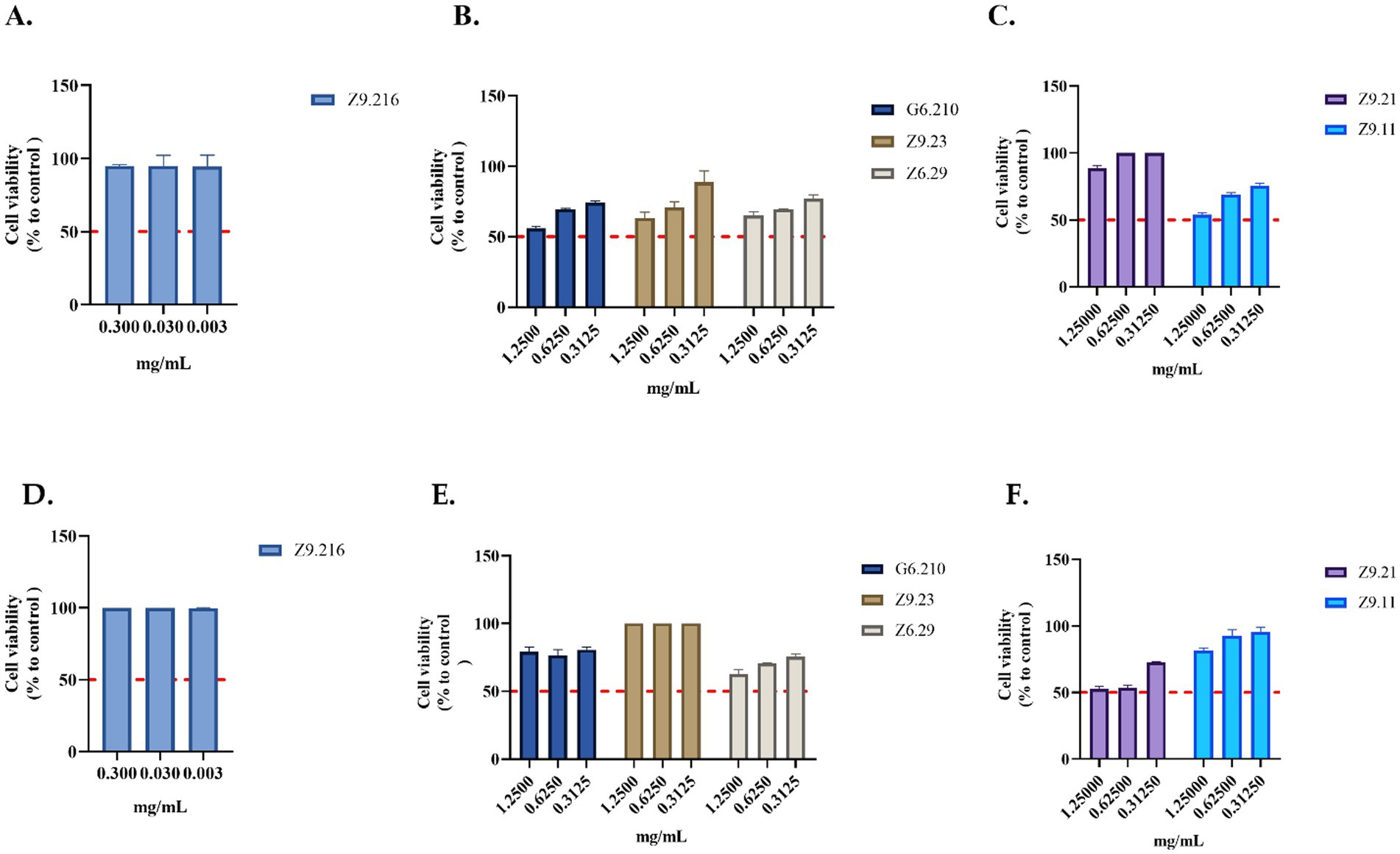
Figure 3. Evaluation of the viability of actinobacterial extracts. (A–C) Extracts evaluated in the HACAT cell line. (D–F) Extracts evaluated in HDFa cells. The crude extracts were evaluated at concentrations that showed activity. Each value represents the mean (n = 2) and error bars represent the standard deviation (SD). Cell viability was calculated by comparing the viability of untreated cells (control).
3.4 Identification of isolates with antibacterial activity by 16S rRNA gene sequencingThe cladogram led to the classification of bioactive isolates among actinobacterial genera, and Z9.216 was identified as a Kocuria, Z9.23 as Nocardia, Z6.29 as Rhodococcus, Z9.11 as Micrococcus, and Z9.21 as Streptomyces (Figure 4). The G6.210 isolate belonged to the Streptomyces genus, as previously reported (Sánchez-Suárez et al., 2021). Consensus sequences of the remaining extracts were deposited in GenBank under accession numbers PP389604, PP741801, PP741804, PP741802, and PP741803. The coding of the end-products and the names of the identified strains are presented in Supplementary Table S1.
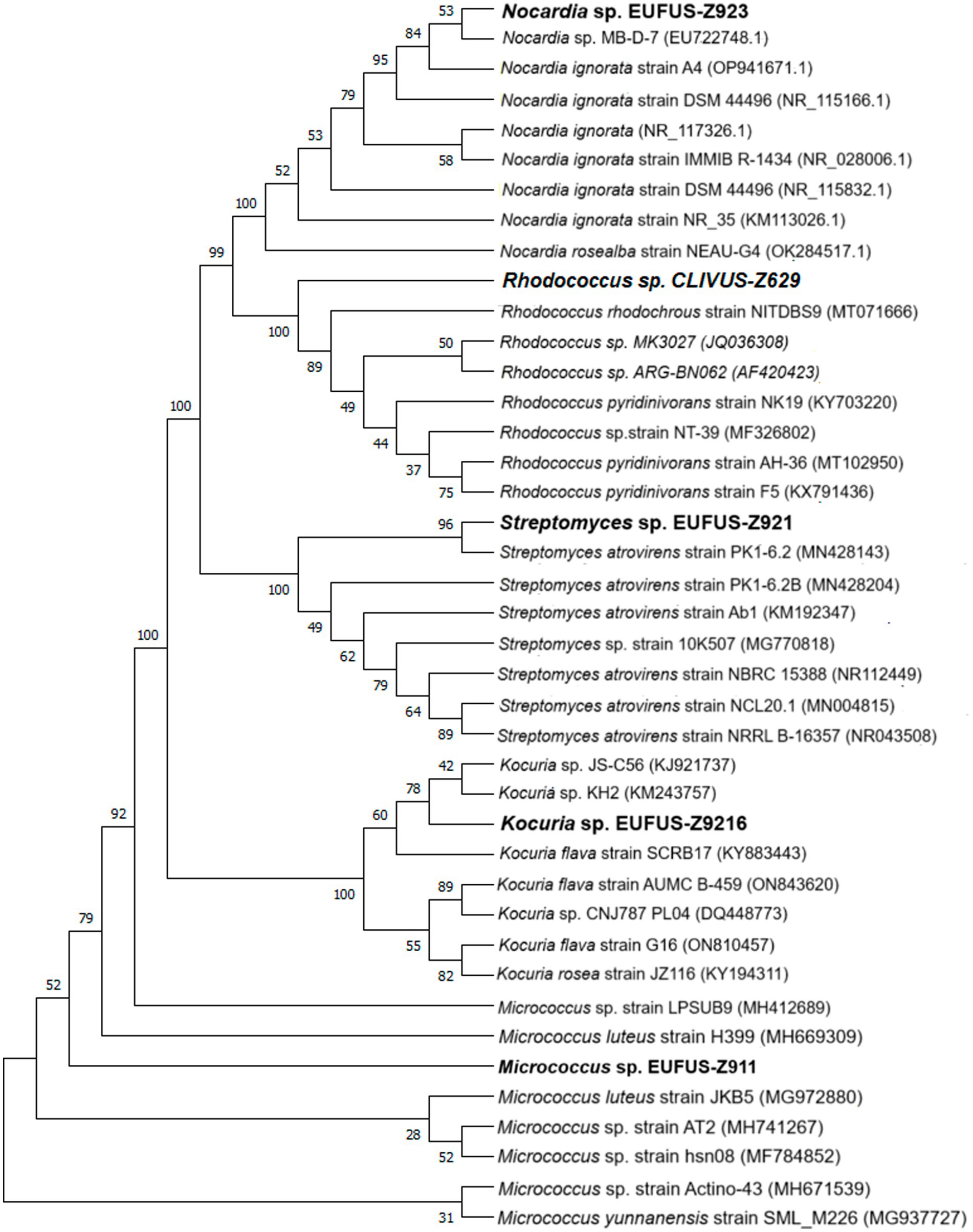
Figure 4. Phylogenetic tree of the bioactive isolates: cladogram for strains of genera Kocuria sp., Nocardia sp., Rhodoccoccus sp., Micrococcus sp., and Streptomyces sp., using 16S rDNA sequences. An optimal tree is shown. The accession codes for the blasted strains are shown in parentheses. The percentage of replicate trees with associated taxa clustered together in the bootstrap test (1,000 replicates) is displayed next to the branches. All ambiguous positions were removed from each pair of sequences (i.e., pairwise deletions).
3.5 Untargeted metabolomics by HPLC-QTOF-MS of promising microbial extractsA total of 468 features were obtained from the analysis, and the comparison between the antibacterial active extracts and non-active extracts yielded 45 statistically significant features. Among these, only 16 were annotated based on the criteria described by Blaženovi (2018). Furthermore, the retention time was considered to match the nature of the compounds for all the features. To assess the quality of the analytical platform, PCA was performed for each analysis.
Clear clustering of QC samples in the unsupervised PCA models (Figure 5A) showed the stability and quality of the acquired data for ESI analysis in the positive mode. Therefore, this result supports that separation between groups is related to biological differentiation. Additionally, PCA revealed distinct clustering of antibacterial (green) and inactive (red) samples (Figure 5B).
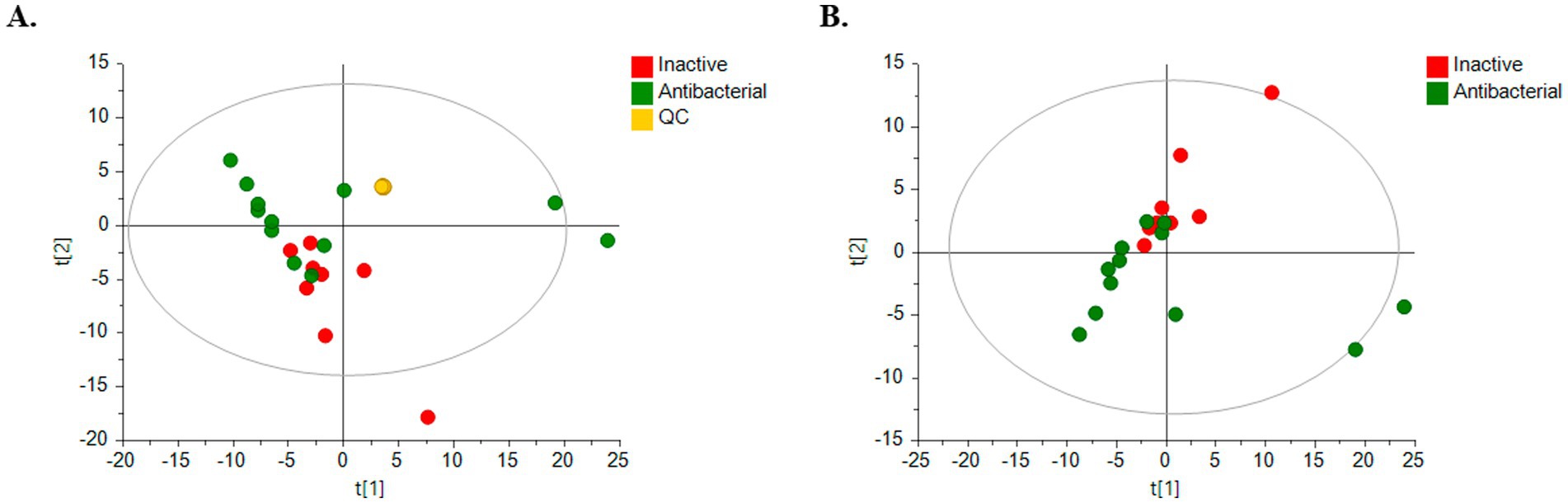
Figure 5. PCA score plots for untargeted metabolomics in positive ESI mode. (Green dots, bioactive samples; red dots, inactive samples; yellow dots, quality control samples, QC.) (A) HPLC-QTOF-MS (+) with QC samples: R2 = 0.733, Q2 = 0.293; (B) HPLC-QTOF-MS (+): R2 = 0.619, Q2 = 0.225.
The OPLS-DA model successfully differentiated samples into active antibacterial and inactive extracts. The OPLS-DA score plots (Figure 6A) clearly show separation, with the active group represented by green circles and the inactive group represented by red circles. The quality and robustness of the OPLS-DA model were validated using permutation tests (n = 100) (Figure 6B). The Q2 intercept value was 0.739, indicating that the original model was statistically effective, the slope of the Q2 values in the permutation test was negative for Y axes, and all predicted Q2 values were lower than those calculated by the Q2 model, indicating not over-fitted model (Triba et al., 2015; Betancur et al., 2020).
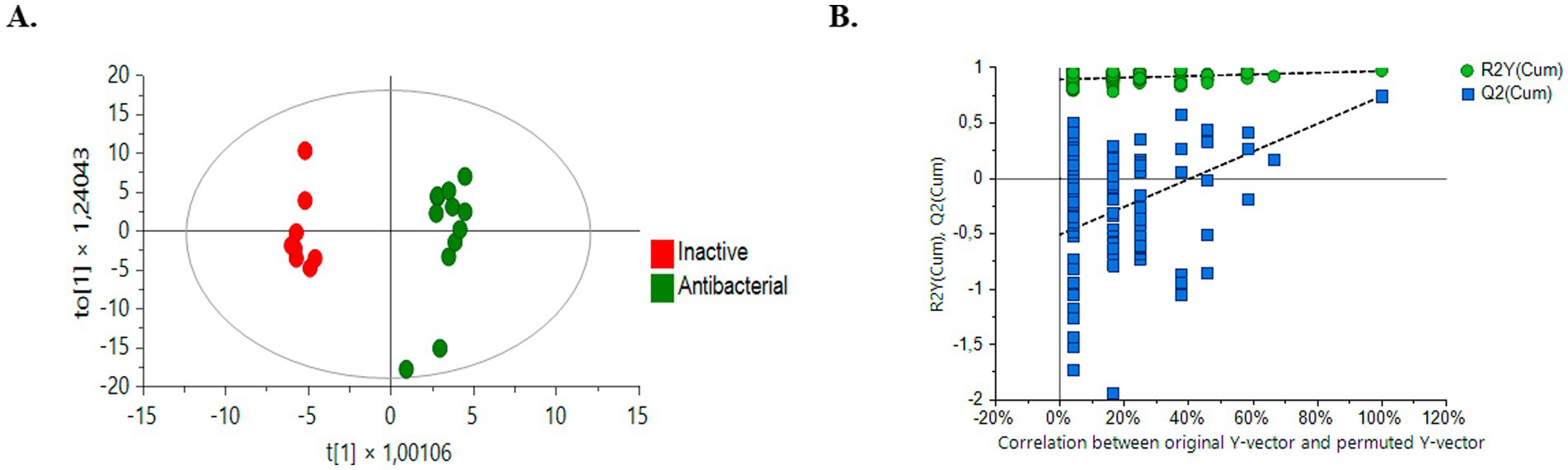
Figure 6. Comparison of supervised OPLS-DA models for untargeted metabolomics of inactive samples and bioactive samples (green dots, bioactive samples; red dots, inactive samples). (A) Positive ESI mode comparison HPLC-QTOF-MS (+): R2Y (cum): 0.966, Q2 (cum): 0.739, CV ANOVA: 1,3–3(B) Permutation test (n = 100) results for the OPLS-DA model.
A univariate analysis (UVA) was performed to assess the significance of each metabolite for comparison. The parameters used to select statistically significant metabolites were those that met the following criteria: p < 0.05, VIP >1. The metabolites that met these requirements were identified as putative, confirmed, or unknown and are presented in Table 2.
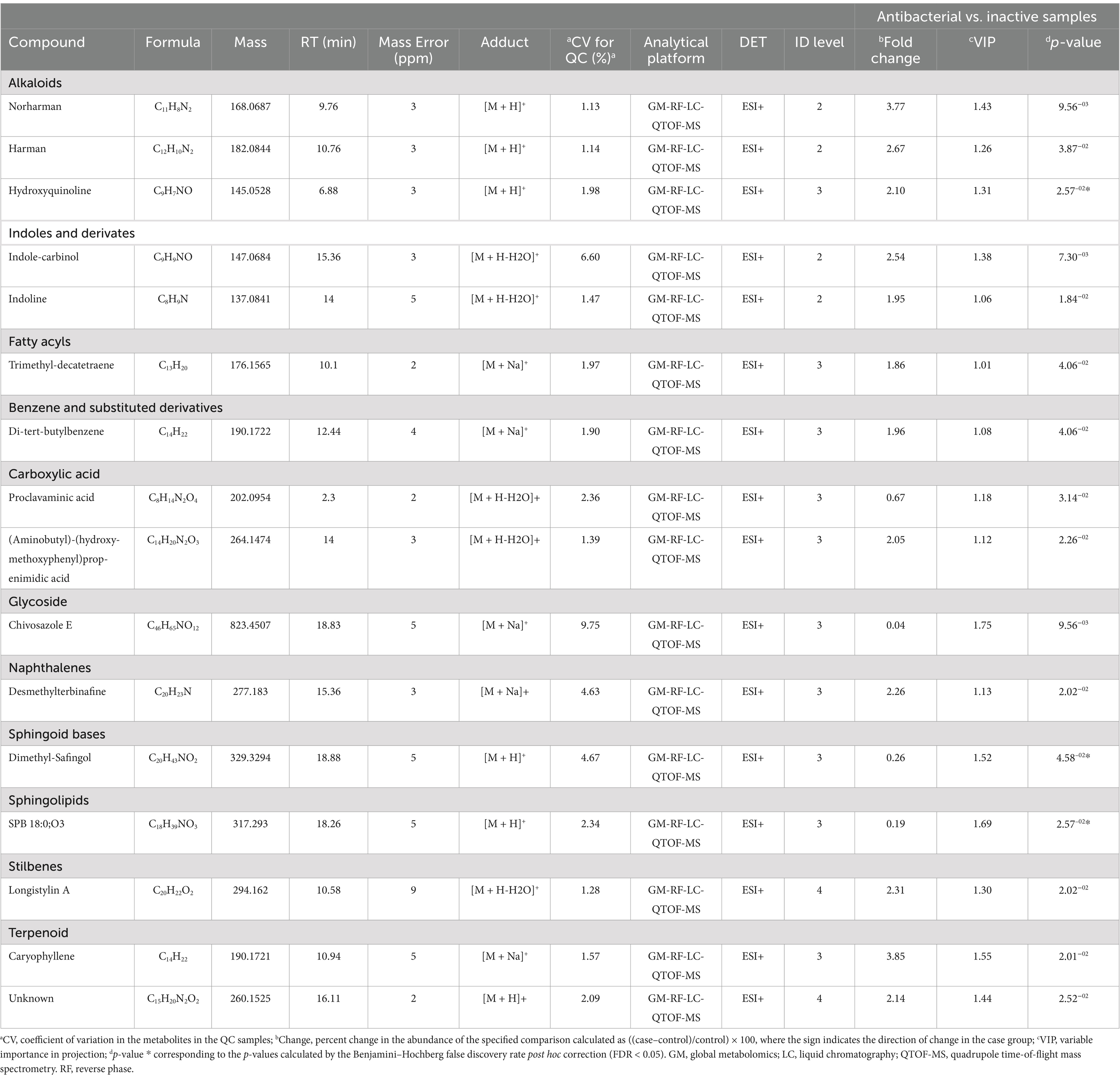
Table 2. Metabolites significantly differentiated between antibacterial active extracts and non-active extracts using ESI modes by untargeted metabolomics via quadrupole time-of-flight mass spectrometry.
Sixteen metabolites were identified as statistically significant when the antibacterial activities of the active and inactive extracts were compared. Among these compounds are alkaloids, such as indoles and their derivatives, quinolines, and other compounds, such as carboxylic acids, naphthalenes, stilbenes, and terpenoids (Figure 7). These features were characterized at different levels: level 2 (probable structure: matched to literature data or databases by diagnostic evidence) utilizing library spectrum matching through MS2 fragmentation with the aid of MSDial software and CFM-ID; level 3 (possible structure or class: most likely structure, isomers possible, substance class, or substructure match Probable structure: matched to literature data or database); and level 4 (an unknown feature of interest) based on the confidence level of compound annotation described by Blaženovi (2018). At level 2, we annotated four compounds (Figure 8), all of which belong to the alkaloid family. Notably, these compounds exhibited a high fold change (Table 2). One such compound, indole-carbinol (1), has been reported in cruciferous vegetables and fungi (Lin et al., 2019; Sung and Lee, 2007). High-resolution time-of-flight mass spectrometry (HPLC-QTOF-MS) data for (1) showed adduct molecular ions at m/z 147.0684 [M + H-H2O]+, which were analyzed using the molecular formula C9H9NO. Moreover, at Level 2, we annotated indoline (2). HPLC-QTOF-MS data for (2) showed adduct molecular ions at m/z 137.0841 [M + H-H2O]+, corresponding to the molecular formula C8H9N. This metabolite has been previously reported in actinobacteria with antibacterial properties, specifically in Streptomyces (Katsuyama, 2019). Harman (3), a β-carboline alkaloid, was also annotated in this level of identification. HPLC-QTOF-MS data for (3) showed adduct molecular ions at m/z 182.0844 [M + H]+, which were analyzed using the molecular formula C12H10N2. Finally, Norharman (4), another β-carboline alkaloid, was annotated with HPLC-QTOF-MS data, showing adduct molecular ions at m/z 168.0687 [M + H] +, corresponding to the molecular formula C11H8N2.
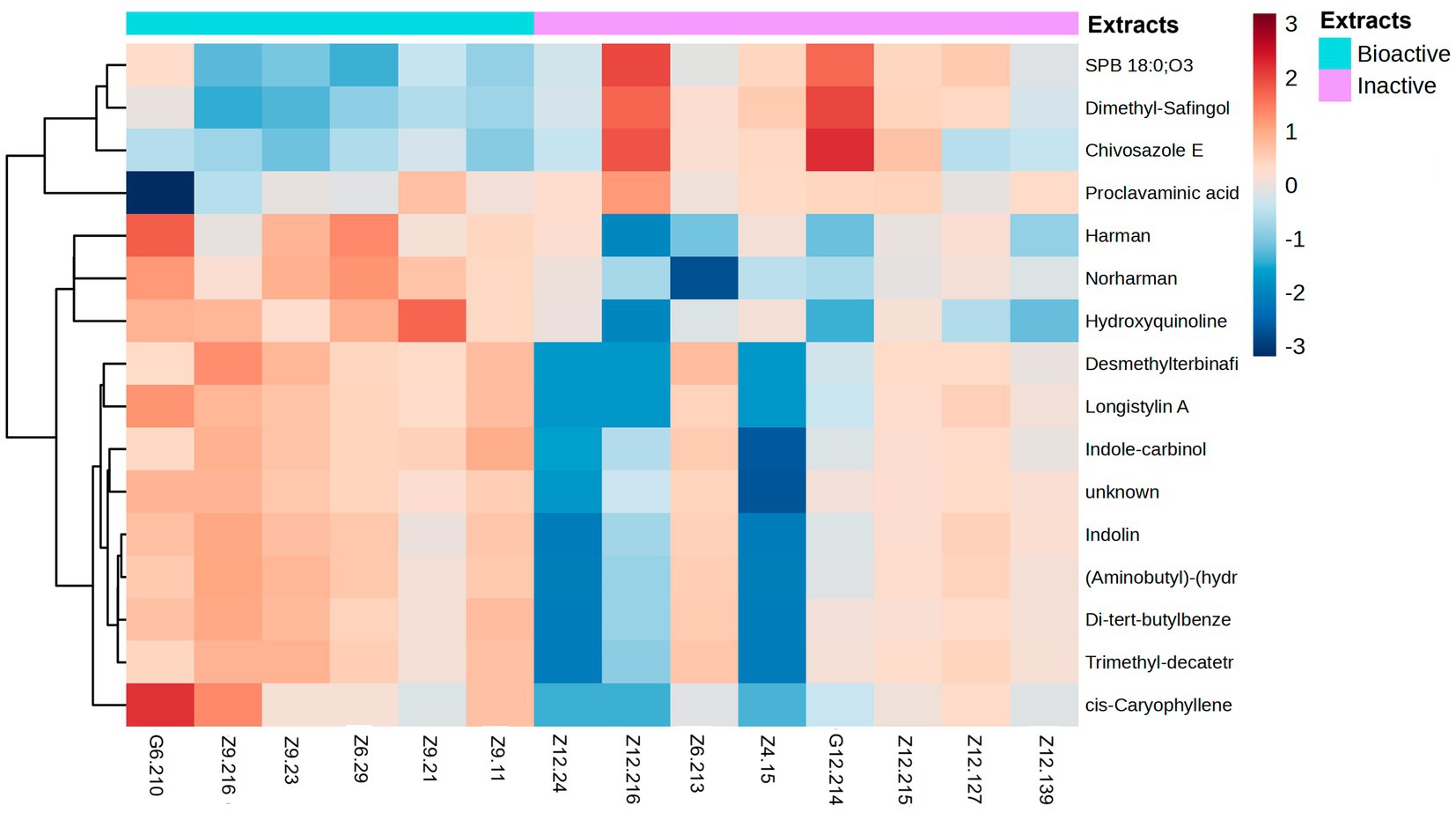
Figure 7. Heatmap of metabolites grouped by fold-change. The heatmap ranges from −3 to 3. Metabolites were identified by selecting the features most responsible for the antibacterial activity using discriminatory analysis (OPLS-DA), which were deemed significant by the linear model (p-value < 0.05). The antibacterial active extracts (denoted as G6.210, Z9.216, Z9.23, Z6.29, Z9.21, and Z9.11) and the non-active extracts (denoted as Z12.24, Z12.216, Z6.213, Z4.15, G12.214, Z12.215, Z12.127, and Z12.139). Hierarchical clustering of the strains was performed using Euclidean distances between the metabolites.
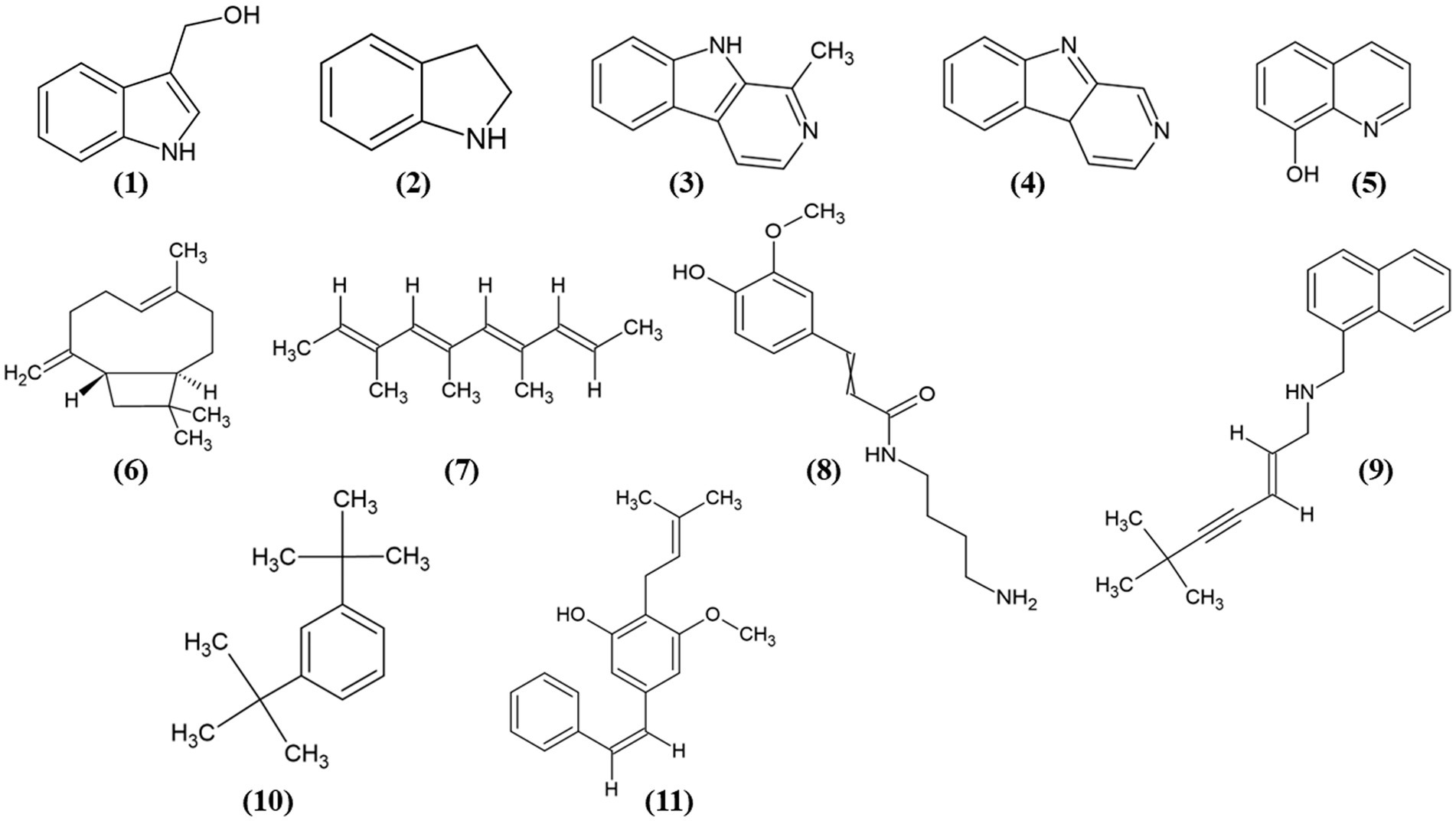
Figure 8. Chemical structures of the identified compounds at different confidence levels. Level 2 compounds (1–4): (1) Indole-3-carbinol, (2) Indoline, (3) Harman, and (4) Norharman. Level 3 compounds (5–11): (5) Hydroxyquinoline, (6) Caryophyllene, (7) Trimethyldecatetraene, (8) (Aminobutyl)-(hydroxy-methoxyphenyl) propenimidic acid, (9) Desmethylterbinafine, (10) Di-tert-butylbenzene, (11) Longistylin A.
Furthermore, we identified other compounds that have been mostly reported in Actinobacteria. Some of these compounds were hydroxyquinoline (5) and caryophyllene (6) (Figure 8). Similarly, we also annotated compounds at level 3 (Figure 8), although they have not been previously reported in actinobacteria, showed a great abundance (Table 2), some of which have been isolated from plants, such as trimethyl-decatetraene (7), a fatty acyl compound, (aminobutyl)-(hydroxy-methoxyphenyl) prop-enimidic acid (8), a compound belonging to the carboxylic acid class, which is a natural product of antimicrobial activity, desmethylterbinafine (9), a naphthalene compound, di-tert-butylbenzene (10), benzene and substituted derivative, and Longistylin A (11), a compound annotated at level 4, but this has been reported to have significant anti-MRSA activity (Wu et al., 2020) belonging to the Stilbenes class of compounds.
The remaining compounds, described in Table 2, were very low in abundance, especially in the inactive samples. Some of these compounds include clavaminic acid, carboxylic acid Chivosazole E, the glycoside compound dimethylsafingol, sphingoid base, SPB 18:0, and O3, a sphingolipid. Moreover, a considerable number of molecules were categorized as ‘unknown.’ However, among these, only one molecule met the criteria for statistical significance and demonstrated a biological relationship consistent with expected antibacterial activity.
4 DiscussionActinobacteria are a relevant source of bioactive compounds primarily in terrestrial ecosystems (Kashfi et al., 2020). However, the repetitive discovery of land-based compounds (Ngamcharungchit et al., 2023; Siddharth and Rai, 2019) has allowed marine actinobacteria to emerge as promising alternative sources, leading to increased interest in marine bioprospecting. Corals, marine sponges, and their associated actinomycetes serve as repositories for novel natural products with diverse and potent biological activities and significant pharmaceutical value (Yang et al., 2015; Balasubramanian et al., 2018).
In our quest for new therapeutic options for acne vulgaris that minimize side effects and address bacterial resistance, we found that six out of 13 isolates studied could inhibit the growth of S. epidermidis, MRSA, and C. acnes. However, these extracts were characterized by a notably low yield, a challenge that has been previously described in similar studies (Lacret et al., 2019; Sujatha et al., 2005). Ethyl acetate was selected as the solvent because of its proven efficacy in isolating bioactive compounds from Actinobacteria, as documented in previous studies (Kurnianto et al., 2021; Sathish Kumar and Kokati Venkata Bhaskara, 2012). Remarkably, the Z9.216 extract displayed significant inhibition of over 95% against MRSA and C. acnes, and over 65% against S. epidermidis at a concentration of 0.003 mg/mL. The antibacterial activity of this extract was comparable to that of the standard antibiotic vancomycin, which was used as a reference, and exhibited 99% inhibition against both MRSA and C. acnes at the same concentration. These results align with those of previous studies, such as Paderog et al. (2020), who reported significant antibacterial activity of marine actinobacterial extracts against MRSA (Paderog et al., 2020).
Additionally, the growth rate assessment revealed no significant differences between the antibiotics and Z9.216 extract for MRSA and C. acnes (p-value < 0.05). Although research on the effects of actinobacterial extracts on C. acnes is limited, our findings are consistent with those of Kim et al. (2023) who demonstrated the antibacterial properties of a marine-derived pigment against C. acnes. The antibiotics currently used for C. acnes treatment, such as clindamycin and erythromycin, are derived from terrestrial actinobacteria, specifically from the Streptomyces genus, or are synthetically derived, such as tetracycline (De La Hoz-Romo et al., 2022). To the best of our knowledge, this study is among the first to demonstrate the therapeutic potential of marine actinobacterial extracts for treating acne vulgaris by targeting C. acnes.
Cytotoxic tests revealed that extract Z9.216 preserved the viability of human keratinocytes and fibroblasts at concentrations at which antibacterial activity was observed. In contrast, the Z9.23 extract caused the highest cytotoxicity at 1.25 mg/mL in HACAT cells, resulting in a viability reduction of up to 40%, a similar result also reported by Dahal et al. (2020).
Streptomyces remains the most prolific genus of actinobacteria (Newaz et al., 2022); however, bioprospecting of rare actinobacterial strains offers a promising source for identifying novel metabolites (Arasu Valan et al., 2016). In our study, four of the six bioactive strains belonged to the rare actinomycete strains Kokuria sp. strain EUFUS-Z9216, Nocardia sp. strain EUFUS-Z923, Rhodococcus sp. strain CLIVUS-Z629, and Micrococcus sp. strain EUFUS-Z911. Recent research has underscored the antibacterial potential of these less-known Actinobacteria genera. For instance, Kocuria species have been reported to produce antimicrobial compounds that are effective against various pathogenic bacteria, including MRSA (Jagannathan et al., 2021; Uzair et al., 2018). Similarly, Nocardiopsis strains have been found to synthesize novel antibiotics with potent activity against gram-positive and gram-negative bacteria (Kamarudheen et al., 2019). The diversity of rare actinobacterial strains examined in this study likely accounts for the range of compounds identified in the extracts.
Metabolomic analyses comparing antibacterial active and non-active extracts revealed diverse alkaloid derivatives as the dominant products, identified at level 2. While these compounds have predominantly been associated with the Streptomyces genus, as evidenced by the bulk of existing literature (Newaz et al., 2022; De Rop et al., 2022; Zhang et al., 2019; Yang et al., 2017; Zhang et al., 2017; Chen et al., 2019), their presence among extracts from other genera suggests a broader biosynthetic potential across various actinobacterial lineages.
In this study, a range of alkaloids were detected, including, hydroxyquinoline, derivatives of indole, Indole-carbinol, Indoline, and β-carboline compounds, such as Harman, and Norharman. These compounds were identified at the highest level and abundance, through metabolomic analysis as indicated by the fold change in Table 2. The indole nucleus, which is prevalent in many of these compounds, is a crucial structural motif in the pursuit of novel drug candidates and has been termed a “privileged structure” (Newaz et al., 2022; de Sa et al., 2009) owing to its versatile interactions with target proteins. Moreover, these compounds exhibit a spectrum of activities, including cytotoxic, antineoplastic, antibacterial, and antifungal (Newaz et al., 2022). In particular, the presence of nitrogen in their molecular architecture appears to play a crucial role in mediating these effects (Netz and Opatz, 2015).
Indole-carbinol, identified in this study, likely contributes to the antibacterial activity of bioactive extracts against MRSA, S. epidermidis, and C. acnes. This naturally occurring alkaloid, commonly found in cruciferous vegetables like broccoli and cauliflower (Netz and Opatz, 2015), is known for its anticancer, anti-inflammatory, and broad-spectrum antibacterial effects, including antibacterial activity against antibiotic-resistant strains (Sung and Lee, 2007; Wu et al., 2019; Wu et al., 2020).
Harman and Norharman, both β-carboline alkaloids, are commonly found in nature from a variety of sources, including plants, insects, and marine organisms (Netz and Opatz, 2015; Suzuki et al., 2018). Harman, known for its antibiotic potential, has previously been identified in marine invertebrates, notably within the tunicate-associated bacterium Enterococcus faecium (Aassila et al., 2003). This aligns with our findings as it supports the presence and potential bioactivity of similar compounds in marine actinobacterial extracts. Similarly, Norharman, an antimicrobial compound isolated from Pseudoalteromonas piscicida associated with the sponge Hymeniacidon perleve, was previously identified by Zheng et al. (2005) as a major antimicrobial agent against Bacillus subtilis, S. aureus, and Escherichia coli (Blockley et al., 2017). The β-carboline structure, common to both compounds, has been associated with the efficacy against certain antibiotic-resistant strains, suggesting its potential as a basis for novel antimicrobial agents (Suzuki et al., 2018). Additionally, the broad-spectrum antimicrobial activity of quaternary ammonium compounds against both gram-positive and gram-negative bacteria is noteworthy. These agents disrupt bacterial cell membranes by interacting with their negative charges, leading to the release of K+ ions and cytoplasmic content, ultimately causing bacterial cell death (Suzuki et al., 2018). Collectively, these insights help to elucidate our results and support the hypothesis that the observed antibacterial activity in actinobacterial extracts may be due to compounds with similar structures and mechanisms.
Among the identified metabolites, hydroxyquinoline (HQ), a quinoline-class alkaloid, demonstrated a statistically significant presence despite its low abundance in metabolomic analysis (Table 2). HQ is well known for its antimicrobial and anticancer activities, and has been frequently reported in actinobacteria, particularly within the Streptomyces genus (De Rop et al., 2022; Balthazar et al., 2022). The diverse biological properties of HQ and i
留言 (0)