Excessive glucose ingested by organisms is stored in the form of inactive polymers, with glycogen serving as such a polymer in bacteria (Wilson et al., 2010; Montero et al., 2011; Liu et al., 2021; Neoh et al., 2024). Glycogen is a polysaccharide with a highly branched structure, primarily composed of glucose residues linked by α-1,4-glycosidic bonds and α-1,6-glycosidic bonds (Li et al., 2023; Neoh et al., 2024). In both prokaryotes and eukaryotes, glycogen can be categorized into three classes based on diameter: protein-rich γ particles less than 3 nm, spherical β particles approximately 20 nm in size, and larger rose-like α particles (Li and Hu, 2020; Liu et al., 2021). There are differences in the utilization and degradation rates among glycogen particles of different levels (Besford et al., 2016). Currently, the process of glycogen synthesis in eukaryotes has been well studied, whereas some synthesis steps of glycogen in prokaryotes remain to be elucidated (Cifuente et al., 2019; Liu et al., 2021; Jenkins et al., 2023).
Glycogen primarily serves as a major energy reserve in prokaryotes, and current research indicates that it is closely associated with the organism’s tolerance to extreme environments, its ability to colonize hosts, and its virulence (Wang et al., 2015; Kwak et al., 2016). The synthesis and metabolism of glycogen in prokaryotes constitute a complex pathway involving a variety of genes responsible for synthesis, metabolism, and regulation (Wilson et al., 2010; Neoh et al., 2024). Among these, five key enzymes have been identified: glucose-1-phosphate adenylyltransferase (AGPase), glycogen phosphorylase (GP), glycogen synthase (GS), glycogen branching enzyme (GBE), and glycogen debranching enzyme (GDE), with both GBE and GDE belonging to the glycoside hydrolase family 13 (GH13) (Seibold et al., 2011; Machtey et al., 2012; Jenkins et al., 2023). In Escherichia coli, these enzymes are encoded by the genes glgB (GBE), glgX (GDE), glgC (AGPase), glgA (GS), and glgP (GP), respectively, and they are organized within an operon named glgBXCAP, collaboratively participating in the synthesis and metabolic processes of glycogen in the bacteria (Wang et al., 2020). The genes glgP and glgX are crucial for the formation of glycogen structure; the absence of glgP and glgX affects the morphology, structural stability, and chain length of glycogen (Yu et al., 1988; Li et al., 2023). Concurrently, the deletion of the two glycogen-degrading enzymes, glgP and glgX, leads to the continuous accumulation of glycogen within the bacterial cell (Wang et al., 2020). Conversely, the deletion of the three glycogen-synthesizing genes, glgB, glgC, and glgA, inhibits the synthesis of glycogen (Wang et al., 2015; Kwak et al., 2016).
Klebsiella pneumoniae, a member of the Enterobacteriaceae family, is a Gram-negative bacterium that colonizes the intestinal tract of the host and can cause life-threatening diseases under certain conditions (Tacconelli et al., 2018; Wyres and Holt, 2022). Consequently, K. pneumoniae, along with other strains such as Acinetobacter baumannii, is included in the World Health Organization’s priority list for the development of new antibiotics (Tacconelli et al., 2018). The virulence of K. pneumoniae is linked to its survival capabilities in extreme conditions, particularly its ability to form biofilms, which can assist the bacterium in colonizing the surfaces of cells and enhances the bacterium’s resistance to antibiotics and its virulence (Sauer et al., 2022; Li and Ni, 2023).
Studies have shown that the development of inhibitors targeting the GlgB protein can effectively suppress the virulence and pathogenicity of Mycobacterium tuberculosis (Dkhar et al., 2015). Additionally, the deletion of glgB and glgX in E. coli results in an increase in biofilm biomass and a reduction in the strain’s viability under extreme conditions, indicating their significant roles in bacterial virulence and survival in harsh environments (Wang et al., 2015, 2020). However, despite ranking as the third most prevalent pathogen responsible for antimicrobial resistance associated deaths, K. pneumoniae has not been extensively studied, particularly in terms of the function of glycogen in the formation of its biofilm and the development of its virulence (Murray et al., 2022). Here, we investigated the GH13 family protein GlgB and GlgX in K. pneumoniae, which are annotated in the UniProt database as glycogen branching enzyme and glycogen debranching enzyme, respectively. Our findings indicate that these enzymes are capable of degrading glycogen. Following this, we conducted a deletion of the glgB and glgX genes in K. pneumoniae to assess the impact on bacterial growth, glycogen accumulation, biofilm formation, and virulence attributes.
2 Methods2.1 Strains and growth conditionsThe strains, plasmids and oligonucleotide primers used in this study are shown in Supplementary Table S1. All strains except those containing the pCaskp plasmid were grown in liquid (shaking at 220 rpm) or solid LB medium at 37°C in incubator or shaker (Zhichu).
2.2 Cloning, expression, and purification of proteinThe DNA sequences for glgB and glgX from K. pneumoniae MGH78578 (GCF_000016305.1) were retrieved from the GeneBank database respectively. Utilizing the genomic DNA of K. pneumoniae MGH78578 as a template, the glgB and glgX genes were amplified via PCR using the PrimerStar DNA Polymerase (Takara). Subsequently, the amplified glgB and glgX sequences were cloned into the pET-28a vector through the In-Fusion Snap Assembly Master Mix (Takara).
E.coli BL21 (DE3) cells (Transgen Biotech) containing pET28a-glgB or pET28a-glgX vector were inoculated into LB medium with kanamycin and incubated at 37°C until OD600 was 0.8, and then incubated at 16°C for 20 h with isopropyl β-D-1-thiogalactoside of 0.5 mM. Bacterial cells were resuspended with binding buffer (50 mM Tris, 300 mM NaCl, pH 7.5) and lysed by FastPrep-24™ 5G homogenizer (MP Biomedicals). Recombinant proteins were enriched by Ni-NTA resins and eluted with elution buffer (50 mM Tris, 500 mM Imidazole, 300 mM NaCl, pH 7.5). Fractions were concentrated by an Amicon Ultra-15 centrifugal filter (Millipore) with a molecular weight cutoff of 10 kDa.
2.3 Activity and properties of enzymesThe assessment of enzyme activity was modified based on previous study (Schumacher et al., 2022). Briefly, the enzyme activity is assessed by measuring the increase in reducing sugars in the reaction solution. After purification, the enzyme is mixed with phosphate buffer and a final concentration of 0.05% glycogen solution, and incubated at 37°C for one hour. Following incubation, an equal volume of bicinchoninic acid (BCA) reagent is added to the reaction mixture and vortexed for 60 seconds on a vortex mixer (IKA), followed by incubation at 80°C for 30 minutes. After the reaction solution has cooled to room temperature, 125 µL of the solution is transferred to a 96-well plate for the measurement of absorbance at 562 nm. D-glucose is used to create the standard curve, and the absorbance values at 562 nm are converted to reducing ends [µM glucose equivalents]. All measurements are performed in triplicate.
To determine the optimal temperature, the enzyme is placed in phosphate buffer at pH 7.5, and its activity is measured at different temperatures in increments of 5°C. For the determination of the optimal pH, the phosphate buffer is replaced with sodium dihydrogen phosphate (100 mM, pH 6.0-7.5) and tris(hydroxymethyl)aminomethane (50 mM, pH 8.5-9.0) buffer solutions, respectively, and the enzyme activity is measured at the optimal temperature. Regarding temperature stability, the enzyme is incubated at the optimal temperature and pH for durations ranging from 4 to 28 hours, after which the activity is assessed.
2.4 Deletion mutantsA dual-plasmid CRISPR/Cas9 system was employed to delete the glgB and glgX genes in K. pneumoniae MGH78578 (Wang et al., 2018). The kanamycin resistance gene in the pSGKP plasmid was replaced with a hygromycin B resistance gene to circumvent the kanamycin resistance of K. pneumoniae MGH78578. The pCasKP plasmid was first electroporated into K. pneumoniae MGH78578 and the correct transformants were selected on LB plates containing apramycin. Subsequently, the pSGKP plasmid carrying spacer sequences along with ssDNA or dsDNA homology arms was co-transformed into K. pneumoniae MGH78578 containing the pCasKP plasmid, which had been induced with L-arabinose. The transformants were then screened on plates containing both apramycin and hygromycin B to further validate the deletion of the glgB or glgX genes. Strains with successful deletion of glgB or glgX were cultured overnight at 37°C on plates containing 5% sucrose to cure the plasmids. Strains that lost the pSGKP plasmid alone were cultured on plates with 5% sucrose at 30°C for subsequent gene deletion, and the strain with the simultaneous deletion of glgB and glgX was named ΔglgBX.
2.5 Growth curvesTo determine the growth rate of the strains, they were cultured overnight in LB medium and then inoculated at a 1% dilution into M63+ medium. The Microscreen HT growth curve analyzer (Jieling Instruments) was used for the measurement of growth curves. The culture medium was added to the 48-well deep-well plate in 1 mL per well and placed in the instrument for incubation at 37°C and 800 rpm. The growth was measured every 20 minutes for a continuous period of 24 hours. All reactions were performed in triplicate.
2.6 Glycogen/protein ratio assayThe M9 minimal medium was employed for the cultivation of various K. pneumoniae. After the bacterial cells were harvested by centrifugation, the glycogen/protein ratio was determined using the method described in previously study (Li et al., 2023). Bacteria cells were lysed by FastPrep-24™ 5G homogenizer (MP Biomedicals). After centrifugation, the supernatant was reacted with 60% KOH at 95°C for 2 hours. After treatment, the supernatant was divided into two equal parts, one of which was used to determine protein concentration using the BCA method, while the other was used for the detection of glycogen content. For the determination of glycogen content, pre-cooled ethanol was added to the samples, and the mixture was thoroughly vortexed to facilitate glycogen precipitation. The mixture was then placed at -20°C for 20 hours. The precipitates containing glycogen granules were collected by centrifugation at 4°C and then treated with ethyl alcohol to wash away impurities, with the washing procedure repeated three times. Thereafter, the supernatant was discarded, and the glycogen granules were placed at 60°C to dry any remaining moisture. All glycogen granules were then resuspended in a sodium acetate solution (50 mM, pH 4.8, containing 0.3 mg/mL amyloglucosidase) and incubated overnight at 37°C to completely convert the glycogen granules into glucose. The glucose content in the reaction system was measured using a glucose detection kit (Solarbio) to estimate the glycogen content and calculate the glycogen/protein ratio.
2.7 Biofilm formation ability assayThe measurement of biofilm biomass was performed using a modified method described in previous study (Haney et al., 2021). K. pneumoniae MGH78578 and three deletion mutants were incubated overnight and then diluted to 1% in M63+ medium. The diluted bacterial solution was incubated with 96-well plate at 37°C for 24h. After incubation, the 96-well plate was washed twice with deionized water to remove floating bacteria and medium. The biofilm of each well was stained with 200 μL of 0.1% (w/v) crystal violet for 15 min, and washed twice with deionized water. It was dissolved with 150 μL of 33% (v/v) glacial acetic acid for 15 min, and the absorbance at 600 nm was measured using BioTek 800 TS microplate detector (Agilent).
2.8 Confocal laser scanning microscopyThe bacterial cells were stained with SYTO-9 prior to observation with the confocal laser scanning microscope (CLSM), as described in previous studies with some modifications (Qiao et al., 2022). K. pneumoniae MGH78578 and deletion mutant strains were incubated overnight and then diluted to 1% in M63+ medium. The diluted bacterial solution was placed in Lab-Tek™ Chamber Slide (Thermo Scientific) at a volume of 400 µL per well and incubated statically at 37°C for 12 hours. After incubation, the medium was removed, and ddH2O was used to wash away the planktonic bacteria, with the washing procedure being repeated once. SYTO-9 dye was then added to wells and incubated in the dark for 20 minutes. After discarding the dye, the wells were washed to remove any residual dye. A Zeiss LSM 710 Confocal Laser Scanning Microscope (CLSM) was utilized to observe the stained biofilms. Subsequent to acquisition, the images were processed with ZEN 2009 software and subjected to analysis with COMSTAT 2.1 software for the quantification of biofilm biomass.
2.9 Determination of virulence of strainsThe virulence of K. pneumoniae is assessed using Galleria mellonella larvae model (Schaefer et al., 2024). G. mellonella larvae were randomly divided into four groups, with ten in each group. Three groups were injected with 5 µL of wild type, ΔglgB and ΔglgX mutants, and the control group was injected with 5 µL of PBS solution. Larvae were incubated in an incubator at 37°C and 60% humidity. The survival of the larvae was observed from day 0 to day 4, with larvae that were unable to right themselves being considered deceased.
2.10 Statistical analysisStatistical analyses were conducted utilizing GraphPad Prism software, wherein a one-way ANOVA was employed to assess the significance of differences. Each experimental was conducted with a minimum of three replicates to ensure reliability of the results.
3 Results3.1 GlgB and GlgX catalyze the degradation of glycogenGlgX and GlgB both belong to the GH13 family, which contains an α-amylase domain, and they can catalyze the degradation of glycosidic bonds. The GlgX and GlgB proteins were expressed in E. coli and purified with Ni-NTA resin to obtain soluble proteins (Figure 1A). The degradation activities of GlgX and GlgB against glycogen, pullulan, amylose, and amylopectin were measured using the BCA method. We observed that GlgB catalyzed the hydrolysis of all substrates to a nearly equivalent extent (Figure 1B). In contrast, GlgX exhibits a pronounced substrate specificity, with the highest degradation activity towards glycogen and pullulan, while its activity towards amylose and amylopectin is significantly reduced (Figure 1C). Previous studies have indicated that Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) enhances the hydrolytic activity of GlgX from E. coli towards glycogen (Schumacher et al., 2022). However, the addition of c-di-GMP at a final concentration of 50 μM did not increase the enzyme activity (Figure 1D). Subsequently, the enzymatic properties were characterized. Notably, both enzymes showed activity in a broad optimal temperature range, with peak activity observed between 35-40°C (Figure 2A). The optimal pH for GlgB was found to be 7.5, whereas for GlgX it was 7.0 (Figure 2B). When pH exceeding 7.5, the activity of both enzymes was markedly reduced (Figure 2B). To evaluate the stability of the enzymes, they were incubated at 37°C for 28 hours. A significant decrease in activity was noted after 4 hours of incubation, followed by a gradual decline in activity thereafter (Figure 2C). Two proteins have been shown to possess glycogenolytic activity in vitro, prompting us to further examine their activity in vivo.
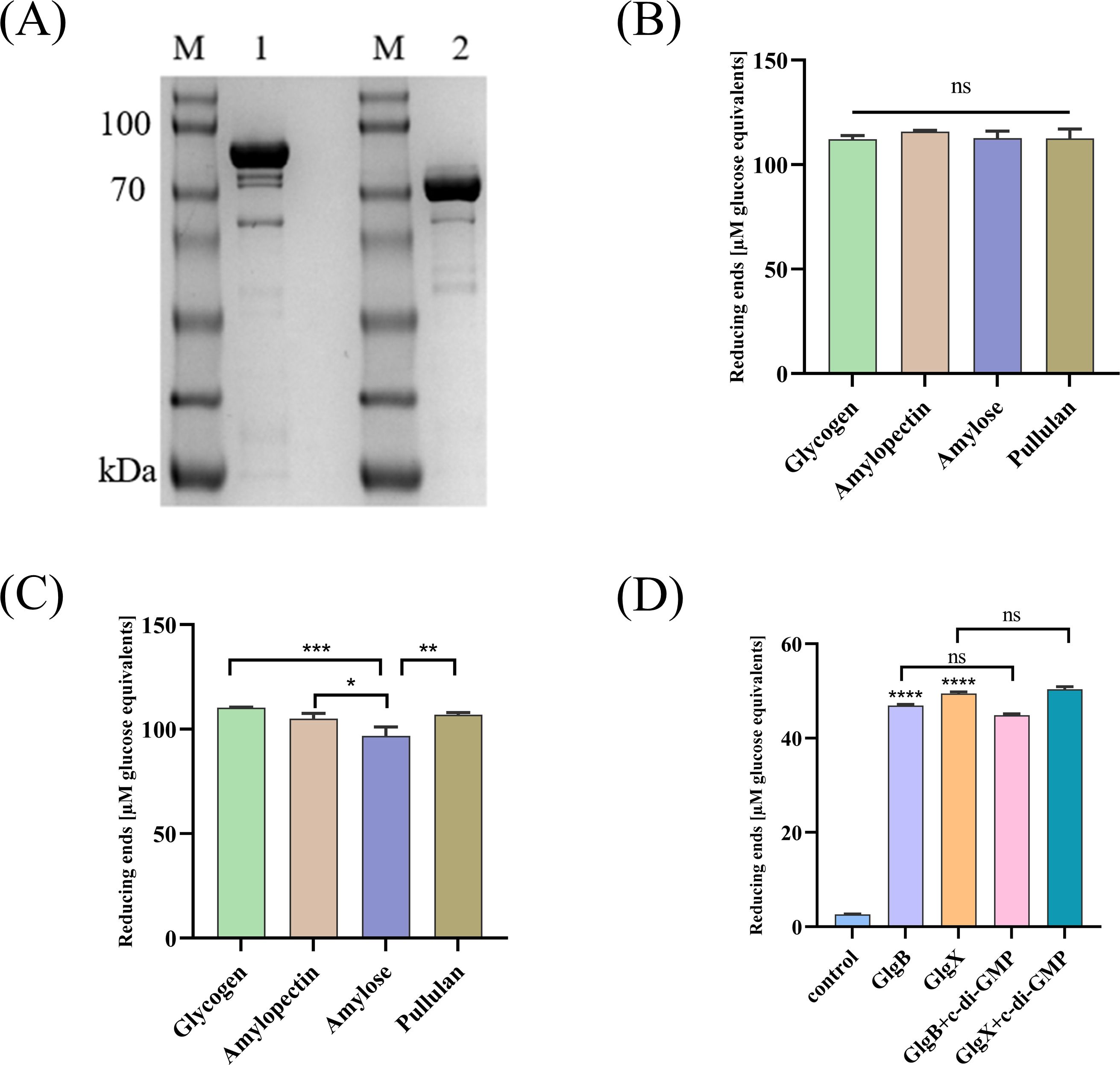
Figure 1. The activity of GlgB and GlgX. (A) Expression of GlgB and GlgX proteins in E coli. M-Protein marker, 1- GlgB protein, 2- GlgX protein. (B) The enzyme activity of GlgB towards different substrates. (C) The enzyme activity of GlgX towards different substrates. (D) Protein activity is not regulated by c-di-GMP. Error bars represent ± S.D of the mean. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns indicates no statistical significance.
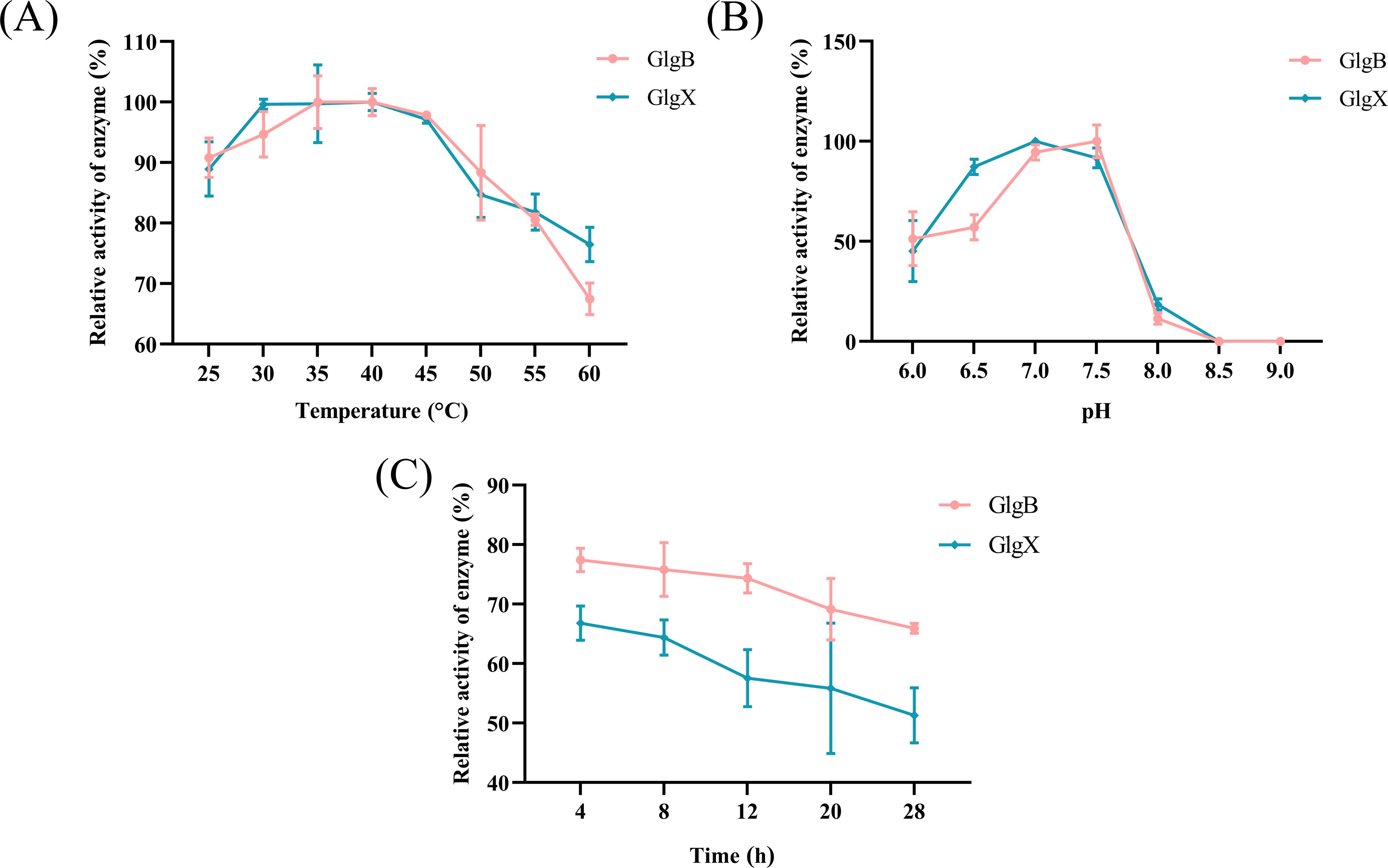
Figure 2. The enzyme properties of GlgX and GlgB. (A) Enzyme activity measured at different temperatures with the maximum activity group as 100%. (B) Enzyme activity measured at different pH values with the maximum activity group as 100%. (C) The residual activity of the enzyme after incubation from 4 to 28h using the enzyme activity at 0 h as 100%. Error bars represent ± S.D of the mean.
3.2 The glgB and glgX genes alter glycogen content and affect the growth rate of the bacterium.GlgB and GlgX are annotated as glycogen branching enzyme and debranching enzyme, respectively, in the UNIPROT database. It has been evidenced that these enzymes play a crucial role in bacteria, such as bacterial growth and the synthesis of glycogen (Wang et al., 2020). To investigate the role of protein in K.pneumoniae, we deleted the active center of GlgX and GlgB, resulting in the ΔglgX, ΔglgB, and ΔglgBX mutants (Supplementary Figure S1). Analysis of growth curves revealed that the ΔglgX mutant exhibited an accelerated growth rate, while the ΔglgB mutants and the ΔglgBX mutant displayed a slower growth pattern compared to the wild-type strain (Figure 3A). As glycogen serves as a crucial energy reserve for bacterial growth, the growth rate of a strain may be influenced by its glycogen content. Therefore, we quantified glycogen levels at different times throughout the bacterial growth cycle. The glycogen content in both the wild-type and ΔglgX mutants reached a peak at 8 hours post-inoculation and subsequently decreased, with the ΔglgX mutant maintaining a slightly higher glycogen level than the wild type (Figure 3B). The glycogen content in the ΔglgB and ΔglgBX mutants was comparable and remained relatively stable during the growth phase, consistently lower than that of the wild type (Figure 3B). The correlation between glycogen content and growth rate among the strains suggests that the amount of glycogen present impacts the growth rate of the bacterial strains. Additionally, since the glgB gene is located upstream of the glgX gene, which leads to the phenotypic similarity between ΔglgB and ΔglgBX mutants, future research will be directed exclusively towards investigating the ΔglgB and ΔglgX mutant.
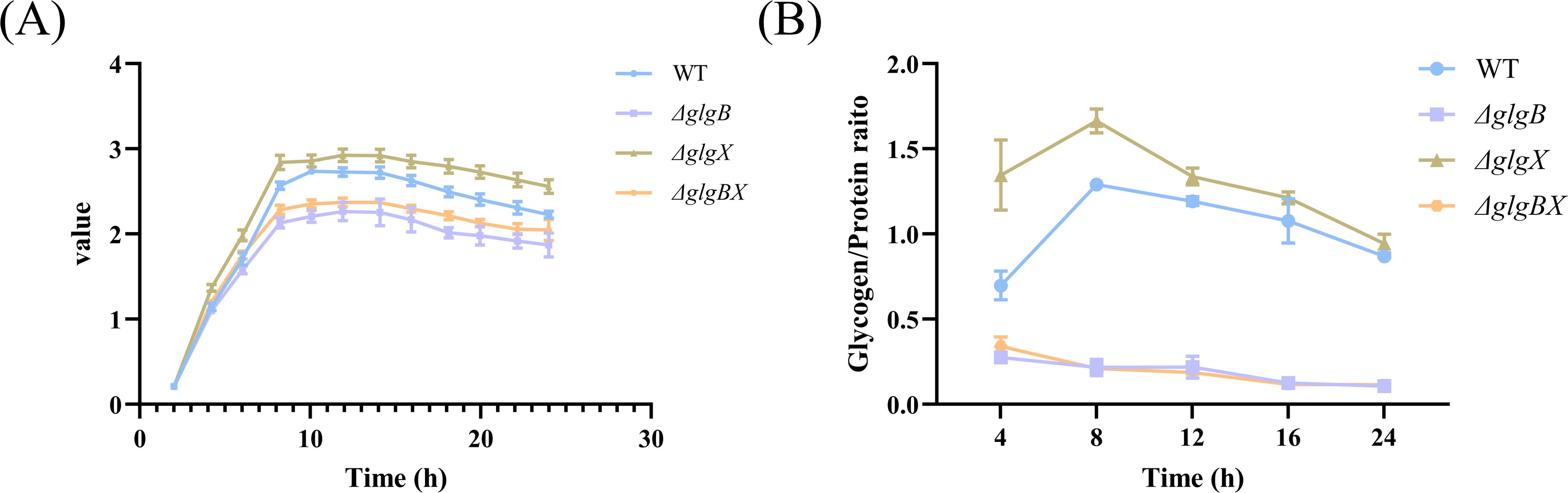
Figure 3. Changes in growth rate and glycogen content of wild-type K. pneumoniae and mutant strains. (A) Growth curves of WT and deletion mutations. (B) The glycogen content of WT and deletion mutations. Error bars represent ± S.D of the mean.
3.3 Only the glgB gene influences biofilm formation of K. pneumoniaePrevious study has indicated that bacterial glycogen can influence biofilm formulation (Wang et al., 2020). To quantify the impact of glycogen on biofilm formation, we utilized crystal violet staining to assess biofilm biomass. The data revealed a reduction in biofilm biomass following the deletion of the glgB gene, while the deletion of the glgX gene did not alter biofilm biomass (Figure 4A). To further investigate the effects of glycogen metabolism on biofilm formation, we employed CLSM and utilized SYTO9 stain the bacterial cells, respectively. In the corresponding images, both ΔglgX and ΔglgB mutants showed a decrease in bacterial biomass compared to the wild type, with the ΔglgB mutant showing a greater decrease in biomass than the ΔglgX mutant (Figure 4B). Subsequently, the COMSTAT software was employed to quantify the bacterial biomass within the images and to assess the significance of the biomass reduction. We found that the biomass of the ΔglgB mutant was significantly lower than that of the wild type, whereas the decrease in biomass of the ΔglgX mutant was not significant, indicating that the deletion of glgB more profoundly affects the biofilm formation capacity of K. pneumoniae (Figure 4C). This indicates that the deletion of glgB reduces the biofilm-forming ability of K. pneumoniae.
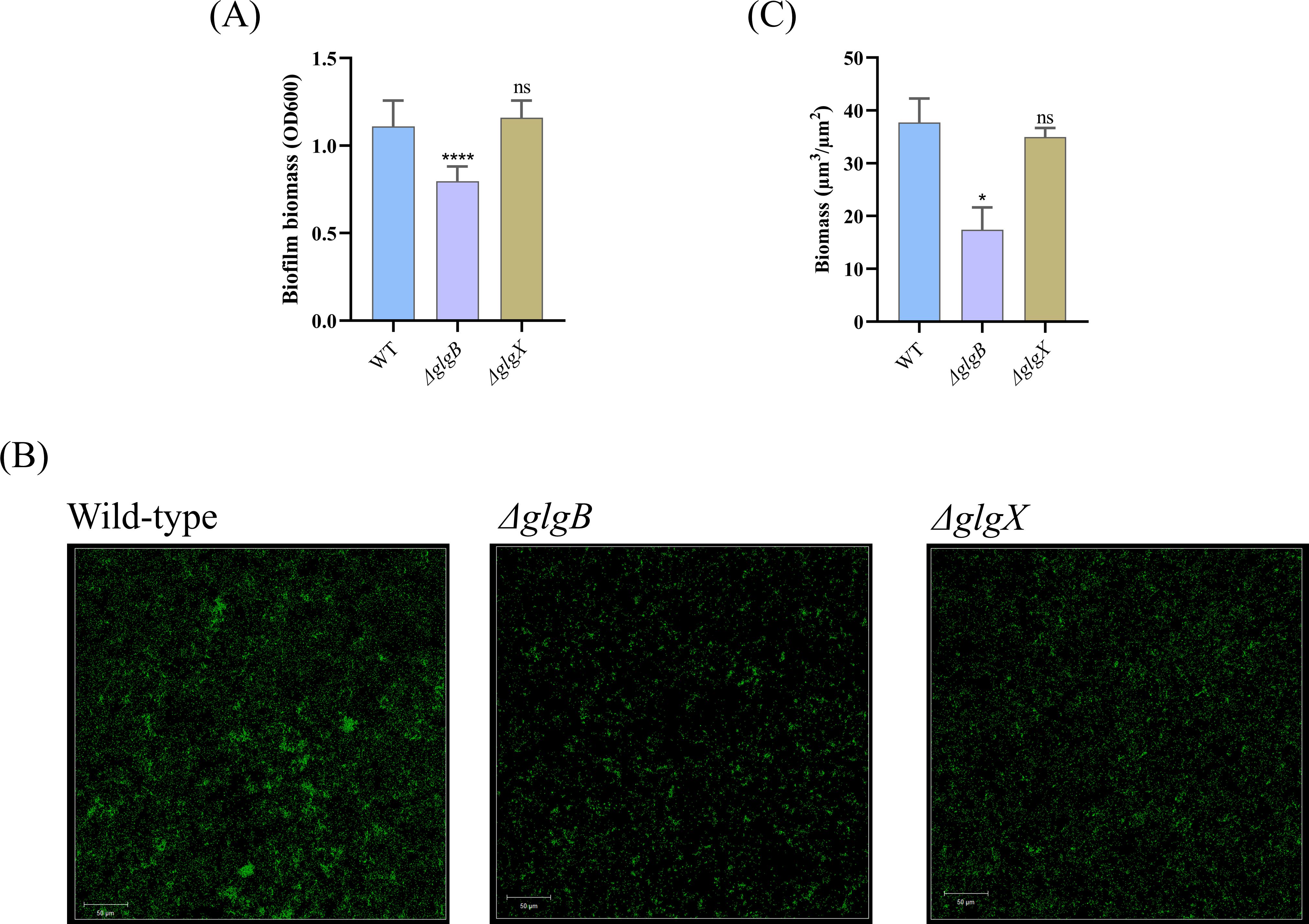
Figure 4. Biofilm biomass of wild-type K. pneumoniae and mutant strains. (A) Determination of biofilm biomass in WT and deletion mutations. (B) Representative confocal images of K. pneumoniae and deletion mutants. Scale bar, 50 μm. (C) The biofilm biomass of confocal images quantified by COMSTAT 2.1 software. Error bars represent ± S.D of the mean. *p<0.05, ****p<0.0001, ns indicates no statistical significance.
3.4 The glgB and glgX genes can influence the virulence of K. pneumoniaeThe deletion of glgB and glgX genes in bacteria resulted in alterations to the growth rate and biofilm biomass compared to the wild type, leading us to hypothesize that these changes might also affect the virulence of the bacteria. Given that the G. mellonella larval model has been widely used in numerous studies to assess bacterial virulence and the efficacy of antimicrobial agents, we adopted this model to assess the virulence profiles of distinct K. pneumoniae isolates (Jin et al., 2024; Firacative et al., 2020). Following the inoculation of larvae with 1×106 CFU of the respective K. pneumoniae strains, a marked difference in survival rates was observed. Larvae injected with the ΔglgB mutant strain exhibited a consistent survival rate of 90% from the initial to the fourth day post-inoculation (Figure 5). In contrast, the survival rate of larvae exposed to the wild-type strain progressively declined from 80% to 60% over the same period, signifying a reduced virulence in the ΔglgB mutant strain compared to the wild-type (Figure 5). Notably, the survival rate of larvae inoculated with the ΔglgX mutant strain decreased from 60% to 50% between the initial day and the fourth day following inoculation, implying a virulence slightly higher than the wild-type strain (Figure 5). Considering that the biofilm formation capacity of the ΔglgB strain is significantly reduced compared to the wild-type strain, this suggests that the absence of glycogen in bacteria leads to a decrease in key factors such as biofilm formation, which in turn diminishes the virulence of the organism.
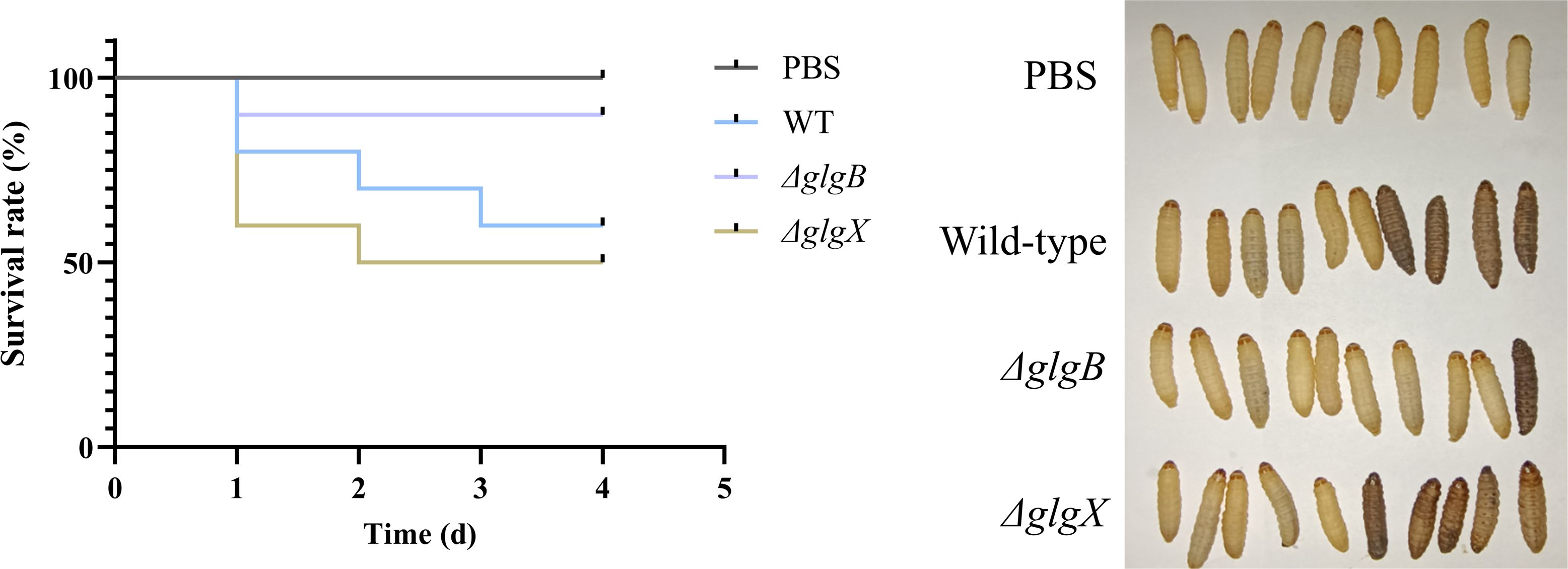
Figure 5. Survival rate of G. mellonella larvae infected with wild-type K. pneumoniae or mutant strains, PBS group as control.
4 DiscussionAccording to the InterPro database, GlgX and GlgB can catalyze the degradation of α-1,6 glycosidic bonds and α-1,4 glycosidic bonds, respectively. To date, GlgX has been reported to degrade various substrates, including glycogen and pullulan (Schumacher et al., 2022). However, the degradation activity of GlgB towards different substrates still requires investigation. After characterizing the GlgB and GlgX proteins from K. pneumoniae, we found that the GlgX protein exhibited the expected significant activity as expected. Concurrently, the GlgB protein also demonstrated considerable activity against various substrates, indicating that GlgB is capable of degrading α-1,4 glycosidic bonds in different substrates and producing glucose residues.
Previous studies have demonstrated that GlgX derived from E. coli and Streptomyces exhibits a significant enhancement in degradation activity towards various substrates in the presence of c-di-GMP (Schumacher et al., 2022; Gallagher et al., 2024). However, we did not observe the promotional effect of c-di-GMP on the activity of GlgX derived from K. pneumoniae. Considering that GlgX proteins with different structures have been shown to exhibit significant differences in activity, this suggests that the promotional effect of c-di-GMP on enzyme activity is limited to specific strains (Song et al., 2010). There are differences in the degradation capabilities of GlgX from various sources towards different polysaccharide substrates. The substrate specificity of GlgX derived from K. pneumoniae is similar to that of GlgX from E. coli, whereas differ with GlgX derived from Vibrio cholerae (Schumacher et al., 2022; Han et al., 2022).
In the synthesis and metabolism of glycogen in prokaryotes, the glycogen branching enzyme GlgB forms branches in the glycogen polymer by linking glucose residues through α-1,6-glycosidic bonds, forming a glucan branch, while the GlgX protein catalyzes the hydrolysis of glucose residues in the α-1,6-glycosidic bonds (Rashid et al., 2016; Strydom et al., 2017; Neoh et al., 2024). The deletion of glgB in E. coli impedes glycogen synthesis, while the deletion of glgX leads to excessive glycogen accumulation, which indirectly affects the growth rate of E. coli and its tolerance to extreme environments (Wang et al., 2020). Studies have demonstrated that excessive glycogen accumulation can enhance the growth rate of bacterial cells (Wang et al., 2020; Okabe et al., 2021). Conversely, the deficiency of glycogen results in a reduced growth rate of the bacterial cells (Lee et al., 2021). The deletion of glgB and glgX genes in K. pneumoniae corresponds to the defects observed in E. coli, leading to deficiencies in glycogen synthesis and degradation in K. pneumoniae, which in turn affects the growth rate of the strain. This evidence confirms that glycogen accumulation is essential for the growth and metabolic processes of K. pneumoniae.
The formation of biofilms is a strategy used by specific pathogens to withstand environmental stresses and resist antibiotics (Rumbaugh and Sauer, 2020; Akbarian et al., 2022; Lin et al., 2022). Mature biofilms provide pathogens with moisture and nutrients, impede the penetration of antibiotics, and facilitate immune evasion by bacteria (Rumbaugh and Sauer, 2020; Vandana and Das, 2022). The formation of biofilms is regulated by numerous factors, including the intake of nutrients, environmental conditions, and specific signaling molecules (Flemming et al., 2023). Glucose has been identified as an influence on the biofilm formation of Staphylococcus epidermidis, while the impact of glycogen on bacterial biofilm formation remains inconclusive (Lerner et al., 2009; Ahmad et al., 2022). It has been reported that the deletion of both glgX and glgB genes enhances the biofilm formation in E. coli, whereas studies have also indicated that inhibiting glycogen synthesis can decrease biofilm formation in Salmonella enteritidis (Bonafonte et al., 2000; Wang et al., 2020). The glycogen synthesis defect caused by the deletion of glgB does indeed inhibit the formation of biofilms in K. pneumoniae. However, the increase in biofilm biomass observed in the ΔglgX mutant of E. coli was not occur in K. pneumoniae, which may be due to differences in the complexity of biofilms between different bacteria (Wang et al., 2020).
The glycogen content in bacterial cells has also been demonstrated to be closely associated with their virulence (Tan et al., 2022; Miao et al., 2023). Multiple studies have shown that inhibiting the function of glgB can effectively reduce the virulence of M. tuberculosis, whereas the deletion of glgX has been associated with an increase in the virulence of V. cholerae (Dkhar et al., 2015; Han et al., 2022). In our evaluation of the virulence of K. pneumoniae using the G. mellonella larval model, we demonstrated that the deletion of glgB reduces the virulence of the bacterium, indicating a positive correlation between glycogen content and the virulence of pathogenic bacteria.
Glycogen has been demonstrated to be closely associated with the virulence of pathogens, and inhibiting glycogen synthesis can effectively reduce the virulence of certain pathogenic bacteria. Our research indicates that the GlgX protein derived from K. pneumoniae is capable of efficiently degrading glycogen. Notably, the GlgB protein also exhibits activity against glycogen substrates, highlighting its potential application prospects. Additionally, this study constructed ΔglgX and ΔglgB mutants of K. pneumoniae, indicating that the deletion of glgB and glgX genes alters the bacterial growth rate by participating in glycogen synthesis and metabolism, thereby affecting the biofilm biomass and the virulence of the K. pneumoniae. Our research reveals the significant role of glycogen synthesis and metabolism in the formation of K. pneumoniae biofilms and its virulence, providing a strategy for addressing severe infections caused by this pathogen.
Data availability statementThe raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statementThe manuscript presents research on animals that do not require ethical approval for their study.
Author contributionsXL: Data curation, Formal analysis, Investigation, Writing – original draft. JL: Formal analysis, Investigation, Methodology, Writing – original draft. RW: Supervision, Writing – review & editing. LB: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-029 & 2017-I2M-1-012) and National Natural Science Foundation of China (82173721).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1507332/full#supplementary-material
ReferencesAhmad, S., Rahman, H., Qasim, M., Nawab, J., Alzahrani, K. J., Alsharif, K. F., et al. (2022). Staphylococcus epidermidis Pathogenesis: Interplay of icaADBC Operon and MSCRAMMs in Biofilm Formation of Isolates from Pediatric Bacteremia in Peshawar, Pakistan. Medicina (Kaunas). 58, 1510. doi: 10.3390/medicina58111510
PubMed Abstract | Crossref Full Text | Google Scholar
Akbarian, M., Chen, S. H., Kianpour, M., Farjadian, F., Tayebi, L., Uversky, V. N. (2022). A review on biofilms and the currently available antibiofilm approaches: Matrix-destabilizing hydrolases and anti-bacterial peptides as promising candidates for the food industries. Int. J. Biol. Macromol. 219, 1163–1179. doi: 10.1016/j.ijbiomac.2022.08.192
PubMed Abstract | Crossref Full Text | Google Scholar
Besford, Q. A., Zeng, X.-Y., Ye, J.-M., Gray-Weale, A. (2016). Liver glycogen in type 2 diabetic mice is randomly branched as enlarged aggregates with blunted glucose release. Glycoconj. J. 33, 41–51. doi: 10.1007/s10719-015-9631-5
PubMed Abstract | Crossref Full Text | Google Scholar
Bonafonte, M. A., Solano, C., Sesma, B., Alvarez, M., Montuenga, L., García-Ros, D., et al. (2000). The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191, 31–36. doi: 10.1111/j.1574-6968.2000.tb09315.x
PubMed Abstract | Crossref Full Text | Google Scholar
Cifuente, J. O., Comino, N., Trastoy, B., D’Angelo, C., Guerin, M. E. (2019). Structural basis of glycogen metabolism in bacteria. Biochem. J. 476, 2059–2092. doi: 10.1042/BCJ20170558
PubMed Abstract | Crossref Full Text | Google Scholar
Dkhar, H. K., Gopalsamy, A., Loharch, S., Kaur, A., Bhutani, I., Saminathan, K., et al. (2015). Discovery of Mycobacterium tuberculosis α-1,4-glucan branching enzyme (GlgB) inhibitors by structure- and ligand-based virtual screening. J. Biol. Chem. 290, 76–89. doi: 10.1074/jbc.M114.589200
PubMed Abstract | Crossref Full Text | Google Scholar
Firacative, C., Khan, A., Duan, S., Ferreira-Paim, K., Leemon, D., Meyer, W. (2020). Rearing and Maintenance of Galleria mellonella and Its Application to Study Fungal Virulence. J Fungi (Basel). 7, 130. doi: 10.3390/jof6030130
PubMed Abstract | Crossref Full Text | Google Scholar
Flemming, H. C., van Hullebusch, E. D., Neu, T. R., Nielsen, P. H., Seviour, T., Stoodley, P., et al. (2023). The biofilm matrix: multitasking in a shared space. Nat. Rev. Microbiol. 21, 70–86. doi: 10.1038/s41579-022-00791-0
PubMed Abstract | Crossref Full Text | Google Scholar
Gallagher, K. A., Tschowri, N., Brennan, R. G., Schumacher, M. A., Buttner, M. J. (2024). How c-di-GMP controls progression through the Streptomyces life cycle. Curr. Opin. Microbiol. 80, 102516. doi: 10.1016/j.mib.2024.102516
PubMed Abstract | Crossref Full Text | Google Scholar
Han, A.-R., Kim, H., Park, J.-T., Kim, J.-W. (2022). Characterization of a cold-adapted debranching enzyme and its role in glycogen metabolism and virulence of Vibrio vulnificus MO6-24/O. J. Microbiol. 60, 375–386. doi: 10.1007/s12275-022-1507-3
PubMed Abstract | Crossref Full Text | Google Scholar
Haney, E. F., Trimble, M. J., Hancock, R. E. W. (2021). Microtiter plate assays to assess antibiofilm activity against bacteria. Nat. Protoc. 16, 2615–2632. doi: 10.1038/s41596-021-00515-3
PubMed Abstract | Crossref Full Text | Google Scholar
Jenkins, D. J., Woolston, B. M., Hood-Pishchany, M. I., Pelayo, P., Konopaski, A. N., Quinn Peters, M., et al. (2023). Bacterial amylases enable glycogen degradation by the vaginal microbiome. Nat. Microbiol. 8, 1641–1652. doi: 10.1038/s41564-023-01447-2
PubMed Abstract | Crossref Full Text | Google Scholar
Jin, Y., Lin, J., Shi, H., Jin, Y., Cao, Q., Chen, Y., et al. (2024). The active ingredients in Chinese peony pods synergize with antibiotics to inhibit MRSA growth and biofilm formation. Microbiol Res. 281, 127625. doi: 10.1016/j.micres.2024.127625
PubMed Abstract | Crossref Full Text | Google Scholar
Kwak, J.-Y., Kim, M.-G., Kim, Y.-W., Ban, H.-S., Won, M.-S., Park, J.-T., et al. (2016). Properties of a glycogen like polysaccharide produced by a mutant of Escherichia coli lacking glycogen synthase and maltodextrin phosphorylase. Carbohydr. Polym. 136, 649–655. doi: 10.1016/j.carbpol.2015.09.091
PubMed Abstract | Crossref Full Text | Google Scholar
Lee, A., Bae, E., Park, J., Choi, K.-H., Cha, J. (2021). Identification of the genes related to the glycogen metabolism in hyperthermophilic archaeon, sulfolobus acidocaldarius. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.661053
PubMed Abstract | Crossref Full Text | Google Scholar
Lerner, A., Castro-Sowinski, S., Lerner, H., Okon, Y., Burdman, S. (2009). Glycogen phosphorylase is involved in stress endurance and biofilm formation in Azospirillum brasilense Sp7. FEMS Microbiol. Lett. 300, 75–82. doi: 10.1111/j.1574-6968.2009.01773.x
PubMed Abstract | Crossref Full Text | Google Scholar
Li, F., Wang, M.-M., Liu, Q.-H., Ma, Z.-W., Wang, J.-J., Wang, Z.-Y., et al. (2023). Molecular mechanisms of glycogen particle assembly in Escherichia coli. Carbohydr. Polym. 299, 120200. doi: 10.1016/j.carbpol.2022.120200
PubMed Abstract | Crossref Full Text | Google Scholar
Lin, Y., Zhou, X., Li, Y. (2022). Strategies for Streptococcus mutans biofilm dispersal through extracellular polymeric substances disruption. Mol. Oral. Microbiol. 37, 1–8. doi: 10.1111/omi.12355
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, Q.-H., Tang, J.-W., Wen, P.-B., Wang, M.-M., Zhang, X., Wang, L. (2021). From prokaryotes to eukaryotes: insights into the molecular structure of glycogen particles. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.673315
PubMed Abstract | Crossref Full Text | Google Scholar
Machtey, M., Kuhn, M. L., Flasch, D. A., Aleanzi, M., Ballicora, M. A., Eglesias, A. (2012). Insights into glycogen metabolism in chemolithoautotrophic bacteria from distinctive kinetic and regulatory properties of ADP-glucose pyrophosphorylase from Nitrosomonas europaea. A. J. Bacteriol. 194, 6056–6065. doi: 10.1128/JB.00810-12
PubMed Abstract | Crossref Full Text | Google Scholar
Miao, J., Regan, J., Cai, C., Palmer, G. E., Williams, D. L., Kruppa, M. D., et al. (2023). Glycogen Metabolism in Candida albicans Impacts Fitness and Virulence during Vulvovaginal and Invasive Candidiasis. MBio 14, e0004623. doi: 10.1128/mbio.00046-23
PubMed Abstract | Crossref Full Text | Google Scholar
Montero, M., Almagro, G., Eydallin, G., Viale, A. M., Muñoz, F. J., Bahaji, A., et al. (2011). Escherichia coli glycogen genes are organized in a single glgBXCAP transcriptional unit possessing an alternative suboperonic promoter within glgC that directs glgAP expression. Biochem. J. 433, 107–117. doi: 10.1042/BJ20101186
PubMed Abstract | Crossref Full Text | Google Scholar
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
留言 (0)