The Gram-negative bacterium Pseudomonas aeruginosa, a common and opportunistic pathogen, causes disease in a variety of hosts (Rahme et al., 1997; C. et al., 2011; Tan et al., 1999). In humans, P. aeruginosa can cause serious complications, form systemic infections in immunodeficient patients and develop into chronic infections (Govan and Deretic, 1996; Lieberman and Lieberman, 2003; Lyczak et al., 2000). P. aeruginosa infections are characterized by antibiotic resistance, limited treatment options and high mortality, and outbreaks caused by multidrug resistant strains are on the rise (Fisher et al., 2005; Obritsch et al., 2005; K. et al., 2001). Pathogenesis and drug resistance of P. aeruginosa depends on its ability to form biofilms, which make infections chronic and untreatable as the biofilm protects against antibiotics and host immunity (Roy et al., 2018). Although biofilms are central to chronic P. aeruginosa infections, research examining P. aeruginosa virulence has largely focused on planktonic bacteria or biofilm studies performed in vitro or ex vivo, reducing translatability and creating a barrier to the development of effective antimicrobials to limit chronic P. aeruginosa infections (Highmore et al., 2022).
Biofilm-related infections are challenging to study and monitor in vivo in because they are typically internal. While e.g. surface wounds can be monitored in real time, analysis of internal biofilms are typically carried out postmortem on ex vivo tissue. There are a few examples of in situ in vivo monitoring of biofilms using advanced imaging techniques, such as microcomputed tomography and micropositron emission tomography but these techniques are highly specialized and expensive (Guzmán-Soto et al., 2021). Developing novel in vivo approaches that allow high-throughput assays to study biofilm-related infections would increase the understanding of bacterial biofilms and interactions between biofilms and host, and could lead to the development of new strategies targeting antimicrobial resistance and biofilms.
The model organism Caenorhabditis elegans offers a valuable tool to study infection that can be developed to perform detailed studies of biofilm formation in vivo, increasing the understanding of bacterial physiology within biofilms and interactions between biofilms and the host. C. elegans has a small size and rapid generation time and offers an extensive research toolkit including functional assays to monitor health outputs and physiology. C. elegans is susceptible to human pathogens and C. elegans-P. aeruginosa infection models are particularly useful, as many P. aeruginosa virulence-related factors are conserved across widely divergent taxa from nematodes to plants to mammals (Kim and Ausubel, 2005; Rahme et al., 1995; Tan and Ausubel, 2000), and the human innate immune system shares many characteristics with that of C. elegans. C. elegans feed on bacteria and C. elegans are infected with bacterial pathogens by simply transferring worms from their normal laboratory food, the Escherichia coli strain OP50, to a lawn of the pathogen of interest growing on agar medium. The ease of C. elegans infection makes it an attractive model for high-throughput screening to identify attenuated and hypervirulent strains and as a first stage for testing novel antibiotic compounds. In addition, C. elegans is transparent, making it possible to image fluorescent reporters and infection in living animals in real time, while bypassing ethical implications of studying pathogenicity in mammalian in vivo models. Thus, C. elegans has potential to be developed as a new model to study pathogenic biofilms in vitro. Here we report that quorum sensing (QS), a signaling process by which P. aeruginosa regulates biofilm formation, is required for P. aeruginosa pathogenicity in C. elegans.
QS enables bacteria to respond to changes in the density and composition of the surrounding bacterial community and synchronize behavioral responses through extracellular molecules, autoinducers, to produce biofilms (Thi et al., 2020). In P. aeruginosa, QS is regulated through the production and secretion of the autoinducers N-3-oxo-dodecanoyl-ʟ-homoserine lactone (3O-C12-HSL) and N-butyryl-ʟ-homoserine lactone (C4-HSL), which are produced by the canonical acylated homoserine lactone synthases LasI and RhlI, respectively. 3O-C12-HSL is sensed by the transcriptional regulator LasR and C4-HSL is sensed by the transcriptional regulator RhlR. Binding of the autoinducers to LasR and RhlR results in transcriptional responses and expression of genes important for biofilm formation and virulence (Mukherjee et al., 2017; O’Loughlin et al., 2013; Thi et al., 2020) (Figure 1A).
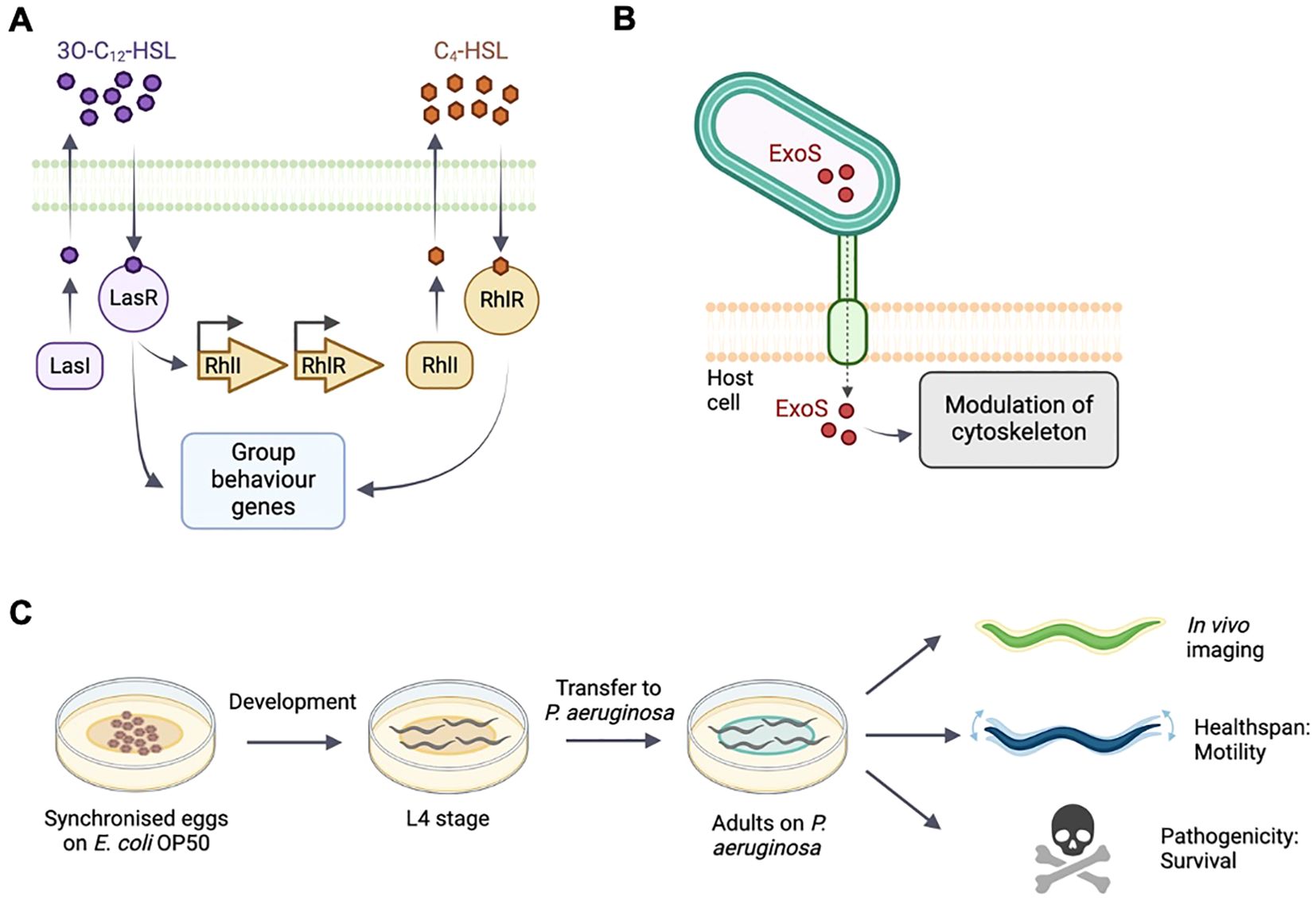
Figure 1. Schematic of quorum sensing and type-III secretion in P. aeruginosa and C. elegans experimental setup. (A) The acyl-homoserine lactone-based quorum sensing network. LasI produces and LasR responds to the autoinducer 3OC12-HSL. The LasR:3OC12–HSL complex activates transcription of many genes including rhlR. RhlI generates the autoinducer C4-HSL. (B) P. aeruginosa injects exotoxin ExoS though a needle complex after contact with the surface of the targeted eukaryotic cell. ExoS modulates the cytoskeleton. (C) C. elegans populations are synchronised and cultivated on E. coli OP50. At L4 stage, animals are transferred onto the different experimental bacterial strains. Bacterial fluorescence, healthspan and survival are monitored throughout adulthood.
In this study, we use fluorescent reporters for the RhlI/R QS system to examine and monitor infection, and the resulting pathogenicity in vivo in C. elegans. We also examine a reporter of the virulence factor exoenzyme S (ExoS), a type III secretion effector which is injected into the host cell during infection, where it reorganizes the cytoskeleton and induces apoptosis (Figure 1B). The type III secretion system is important for acute phases of infection but a role in biofilm formation has not been clearly established (Jouault et al., 2022).
Using fluorescent P. aeruginosa reporters, we confirm that QS is required for P. aeruginosa to form biofilms in vitro. We combine in vivo epifluorescence microscopy and functional readouts using automated tracking systems (Figure 1C) to demonstrate that RhlR signaling is required for P. aeruginosa to invade C. elegans tissues, leading to a reduction in healthspan and survival. Our findings suggest that P. aeruginosa infects C. elegans through QS and biofilm formation, and that methods combining C. elegans, functional assays and bacterial fluorescent reporters can be developed into high-throughput approaches to investigate pathogenic biofilms and evaluate anti-biofilms strategies in vivo.
2 Materials and methods2.1 Bacterial strains and growth conditionsBacterial strains used in this study are listed in Table 1. Bacterial strains were grown in Luria-Bertani broth (LB) and on LB plates fortified with 1.5% Bacto agar at 37°C. Single bacterial colonies were inoculated and incubated at 37°C for 16 hours at 250rpm. When appropriate, antimicrobials were included at the following concentrations: 200 μg/mL carbenicillin, 25 μg/mL gentamycin, 50 μg/mL tetracycline. For assays using 4-Nitropyridine-N-oxide (NPO), the compound was dissolved in DMSO and added to bacterial cultures, keeping DMSO at a maximum of 1% (v/v).
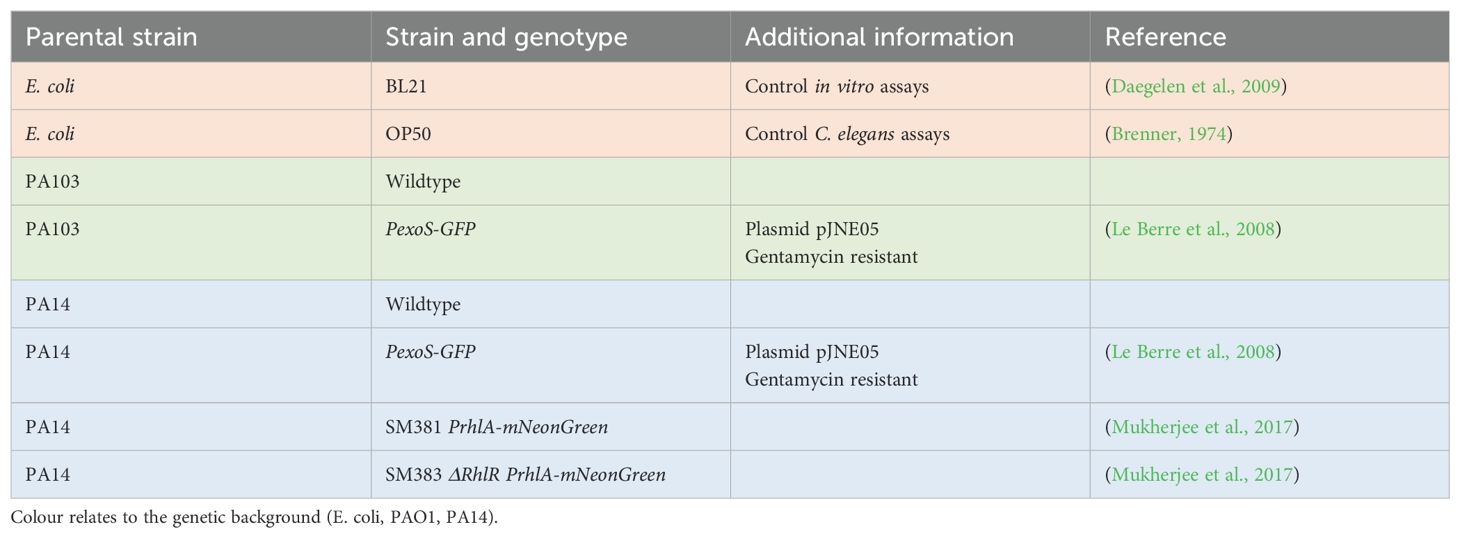
Table 1. Bacterial strains used in study.
2.2 Crystal violet staining assayStatic biofilm formation was evaluated using the crystal violet staining method following established procedure (Campo-Pérez et al., 2023). Briefly, bacterial cultures were washed with PBS three times to remove unbound cells, resuspended in Mueller Hinton broth (M-HB) and diluted to OD600 0.2. 10 µl of each sample was mixed with 190 µl of M-HB in 96-well microplate. After 2 hours of static incubation at 37°C media and non-adhered cells were removed and replaced with fresh M-HB and further incubated for 24 hrs. Adherent cells were quantified by staining with crystal violet and measurement of A550. The assay was performed with three biological replicates consisting of three technical replicates for each condition.
2.3 Fluorescence biofilm assayFluorescence biofilm assays were adapted from previously described methods (Alam et al., 2023). 96-well microplates with bacterial cultures were prepared as for the Crystal Violet assay but instead of staining GFP fluorescence was measured using a microplate spectrofluorometer reader. The assay was performed with three replicates consisting of three technical replicates for each condition.
2.4 C. elegans culture and strainsC. elegans were maintained at 15˚C on media plates seeded with E. coli OP50 as previously described (Brenner, 1974). For epifluorescence imaging and survival experiments, animals were cultivated using plates with 10 mL Nematode Growth Medium. For healthspan assays, plates with 15 mL defined agar were used (Maynard et al., 2018). The C. elegans strains used in this study was SS104 glp-4(bn2), a temperature sensitive mutant which limits germline development at 20°C or above, leading to sterility.
2.5 Preparation of cohorts for imaging, survival and healthspan assaysAn overview of preparation of C. elegans cohorts is shown in Figure 1C. For imaging and survival assays, animals were synchronized by bleaching gravid adults and placing eggs on NGM seeded with E. coli OP50, and incubated at 15°C. Infection by P. aeruginosa was conducted as described previously (Liu et al., 2024) with the exception that assay plates were kept at RT before starting the experiment. At L4 stage animals were transferred to experimental plates seeded with P. aeruginosa wildtype, P. aeruginosa reporter strains or E. coli OP50 control and shifted to 25°C. For healthspan assays, animals were synchronized by egg lay and transferred to experimental plates and 24°C at L4 stage. All experimental plates were seeded with 250 μL of bacterial culture and used the following day.
2.6 Imaging and image analysisAnimals were imaged at 24 and 96 hours after shifting L4s to experimental plates and 25°C. At each time point, 10-15 animals were mounted onto 2.5% agar pads and anesthetized with 25 mM tetramisole. DIC and fluorescence (488nm Exc./505-575nm Em) images were acquired with a CellCam Rana 200CR camera and Leica DMR compound microscope driven by Micro-Manager Studio Version 2.0.0. Brightfield and fluorescent images were stitched together in ImageJ using the Pairwise Stitching tool. The brightfield and fluorescent images were converted into an image stack. Outlines of the animals were drawn from the start to the end of the intestine (just before terminal bulb until the anus) based on the brightfield image using the Polygon Selection tool. This region was then measured in the corresponding fluorescent channel for area (pixels) and brightness (mean brightness). Size was quantified by measuring the area from neck to tail. Two biological replicates with 10-15 animals in each were performed.
2.7 Killing assaysAnimals were shifted to experimental plates and 25°C at L4 stage and scored dead or alive at 48, 96, and 192 h (Liu et al., 2024; Mylonakis et al., 2002). Animals were considered dead if not responsive to prodding with a platinum wire. Three replicates were performed with 30 worms per replicate. Animals with internal hatching or that went missing from the plates were censored.
2.8 C. elegans healthspan assaysHealthspan was assessed using the WormGazer™ automated imaging technology (Magnitude Biosciences) (Zavagno et al., 2024). 60 mm plates were imaged using Raspberry Pi Version 2 cameras at a distance of 60 mm from the plate using white transmission illumination from a generic LED light panel. The cameras were located inside a temperature controlled laboratory set to 24°C. For each dish, a sequence of 200 images were taken over a 160 secs, with recording performed every 5 minutes until day 10 of adulthood. From these images, the number of moving objects is calculated by applying a threshold of the minimum speed of each object of 10 µm s−1. The speed is derived from the length of the object divided by the 160-s time interval of the imaging window. Plates were censored if they failed a quality control inspection after the experimental runtime, for example if they were contaminated with another microbe or the worms had burrowed into the agar. Censored plates were omitted from data analysis. Animals were imaged continuously using a minimum of 6 plates per condition.
2.9 Transmission electron microscopySamples were prepared as previously described (Tataridas-Pallas et al., 2021). Briefly, C. elegans were first fixed using 3.2% formaldehyde, 0.2% glutaraldehyde in 100mM sodium cacodylate buffer (CAB; pH 7.2). Bodies were separated from heads and tails using a scalpel and left overnight in 2.5% glutaraldehyde at 4 °C. Fixative was removed by washing, followed by re-suspension in 2% low melting-point agarose in CAB. Bodies were excised, transferred to glass vials and they were stained with 1% osmium tetroxide in CAB for 1 h at room temperature. After removal of excess stain by washing bodies were dehydrated in an ethanol series (50%, 70%, 90%, and 100%). Dehydration was followed by t washing in propelyne oxide, after which the samples were treated with a 1:1 mixture of low-viscosity (LV) resin and propelyne oxide for 30 minutes at room temperature. Samples were incubated in fresh LV resin twice for 2 hours each before being embedded in LV resin by polymerisation at 60 °C for 24 h. Embedded samples were sectioned to generate 70 nm-thick transverse sections, approximately equidistant from the vulva and the anterior tip of the worm body, using a Diatome diamond knife and an EM UC7 ultramicrotome. Sections were collected onto 400-mesh copper grids (Agar Scientific) and counterstained with 4.5% uranyl acetate for 45 minutes, followed by Reynolds’ lead citrate for 7 minutes. Sections were imaged using JEOL F200 STEM at 200 kV.
2.10 Statistical analysisFor healthspan measures, the difference between the means were analyzed using Gaussian error statistics and setting significance thresholds with reference to the difference expressed in terms of the standard error. Non significance is defined as a difference of less than 1.64 standard errors and significance defined as a difference of 1.64 or more. A difference between 1.64 and 2.33 standard errors is defined as one star (*), corresponding to p < 0.05 on a one-sided test. A difference between 2.33 and 2.83 standard errors is defines as two stars (**), corresponding to p < 0.01 on a one-sided test. A difference greater than 2.83 standard errors is defined as three stars (***) corresponding to p < 0.002 on a one-sided test. Crystal Violet and fluorescence measurements were analyzed using One-way ANOVA, and survival was analyzed using Log-rank (Mantel-Cox) tests using GraphPad Prism 10.2.3.
3 Results3.1 P. aeruginosa biofilm formation in vitro is dependent on QS systemsWe assessed the biofilm forming capacity of bacterial fluorescent reporters of the RhlI/R QS system and the virulence factor ExoS (Table 1). For RhlI/R we used a fluorescent transcriptional RhlA reporter fusion (PrhlA-mNeonGreen) in the PA14 background that targets expression during biofilm formation and infection. We also used the same reporter carrying a deletion in RhlR (ΔrhlR PrhlA-mNeonGreen). The ΔrhlR mutant has been reported to have biofilm abnormal morphology phenotypes and attenuated virulence (Mukherjee et al., 2017). For ExoS, we used a transcriptional reporter fusion to the exoS promoter (PexoS-gfp) in PA14 and PA103 backgrounds. As controls we used E. coli BL21, which does not form biofilms (Wang et al., 2020) and the PA14 and PA103 wildtype strains. PA14 is a highly virulent clinical isolate and PA103 is a QS defective LasR mutant strain producing defective biofilms (Gambello and Iglewski, 1991; Haley et al., 2012; Le Berre et al., 2008).
We first tested biofilm formation in vitro using the peptidoglycan stain Crystal Violet, a classical staining method used to measure biofilms (Campo-Pérez et al., 2023). The bacterial strains were inoculated in microtiter plates. After 24 hours, biofilm formation was evaluated by removing non-adherent bacteria, staining the adherent cells using Crystal Violet, and measuring absorbance at 550 nm. PA14 showed a 4.2-fold increase in absorbance in comparison to the biofilm incompetent E. coli BL21, in agreement with good biofilm forming ability. In contrast, PA103 had similar staining levels as E. coli BL21, confirming a low biofilm forming capacity (Figure 2A). The PexoS-gfp and PrhlA-NeonGreen reporters in PA14 background showed absorbance levels similar to PA14 wildtype. ΔrhlR PrhlA-NeonGreen had a 1.7-fold lower absorbance compared to PrhlA-NeonGreen, and PA103 and PA103 PexoS-gfp had absorbance levels similar to E. coli BL21, suggesting compromised biofilm-forming abilities in these strains (Figures 2B, C). These findings confirm the biofilm forming capacity of PA14 and central role of the RhlI/R quorum sensing systems for the formation of P. aeruginosa biofilms.
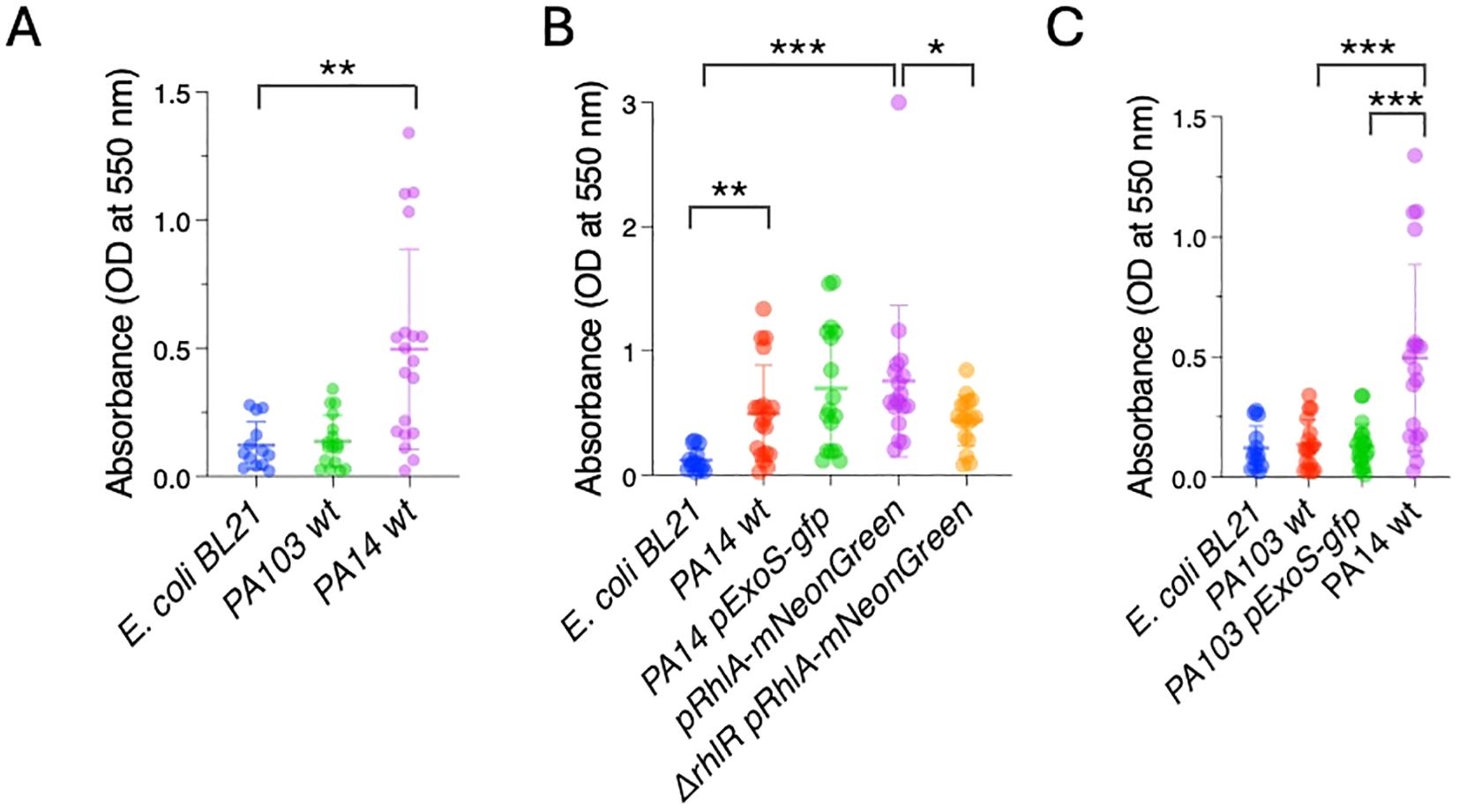
Figure 2. P. aeruginosa PA14 forms biofilms in vitro. Crystal violet staining was measured by absorbance at OD 550 nm in microtiter plates to quantify biofilm formation. (A) PA14 has high biofilm-forming capacity compared to E. coli BL21. (B) Reduced biofilm-forming capacity in ΔrhlR PrhlA-NeonGreen. (C) Low biofilm-forming capacity in PA103 and PA103 PexoS-gfp. Three biological replicates consisting of three technical replicates each were performed. Error bars represent SD of three replicates. One-way ANOVA, *p <0.05, **p <0.001, ***p <0.0001.
3.2 P. aeruginosa reporters form fluorescent biofilms in vitroNext, we tested if the biofilms can also be detected by measuring fluorescence produced by the reporters. Bacterial strains were inoculated in microtiter plates, and after 24 hours planktonic cells were washed off. Biofilms were quantified by measuring fluorescence at 488 nm/507 nm. E. coli BL21 and PA103 had low levels fluorescence relative to PA103 PexoS-gfp, which showed a 13.14-fold increase in fluorescence compared to PA103 background (Figure 3C). PA14 had significantly higher levels of fluorescence compared to E. coli BL21 (Figure 3B), likely due to autofluorescence resulting from pyocyanin production (Mukherjee et al., 2017). PA14 PexoS-gfp and PrhlA-NeonGreen fluorescence was 3.1 and 3.7-fold higher than E. coli BL21, respectively, but not significantly different from PA14 wildtype. PrhlA-NeonGreen fluorescence was reduced by 3.5-fold in the ΔrhlR mutant (Figure 3B).
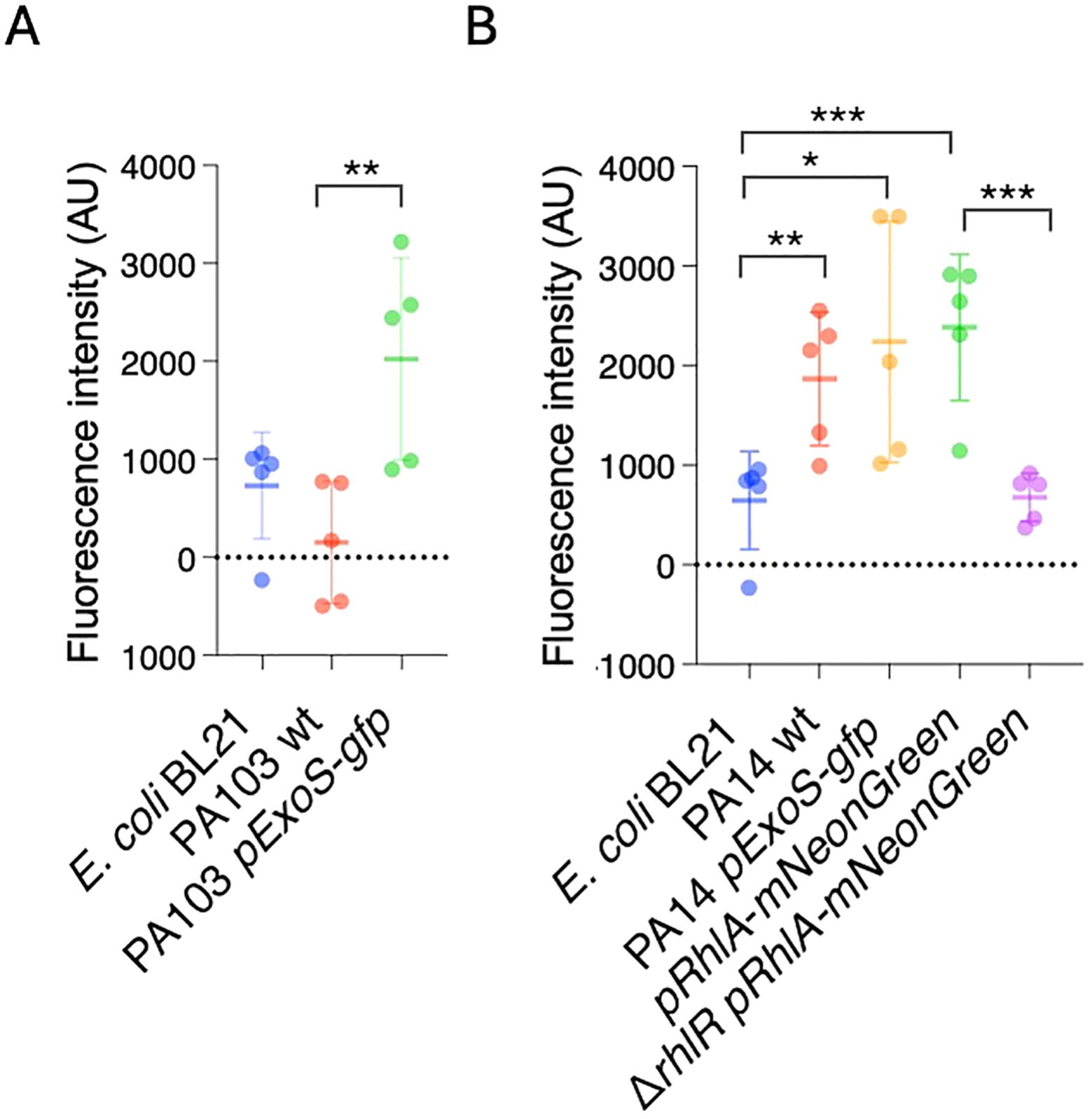
Figure 3. P. aeruginosa reporters form fluorescent biofilms in vitro. GFP fluorescence was measured in microtiter plates to quantify biofilm formation. (A) Increased fluorescence in PA103 PexoS-gfp compared to PA103. (B) Fluorescence in PrhlA-NeonGreen reduced by rhlR mutation. Three biological replicates consisting of three technical replicates each were performed. Error bars represent SD of three replicates. One-way ANOVA, *p <0.05, **p <0.001, ***p <0.0001.
Except for PA103 PexoS-gfp, the reporters yielded similar results across both biofilm assays. The PA103 PexoS-gfp showed high levels of fluorescence compared to PA103, but similar levels of absorbance in the Crystal Violet assay, indicating discrepancies between the two methods for this strain. The fluorescence assay was not able to detect differences in between reporters and PA14 wildtype, likely due to overlapping spectra between the reporters and background fluorescence arising from autofluorescence.
3.3 Infection by PA14 can be visualized using of ExoS and RhlA reportersAfter measuring biofilms produced by the reporters in vitro, we asked if the reporters could be used to visualize bacterial infection in vivo in C. elegans. C. elegans is transparent, allowing imaging living animals without killing or dissection, making it ideal to monitor infection using fluorescent reporters. We cultivated C. elegans with the laboratory standard, non-pathogenic bacteria E. coli OP50 during development and exposed animals to the P. aeruginosa strains at L4 stage (end of development). Fluorescence was monitored in vivo using epifluorescence imaging 24 and 96 hours after exposure to P. aeruginosa. The later timepoint was chosen to capture early phases infection. We also attempted to image 6 days after exposure but found that these timepoints were not suitable, since for some of the conditions the surviving animals were very fragile, leading to bursting and death when mounted on slides.
In C. elegans the primary entry point for bacteria and site of infection is the intestine via the upper part of the gastro-intestinal tract, which consists of the mouth and pharynx. The intestine contains lysosome-related granules that generate autofluorescence. We observed fluorescence within the gut granules in all experimental conditions. In animals exposed to E. coli OP50 fluorescence was not seen in the intestinal lumen nor in other tissues. At 24 hours after exposure, there were only minor differences in fluorescence between the experimental conditions (Figures 4A, D). In contrast, after 96 hours different patterns of fluorescence could be observed. In animals infected with PA14 PexoS-gfp, fluorescence could be seen in the intestinal lumen, consistent with it coming from GFP derived from bacteria proliferating in the lumen. In some animals’ widespread fluorescence, not only localized to gut granules, was present in the intestine as well as in the body cavity, indicating GFP-expressing bacteria had crossed the intestinal barrier and entered other tissues. Animals exposed to PexoS-gfp in PA103 background showed fluorescence contained to gut granules, with a subset of animals also having fluorescence present in the intestinal lumen. No fluorescence was observed in the body cavity, suggesting a reduced ability of PA103 to translocate through the intestine and infect the animal compared to PA14. Animals exposed to PA14 pRhlA-NeonGreen had fluorescence present in the intestinal lumen, intestinal cells and body cavity, while in inanimals exposed to ΔrhlR PA14 pRhlA-NeonGreen fluorescence was not observed in the body cavity (Figures 4B, E).
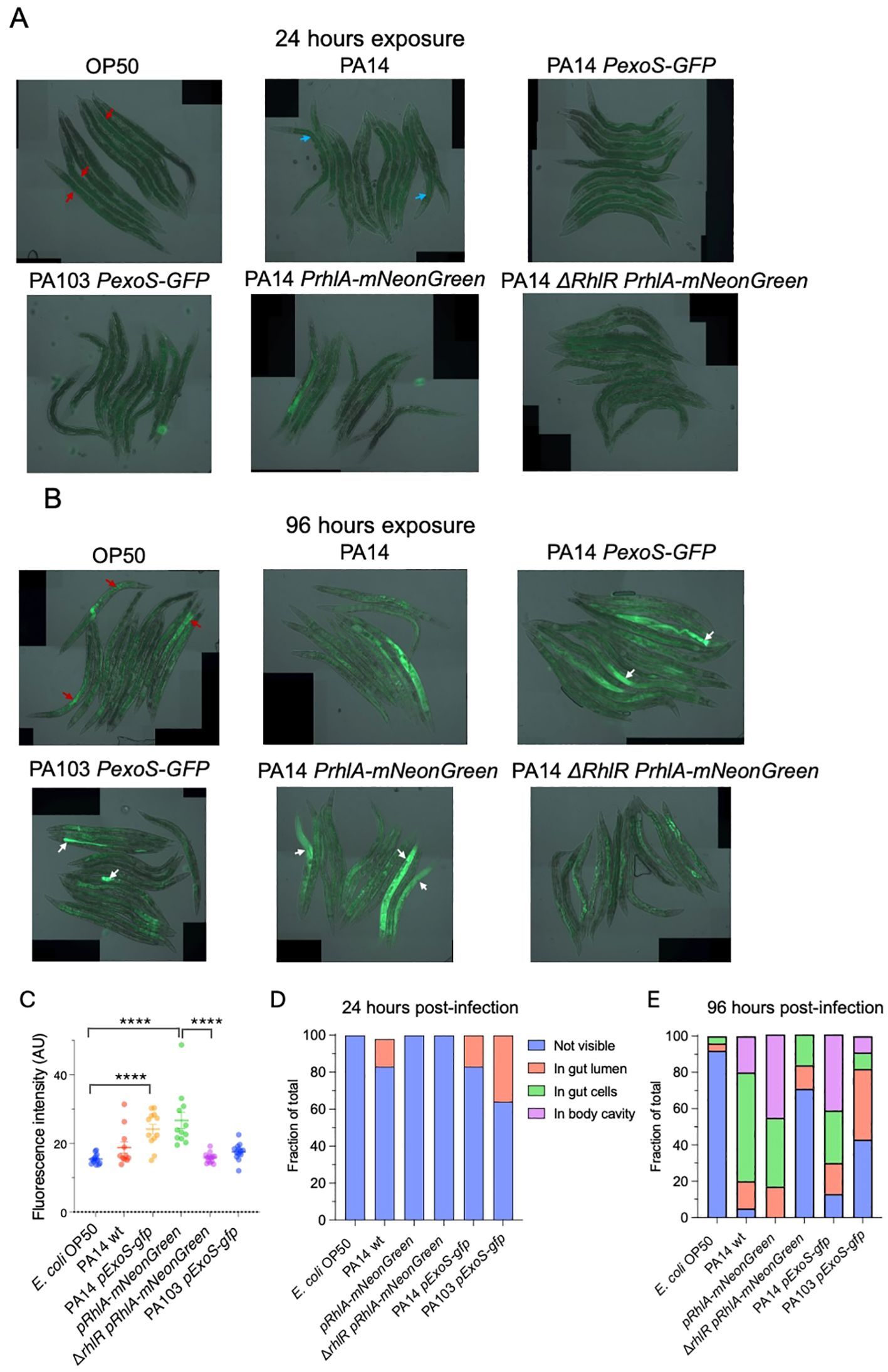
Figure 4. In vivo imaging of C. elegans exposed to P. aeruginosa reporter strains. (A) Overlay of DIC and epifluorescence images taken after 24 hours of exposure. Autofluorescence from C. elegans gut granules within the intestinal cells (red arrows). Autofluorescence from P. aeruginosa pyocyanin within the intestinal lumen (blue arrows). (B) Overlay of DIC and epifluorescence images taken after 96 hours of exposure. Widespread fluorescence observed in animals exposed to PA14 PexoS-gfp and pRhlA-NeonGreen (white arrows). Widespread fluorescence absent in ΔrhlR mutant background. PA103 PexoS-gfp fluorescence is contained within the intestinal lumen (white arrows). Autofluorescence from C. elegans gut granules within the intestinal cells (red arrows). (C) Quantification of mean fluorescence intensity. Two-way ANOVA was used for statistical comparisons. (D, E) Fraction of animals where bacterial fluorescence was not visible, visible in gut lumen, visible in gut cells or visible in body cavity at (D) 24 hours and (E) 96 hours post-infection. Fischer exact probability test was used for statistical comparisons (results described in main text). Two biological trials with 10-15 animals in each were conducted. ****p <0.0001.
Quantification of mean fluorescence at 96 hours post-infection showed that fluorescence intensity was increased in animals infected with PA14 and PA103 strains compared to OP50 controls. The ΔrhlR mutation significantly decreased mean intensity compared to PA14 pRhlA-NeonGreen and significantly altered the distribution of fluorescence between PA14 pRhlA-NeonGreen and ΔrhlR PA14 pRhlA-NeonGreen (Figures 4C, E). Distribution was also significantly different when comparing OP50 to PA14 wt, PA14 PexoS-gfp and PA103 PexoS-gfp but not compared to ΔrhlR PA14 pRhlA-NeonGreen, and significantly different between PA14 PexoS-gfp and PA103 PexoS-gfp (Figure 4E). Thus we could observe differences both in distribution and intensity of the fluorescent reporters in vivo.
Together our imaging data show that by 96 hours of exposure, PA14 can cross the intestinal epithelial barrier to invade tissues, while for PA103 background the bacteria remain within the lumen. Translocation by PA14 can be visualized using the PexoS-gfp and pRhlA-NeonGreen reporters and is dependent on RhlR, confirming quorum sensing is important for PA14 virulence in C. elegans.
3.4 Reduction of RhlR signaling decreases PA14 pathogenic effects on lifespan and healthspanTo further visualize how host-pathogen interactions are affected by RhlR we used transmission electron microscopy (TEM). C. elegans were fixed after 96 hours of exposure to E. coli OP50, PA14 pRhlA-NeonGreen or ΔrhlR PA14 pRhlA-NeonGreen. In animals exposed to E. coli OP50, only very few bacterial cells were observed, consistent with OP50 not being pathogenic, and cells being digested by the host. In contrast, in animals exposed to both PA14 strains bacterial cells were numerous, distending the gut (Figures 5A, B). Noticeably, in animals exposed to PA14 pRhlA-NeonGreen, intestinal microvilli were degraded, shortened and broken, while animals exposed to PA14 ΔrhlR had intact microvilli. PA14 pRhlA-NeonGreen cells appeared less densely packed compared to PA14 ΔrhlR mNeongreen, suggesting the presence of an extracellular matrix separating the cells and consistent reduced biofilm-forming capacity resulting from ΔrhlR. In addition, we observed considerable amounts of extracellular material between the bacterial PA14 pRhlA-NeonGreen cells within the C. elegans gut, e.g. membrane vesicles and cellular debris. The extracellular material between PA14 ΔrhlR pRhlA-NeonGreen appeared qualitatively distinct and with fewer vesicles present (Figure 5B), consistent with the abnormal biofilm morphology phenotypes reported for ΔrhlR mutants (Mukherjee et al., 2017). Although degraded microvilli were not observed in PA14 ΔrhlR pRhlA-NeonGreen infected animals, bacterial cells were in direct contract to the microvilli surface, appearing to be in the process of penetrating the intestinal wall (Figure 5A).
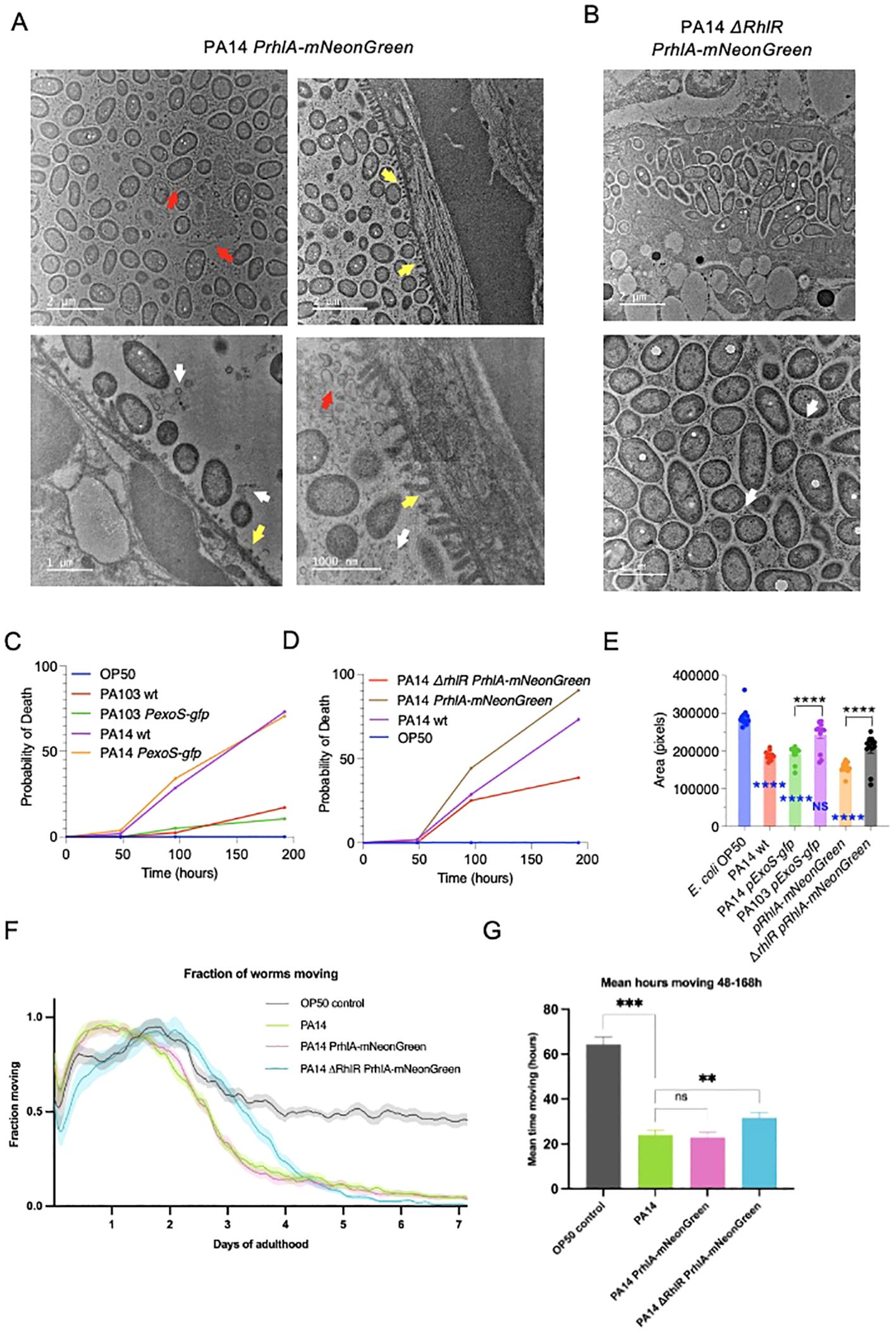
Figure 5. Decreased RhlR signalling in P. aeruginosa reduces pathogenicity in C. elegans. (A, B) Representative cross-sectional transmission electron micrographs of C. elegans infected with PA14 pRhlA-NeonGreen and ΔrhlR pRhlA-NeonGreen (A) Degraded and broken microvilli (yellow arrows), bacterial vesicles (white arrows) and extracellular material (red arrows) visible in PA14 pRhlA-NeonGreen infected animals. (B) In PA14 ΔrhlR pRhlA-NeonGreen infected animals broken microvilli are not visible. Some vesicle observed (white arrows). Extracellular material is present but appears structurally different. Bacterial cells are more densely packed and are penetrating the intestinal brush border. N=2 per condition. (C, D) C. elegans survival was monitored up to 192 hours from L4 stage. Exposure to PA14 strains resulted in decreased survival, which was reduced by ΔrhlR. Exposure to PA103 did not affect survival. Three biological replicates with n=30 were performed. Log-rank (Mantel-Cox) test was performed for comparison of survival. (E) Measurement of C. elegans body area after 96 hours of exposure to bacteria. Two biological trials with 10-15 animals in each were conducted. Two-way ANOVA. Blue indicates comparison with OP50. *p <0.05, *p <0.01, **p <0.001, ****p <0.0001. (F, G) C. elegans healthspan assays were performed by monitoring motility for 7 days starting at L4 stage. Exposure PA14 and PA14 PrhlA-mNeonGreen reduced motility compared to E. coli OP50. Animals exposed with to ΔrhlR PrhlA-mNeonGreen remained mobile for longer. (F) Fraction of animals moving in healthspan assay. Shading shows SEM. (G) The area under the curve integration for hours 48-168 of the healthspan assay. Error bars show SEM. Two biological replicates with n=180 were performed. **p<0.01, ***p<0.002, one-tailed t-test.
The differences we observed between the reporters in biofilm-formation capacity, ability to cross the intestinal barrier and bacteria-host interactions, prompted us to examine virulence resulting from infection with the strains. We first conducted killing assays to determine if strains that translocate into the body cavity are more lethal than those that do not. Animals were cultivated on E. coli OP50 and transferred to P. aeruginosa strains at L4 stage and survival was monitored at 48, 96 and 192 hours (8 days). Survival of control animals kept on E. coli OP50 was not affected during the experiment, and exposure to PA103 and PA103 PexoS-gfp resulted in a small, non-significant reduction in survival. In contrast, infection with wildtype PA14 and PA14 PexoS-gfp resulted in 73.2% and 71.6% of animals dead at 192 hours (Figure 5C). Exposure to PA14 pRhlA-NeonGreen resulted in 91.6% of animals dead while ΔrhlR reduced lethality of PA14 PrhlA-mNeonGreen, animals exposed to this strain had a survival of 61.4% at 196 hours (Figure 5D). These data are consistent with observations in our imaging experiments; PA103 is does not invade C. elegans tissues and has limited pathogenicity, while PA14 is capable of crossing the intestinal barrier and is highly pathogenic. PA14 pathogenicity is dependent on RhlR, consistent with RhlR signaling being important for virulence. Pathogenicity of PA14 PexoS-gfp and PA14 PrhlA-mNeonGreen were not different from PA14, thus expression of the reporters does not affect PA14 virulence.
C. elegans infection models typically test the ability of pathogens to kill the host, while effects on overall physical fitness are usually not assessed. We asked if PA14 pathogenicity in addition to affecting survival, also impacts on fitness. We first compared body size in animals exposed to OP50 and PA14, and found that PA14 reduces body area by 34.6% (Figure 5E), suggesting effects on host fitness. Comparison of the reporter strains showed that PA14 pExoS-gfp and PA103 pExoS-gfp reduced body size by 31.7% and 11.7% respectively compared to OP50. Body area in animals exposed to PA14 pExoS-gfp and PA103 pExoS-gfp were significantly different from each other (22.6%), and compared to ΔrhlR pRhlA-mNeonGreen, pRhlA-mNeonGreen reduced body size by 25.7% (Figure 5E). The changes in body size mirror the effects we observed in survival and are consistent with PA14 biofilms affecting host fitness.
Next we used automated tracking and image analysis (Zavagno et al., 2024) to assess healthspan. We profiled motility of animals following infection with P. aeruginosa, comparing PA14 wildtype, PA14 PrhlA-mNeonGreen and PA14 ΔrhlR PrhlA-mNeonGreen. Animals were tracked continuously from L4 stage until day 7 of adulthood to measure fraction animals moving, and hours spent moving and compared to E. coli OP50 controls. Animals in all conditions were active and mobile during the first two days of tracking, as expected in young, healthy animals (Figure 5F). Animals grown on OP50 showed a partial reduction in movement between days 2 and 4 of adulthood due to normal age-related decline in muscle function (Huang et al., 2004; Newell Stamper et al., 2018). PA14 and PA14 PrhlA-mNeonGreen reduced overall movement and the time spent moving (Figures 5F, G). Animals exposed to PA14 ΔrhlR PrhlA-mNeonGreen exhibited a prolonged healthspan compared to PA14 and PA14 PrhlA-mNeonGreen, increasing both the distance moved and time spent moving (Figures 5F, G). Thus P. aeruginosa negatively affects both survival and healthspan and these effects are reduced when QS is compromised.
4 DiscussionP. aeruginosa biofilms have been studied extensively on glass or plastic surfaces in vitro, but much less is known about how biofilms form and develop on an epithelial barrier, the most common site of infection. In our study we infected the genetically tractable and transparent model organism C. elegans with fluorescent P. aeruginosa reporters to monitor infection in vivo while providing readouts of host health. By combining in vivo microscopy methods with new automated tracking technology, we show that the transcriptional RhlR receptor is important for the ability of P. aeruginosa to invade host tissues, and for pathogenic effects on survival and healthspan. As P. aeruginosa uses the same invasion mechanism across different host species, and mammals and invertebrates use conserved signal transduction pathways to activate defense-related genes (Tan et al., 1999), C. elegans is able to model certain aspects of mammalian pathogenesis. Our study is consistent with other studies examining P. aeruginosa infections and showing that RhlR plays central role in biofilm formation and virulence (Mukherjee et al., 2018; Kumar et al., 2021). For example, mutations in the RhlR transcription factor receptor alter biofilm morphology and reduce the activation of host anti-pathogen defenses (Peterson et al., 2023). In another study, comparing wildtype P. aeruginosa with an ΔrhIR mutant, mice were exposed to an intratracheal challenge and bacterial colonization was monitored in real time. The study showed that the ΔrhIR mutation attenuates virulence and bacterial load while also reducing pathogenicity in C. elegans (Mukherjee et al., 2017). Thus, there are parallels between findings in the murine lung infection and C. elegans models and using C. elegans as pre-clinical model has the potential to accelerate the development of new antimicrobial strategies.
Click or tap here to enter text. Click or tap here to enter text.
In vivo imaging infection of C. elegans with the PrhlA-mNeonGreen reporters showed that the widespread infection in wildtype background was absent in the animals exposed to the mutant, consistent with the compromised biofilm forming capacity in rhlR mutants resulting in reduced virulence. Using standard biofilm assays, we confirmed that the rhlR deletion reduces P. aeruginosa biofilms in vitro. In vitro the autofluorescence generated by pyocyanin production meant there were no differences in overall fluorescence levels between PA14 and the PrhlA-mNeonGreen reporter. In contrast, we could observe clear differences in pattern and distribution of bacterial fluorescence in vivo.
Also, the PexoS-gfp reporter in PA14 background showed a high degree of biofilm formation capacity combined with pathogenicity. As with the mNeonGreen reporter, we could not detect differences in fluorescence intensity between wildtype PA14 and PA14 PexoS-gfp in vitro due to autofluorescence, but found major differences in bacterial fluorescence in vivo, with widespread tissue fluorescence following PA14 PexoS-gfp infection. This fluorescence was not observed in animals exposed to PA103 PexoS-gfp, and PA103 was also less pathogenic. In mice models of acute pneumonia, PA103 is highly virulent, severely reducing survival rates within 24 hours of exposure (MaChado et al., 2010). The moderate pathogenicity of PA103 and lack of intestinal infection in our C. elegans assays highlight species-specific differences and raise the question of why C. elegans is able to avoid infection. A study using two different models of infection in Drosophila showed that PA103 results in high lethality in a fly nicking model but not in a feeding model (L. et al., 2008). As PA103 lacks a functional QS system, QS might be required for lethality by feeding, e.g. to successfully attach to and infect intestinal cells. This can explain the lack of virulence in our C. elegans experiments were we also used feeding as means to expose the animals to the bacteria.
P. aeruginosa is listed as a high-priority pathogen in the 2024 WHO Bacterial Priority Pathogens List, due to its growing antibiotic resistance and global threat, especially in health-care settings (WHO Bacterial Priority Pathogens List, 2024). Currently very few treatments targeting P. aeruginosa biofilms are in development (Reynolds and Kollef, 2021) and the need for developing effective treatments against antibiotic resistant pathogens calls for in vivo models that are rapid and can be used in high throughput. Our work provides insight into how C. elegans combined with low-cost epifluorescence in vivo imaging and automated tracking technologies can be used to study P. aeruginosa infection. We envision the approaches described here being developed into high-throughput in vivo methods using e.g. 96-well plates and compound libraries, to screen for novel antimicrobials targeting P. aeruginosa biofilms, improving efficiency of pre-clinical antimicrobial testing and reduce costs, timelines and ethical burdens.
Future studies could make use of fluorescent reporters with emission wavelengths that do not overlap with those of pyocyanin and gut granules to more clearly differentiate between expression of the reporter and background. It would also be useful to develop methods to assess bacterial viability in the C. elegans gut and body cavity, such as BacLight, and to confirm the presence of biofilms, e.g. biofilm staining methods. Overall, our work suggests that using C. elegans to study QS and biofilms could provide an exciting path forward for the development of novel antimicrobials.
Data availability statementThe raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statementThe manuscript presents research on animals that do not require ethical approval for their study.
Author contributionsFX: Formal analysis, Investigation, Writing – review & editing. MR: Formal analysis, Investigation, Writing – review & editing. SB: Formal analysis, Investigation, Writing – review & editing. MF: Formal analysis, Investigation, Writing – review & editing. SM: Formal analysis, Investigation, Writing – review & editing. ML: Writing - review & editing, Methodology, Investigation. SR: Writing – review & editing, Investigation. GM: Writing – review & editing, Investigation. FT: Formal analysis, Project administration, Writing – review & editing. RT: Investigation, Writing – review & editing. LS: Conceptualization, Funding acquisition, Project administration, Resources, Writing – review & editing. RH: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing. CS: Methodology, Writing – review & editing. DW: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing. ME: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Biofilms Innovation Centre 04POC21-188 and BBSRC BB/V011243/1 and BB/W014165/1. C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
AcknowledgmentsWe thank Bonnie Bassler (Princeton University) and Gary Robinson (University of Kent) for bacterial strains used in the study.
Conflict of interestAuthors MF, SM, FT, CS, and DW were employed by Magnitude Biosciences Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAlam, F., Blackburn, S. A., Davis, J., Massar, K., Correia, J., Tsai, H.-J., et al. (2023). Pseudomonas aeruginosa increases the susceptibility of Candida albicans to amphotericin B in dual-species biofilms. J. Antimicrobial Chemotherapy 78, 2228–2241. doi: 10.1093/jac/dkad228
PubMed Abstract | Crossref Full Text | Google Scholar
C., N. S., See-Wai, L. J., C., A. E., G., A. J., S., R. S., H., D. T. (2011). The sensor kinase kinB regulates virulence in acute Pseudomonas aeruginosa infection. J. Bacteriology 193, 2989–2999. doi: 10.1128/jb.01546-10
PubMed Abstract | Crossref Full Text | Google Scholar
Campo-Pérez, V., Alcàcer-Almansa, J., Julián, E., Torrents, E. (2023). A high-throughput microtiter plate screening assay to quantify and differentiate species in dual-species biofilms. Microorganisms 11. doi: 10.3390/microorganisms11092244
PubMed Abstract | Crossref Full Text | Google Scholar
Daegelen, P., Studier, F. W., Lenski, R. E., Cure, S., Kim, J. F. (2009). Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3). J. Mol. Biol. 394 (4), 634–643. doi: 10.1016/j.jmb.2009.09.022
PubMed Abstract | Crossref Full Text | Google Scholar
Fisher, J. F., Meroueh, S. O., Mobashery, S. (2005). Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105, 395–424. doi: 10.1021/cr030102i
PubMed Abstract | Crossref Full Text | Google Scholar
Gambello, M. J., Iglewski, B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriology 173, 3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991
PubMed Abstract | Crossref Full Text | Google Scholar
Govan, J. R., Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiological Rev. 60, 539–574. doi: 10.1128/mr.60.3.539-574.1996
PubMed Abstract | Crossref Full Text | Google Scholar
Guzmán-Soto, I., McTiernan, C., Gonzalez-Gomez, M., Ross, A., Gupta, K., Suuronen, E. J., et al. (2021). Mimicking biofilm formation and development: Recent progress in in&xa0;vitro and in&xa0;vivo biofilm models. IScience 24. doi: 10.1016/j.isci.2021.102443
PubMed Abstract | Crossref Full Text | Google Scholar
Haley, C. L., Colmer-Hamood, J. A., Hamood, A. N. (2012). Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol. 12, 181. doi: 10.1186/1471-2180-12-181
PubMed Abstract | Crossref Full Text | Google Scholar
Highmore, C. J., Melaugh, G., Morris, R. J., Parker, J., Direito, S. O. L., Romero, M., et al. (2022). Translational challenges and opportunities in biofilm science: a BRIEF for the future. NPJ Biofilms Microbiomes 8, 68. doi: 10.1038/s41522-022-00327-7
PubMed Abstract | Crossref Full Text | Google Scholar
Huang, C., Xiong, C., Kornfeld, K. (2004). Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 101 (21), 8084–8089. doi: 10.1073/pnas.0400848101
PubMed Abstract | Crossref Full Text | Google Scholar
Jouault, A., Saliba, A. M., Touqui, L. (2022). Modulation of the immune response by the Pseudomonas aeruginosa type-III secretion system. Front. Cell. Infection Microbiol. 12. doi: 10.3389/fcimb.2022.1064010
留言 (0)