Proper morphogenesis during the embryonic development is crucial for the heart to function effectively as a pump. The internal structure of the heart undergoes dramatic transformations over a short period, with remodeling continuing into the neonatal period. Following cardiac looping, endocardial cells in the outflow tract and atrioventricular canal regions undergo endothelial-to-mesenchymal transformation (EndoMT) to form cushion mesenchyme that eventually remodel into cardiac valves and septum. Recent studies have expanded this understanding, demonstrating that endocardial cells also undergo endothelial-to-hematopoietic transformation (EHT), contributing to cushion remodeling (Nakano et al., 2013; Shigeta et al., 2019; Liu et al., 2023b). These findings challenge the conventional view that embryonic hematopoiesis occurs exclusively in the yolk sac and aorta-gonad-mesonephros (AGM) region in mammals. This opinion article summarizes existing research on endocardial hematopoiesis and its role in cardiac morphogenesis.
In Drosophila embryos, hematopoiesis is closely linked to heart development. Both the heart and hematopoietic systems share developmental origins and molecular mechanisms, including Tinman (the orthologue of Nkx2-5), GATA factors, and Notch signaling (Mandal et al., 2004; Han and Olson, 2005). Specifically, the dorsal vessel, which serves as the heart tube in flies, is also integral to the development of hemocytes, blood cells that play roles analogous to mammalian macrophages in immune responses and tissue maintenance (Lebestky et al., 2000).
Studies have shown that endocardial hematopoiesis observed in mouse embryos is conserved in Drosophila. In mice, hematopoietic cells derived from endocardial cells are produced in an Nkx2-5-dependent manner (Nakano et al., 2013; Liu et al., 2023b), with macrophages as the predominant cell lineage involved in cardiac cushion remodeling (Shigeta et al., 2019; Liu et al., 2023b). However, the notion that endocardial cells give rise to macrophages via de novo hematopoiesis remains controversial (Liu et al., 2022; Liu et al., 2023a). Despite these debates, endocardial hematopoiesis has also been observed in zebrafish (Gurung et al., 2024; Bornhorst et al., 2024), supporting its evolutionary conservation.
This article addresses the ongoing controversies surrounding endocardial hematopoiesis and explores potential directions for future research in endocardial hematopoiesis, aiming to advance our understanding of its role in cardiac development.
Discovery of endocardial hematopoiesisIn early mammalian cardiac primordia, cardiac progenitor cells marked by Flk1, Isl1, and Nkx2-5 differentiate into cardiomyocytes, smooth muscle cells, and endothelial/endocardial cells (Moretti et al., 2006). Researchers discovered that these progenitors also express hematopoietic transcription factors, including Gata1, Lmo2, Runx1, and Tal1 (Masino et al., 2004). Despite this finding, the significance of hematopoietic signatures in cardiac progenitor cells remained unclear for many years. Interestingly, an earlier study identified hematopoietic-like cells in the endocardial layer of zebrafish (Al-Adhami and Kunz, 1977), suggesting a possible evolutionary link between hematopoiesis and cardiogenesis.
While the plasticity of endocardial cells has primarily been studied in the context of their contributions to mesenchymal cells and coronary endothelial cells, their hematopoietic potential has remained unexplored (Gise and Pu, 2012; Nakano et al., 2016; Zhang et al.,2018; Tian et al., 2015; D’Amato et al., 2022). Endocardial cells, lining the inner surface of the heart, are typically squamous in shape. However, upon hematopoietic transformation, they adopt a rounded morphology and begin to express early hematopoietic markers such as CD41 and Tal1 (Nakano et al., 2013; Collart et al., 2021). Our studies and others have identified endocardial cells expressing hematopoietic markers in the outflow tract, atrioventricular canal, and inflow tract of the mouse embryonic heart (Nakano et al., 2013; Yzaguirre and Speck, 2016). This localization pattern overlaps with distribution of Nkx2-5 lineage endocardial cells and endocardial cushion (Figure 1A). Nkx2-5 knockout (KO) mice die in mid-gestation due to lack of endocardial cushion formation and hypoplastic cardiomyocytes (Lyons et al., 1995; Tanaka et al., 1999). Notably, the KO mice also develop hematopoietic defects in yolk sac and endocardium (Lyons et al., 1995; Nakano et al., 2013). Therefore, Nkx2-5 is not only expressed in the hemogenic endocardial cells but also required for the hematopoiesis.
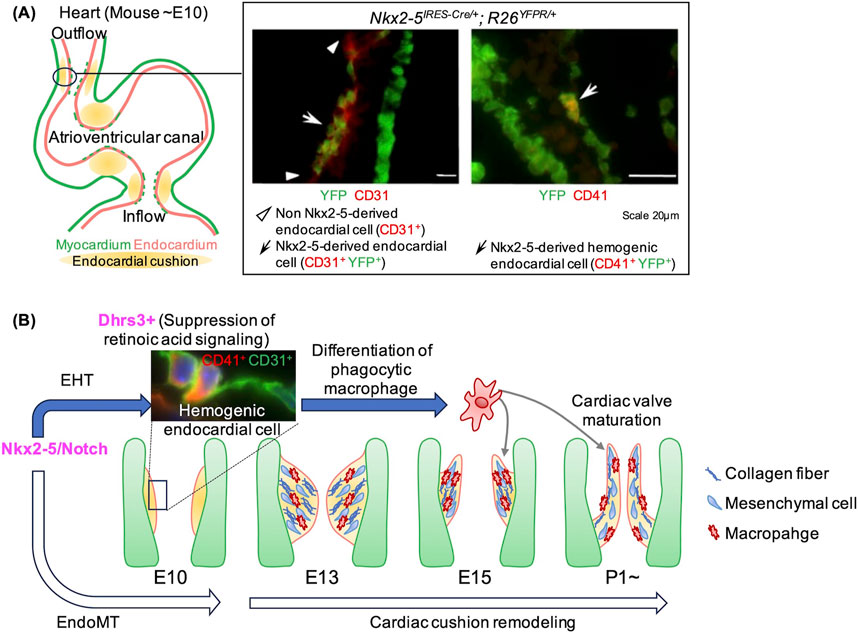
Figure 1. Endocardial hematopoiesis and its role in cardiac morphogenesis during mouse development. (A) Hemogenic endocardial cells are localized in regions of endocardial cushion formation and originate from the Nkx2-5 lineage. The immunofluorescent image depicts the outflow tract cushion in an Nkx2-5IRES-CRE/+; R26YFPR/+ embryo, where Nkx2-5-derived cells are marked by YFP expression. Most ventricular cardiomyocytes and a subset of endocardial cells are positive for YFP (left). A subset of Nkx2-5-derived endocardial cells express CD41 (right). (B) The schematic illustrates our findings on the molecular mechanisms and the role of endocardial hematopoiesis. Nkx2-5/Notch signaling drive the transformation of endocardial cells into both mesenchymal cells (via EndoMT) and hematopoietic cells (via EHT). Hematopoietic cells derived from the endocardium express Dhrs3, which suppress retinoic acid signaling. This suppression promotes the differentiation of these cells into macrophages, which display enhanced phagocytic activity. These macrophages play a crucial role in remodeling the cardiac cushion, ultimately contributing to the formation of mature heart valves. Immunofluorescent staining images are adapted from Nakano et al. (2013).
Using in vivo and single-cell RNA-sequencing (scRNA-seq) analysis (GSE76118 (Li et al., 2016)), we found that Nkx2-5 KO mouse endocardium lacks both cushion endocardial cells and hematopoietic progenitor cells (Nakano et al., 2013; Liu et al., 2023b). This finding aligns with the tinman-dependent hematopoiesis observed in Drosophila larva (Mandal et al., 2004; Han and Olson, 2005), suggesting that Nkx2-5-dependent hematopoiesis is across species. Further scRNA-seq analysis revealed two key signaling pathways involved in Nkx2-5-dependent endocardial hematopoiesis: Notch signaling and retinoic acid (RA) signaling (Liu et al., 2023b). Forced activation of Notch signaling in Nkx2-5-lineage cells restored both endocardial cushion and hematopoietic cell deficits in Nkx2-5-null background, demonstrating that Notch signaling promotes endocardial cushion formation and hematopoiesis downstream of Nkx2-5. RA signaling also plays a critical role. Dhrs3 (dehydrogenase/reductase 3) encoding an enzyme that reduces all-trans RA (atRA) levels was significantly downregulated in Nkx2-5 KO endocardial cells. Ex vivo hematopoietic colony formation assays showed that excessive RA signaling inhibits hematopoietic progenitor differentiation, including macrophage differentiation, suggesting that RA suppression is essential for these processes. Forced activation of Notch signaling in Nkx2-5-lineage cells enhanced macrophage production with an increase in Dhrs3-positive proportions, linking the Nkx2-5-Notch signaling axis to Dhrs3-mediated RA regulation and macrophage differentiation (Liu et al., 2023b) (Figure 1B). It remains unclear whether Nkx2-5 expression is directly required for EHT. As genome-wide ChIP-seq study has identified Nkx2-5 binding sites in the conserved regulatory regions of Notch1, Jag1, Rbpjk, and Runx1, some of these established regulators may be direct target of Nkx2-5 (He and Pu, 2010). Further studies are required for establishing the precise mechanism of Nkx2-5-dependent hematopoiesis and EHT.
Flow cytometric analysis using Nfatc1-lineage tracing revealed that a small fraction of endocardial-derived tissue macrophages (2.6%–17.4%) persists in fetal hearts and into adulthood (Shigeta et al., 2019). However, the hemogenic activity of endocardial cells remains controversial. Two studies from Dr. Zhou’s group identified Nfatc1-labeled cells in yolk sac and failed to confirm hemogenic activity in mammalian endocardial cells (Liu et al., 2022; Liu et al., 2023a). These issues and implications are discussed elsewhere (Nakano and Liu, 2023).
Recently, live imaging studies in zebrafish have provided new insights into endocardial hematopoiesis. Gurung et al. observed EHT of endocardial cells as early as 24 h post-fertilization (hpf), corresponding to mouse E8.0, before the onset of heartbeat (Gurung et al., 2024). This process depends on gata5/6 and hedgehog signaling rather than canonical hematopoietic transcription factors like etv2/etsrp and scl/tal1, with neutrophils as the primary outcome (Gurung et al., 22024). On the other hand, Bornhorst et al. reported increased endocardial hematopoiesis starting at 74 hpf, corresponding to mouse E10.5, when endocardial cushion formation is more advanced (Bornhorst et al., 2024). Their study utilized live imaging, lineage-tracing, and scRNA-seq analysis with a photoconversion-based approach to demonstrate that hemogenic endocardial cells give rise to hematopoietic stem/progenitor cells (HSPCs) by maintaining their adhesion to the endocardium via itga4 and vcam1 (Bornhorst et al., 2024). Together, these findings suggest that endocardial cells may also influence systemic hematopoiesis by serving as a source of neutrophils and an HSPC niche.
Further investigations using advanced live imaging and more sophisticated tracing techniques are needed to resolve ongoing controversies and clarify the contribution of hematopoietic endocardium to cardiac development and systemic hematopoiesis.
Physiological significance of endocardial hematopoiesisThe physiological relevance of endocardial hematopoiesis is an emerging area of study. We have demonstrated that hematopoietic cells derived from endocardial cells differentiate into tissue macrophages that reside within the cardiac cushion mesenchyme (Nakano et al., 2013; Shigeta et al., 2019; Liu et al., 2023b). Bulk RNA-seq analysis revealed that endocardial macrophages are enriched in genes involved in antigen presentation, lysosome activity, and phagosomes function as compared with other tissue macrophage populations (Shigeta et al., 2019). Functional phagocytosis assays corroborated with these findings highlighting these macrophages’ phagocytic capabilities (Shigeta et al., 2019). To further elucidate the physiological role of endocardial-derived macrophages, we genetically ablated these cells by crossing Nfatc1-cre or Nkx2-5-cre mice with Csf1r-flox/flox mice, where the colony-stimulating factor 1 receptor (Csf1r), crucial for macrophage differentiation, was deleted specifically in the endocardium. Endocardial-derived macrophage-depleted mice exhibited cardiac valve anomalies characterized by excessive extracellular matrix (ECM) accumulation and increased cellularity. These findings indicate that endocardial-derived macrophages play a crucial role in proper valve remodeling (Shigeta et al., 2019; Liu et al., 2023b). Notably, despite the compensation of the total number of macrophages by the compensatory recruitment of monocyte-derived macrophages, the cardiac valve phenotypes persisted. This highlights that their unique, non-redundant role in cardiac cushion remodeling and valve formation (Figure 1B).
As discussed earlier, studies in zebrafish have reported distinct lineage contributions of endocardial-derived hematopoietic cells: At 24 hpf, Gurung et al. observed that hematopoietic cells detach from the endocardium and express neutrophil markers following EHT, suggesting that endocardial-derived cells may serve as a major source of neutrophils during early development (Gurung, et al., 2024). In contrast, at 74 hpf and later stages, Bornhorst et al. found that de novo EHT in the endocardium maintain cell adhesion to the endocardial layer while differentiating into HSPCs (Bornhorst et al., 2024). Unlike our findings in mice, both zebrafish studies reported minimal contributions of endocardial-derived cells to macrophage populations in cardiac valves. This discrepancy likely reflects differences in developmental stages and species-specific physiological requirements. Endocardial cushion remodeling is not as extensive in zebrafish valve formation. Zebrafish valve mesenchyme cells form valve cusps that are thin and simple in structure (Pestel et al., 2016; Gunawan et al., 2020), whereas mammalian valve formation involves extensive ECM remodeling and sculpting to generate structurally complex and durable valves [Reviewed in (MacGrogan et al., 2014; O’Donnell and Yutzey, 2020)].
These distinctions underscore the unique and indispensable role of endocardial-derived macrophages in mammalian cardiac development, particularly in the context of the more intricate and mechanically demanding architecture of mammalian valves. Their specialized functions in remodeling the cardiac cushion mesenchyme are vital for ensuring proper valve formation, highlighting their evolutionary significance in adapting to the higher mechanical stresses of the mammalian circulatory system.
DiscussionOur studies demonstrate that endocardial cells undergo both EHT and EndoMT in an Nkx2-5/Notch-dependent manner. These processes generate hematopoietic cells that differentiate into macrophages through the inhibition of RA signaling. These results reveal a previously underexplored role of endocardial hematopoiesis in local tissue remodeling during heart development. However, significant questions remain. The ultimate fate of endocardial-derived hematopoietic cells, such as their potential contributions to other hematopoietic lineages or their broader roles in cardiac or systemic physiology, is still unclear. Additionally, the mechanisms that govern the balance between EHT and EndoMT in these cells and their interactions with other macrophage populations warrant further investigation. Addressing these gaps will be crucial for a comprehensive understanding of endocardial hematopoiesis and its implications for cardiovascular development and homeostasis.
Overcoming current technical limitations, such as live imaging of these rare cell populations and their dynamic transitions, will be essential for advancing our understanding of endocardial hematopoiesis. Advanced methodologies, including single-cell multiomics and cutting-edge lineage-tracing approaches, hold the potential to unravel their developmental trajectories and physiological significance. Future studies aimed at addressing these questions will provide critical insights into the unique contributions of endocardial hematopoiesis in heart development and its potential relevance to other organ systems. Such knowledge could have profound implications for understanding both normal physiology and disease processes across multiple biological contexts.
Author contributionsNL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. AN: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Validation, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was supported by R01HL127427 from NIH, and the Fund for Joint International Research (19K24689) and the Fund for Early-Career Scientists (21363933) from Japan Society for the Promotion of Science (JSPS KAKENHI).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAl-Adhami M. A., Kunz Y. W. (1977). Ontogenesis of haematopoietic sites in brachydanio rerio (Hamilton-buchanan) (teleostei). Dev. Growth and Differ. 19 (2), 171–179. doi:10.1111/j.1440-169X.1977.00171.x
CrossRef Full Text | Google Scholar
Bornhorst D., Hejjaji A. V., Steuter L., Woodhead N. M., Maier P., Gentile A., et al. (2024). The heart is a resident tissue for hematopoietic stem and progenitor cells in zebrafish. Nat. Commun. 15 (1), 7589. doi:10.1038/s41467-024-51920-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Collart C., Ciccarelli A., Ivanovitch K., Rosewell I., Kumar S., Kelly G., et al. (2021). The migratory pathways of the cells that form the endocardium, dorsal aortae, and head vasculature in the mouse embryo. BMC Dev. Biol. 21 (1), 8. doi:10.1186/s12861-021-00239-3
PubMed Abstract | CrossRef Full Text | Google Scholar
D’Amato G., Phansalkar R., Naftaly J. A., Fan X., Amir Z. A., Coronado P. E. R., et al. (2022). Endocardium-to-Coronary artery differentiation during heart development and regeneration involves sequential roles of Bmp2 and cxcl12/cxcr4. Dev. Cell 57 (22), 2517–2532.e6. doi:10.1016/j.devcel.2022.10.007
PubMed Abstract | CrossRef Full Text | Google Scholar
Gise A. von, Pu W. T. (2012). Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circulation Res. 110 (12), 1628–1645. doi:10.1161/CIRCRESAHA.111.259960
CrossRef Full Text | Google Scholar
Gunawan F., Gentile A., Gauvrit S., Stainier D. Y. R., Bensimon-Brito A. (2020). Nfatc1 promotes interstitial cell formation during cardiac valve development in zebrafish. Circulation Res. 126 (8), 968–984. doi:10.1161/CIRCRESAHA.119.315992
PubMed Abstract | CrossRef Full Text | Google Scholar
Han Z., Olson E. N. (2005). Hand is a direct target of tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Dev. Camb. Engl. 132 (15), 3525–3536. doi:10.1242/dev.01899
PubMed Abstract | CrossRef Full Text | Google Scholar
He A., Pu W. T. (2010). Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr. Protoc. Mol. Biol. Chapter 21 (October), Unit 21.20. doi:10.1002/0471142727.mb2120s92
PubMed Abstract | CrossRef Full Text | Google Scholar
Lebestky T., Chang T., Hartenstein V., Banerjee U. (2000). Specification of Drosophila hematopoietic lineage by conserved transcription factors. Sci. (New York, N.Y.) 288 (5463), 146–149. doi:10.1126/science.288.5463.146
PubMed Abstract | CrossRef Full Text | Google Scholar
Li G., Xu A., Sim S., Priest J. R., Tian X., Khan T., et al. (2016). Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39 (4), 491–507. doi:10.1016/j.devcel.2016.10.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu K., Jin H., Tang M., Zhang S., Tian X., Zhang M., et al. (2022). Lineage tracing clarifies the cellular origin of tissue-resident macrophages in the developing heart. J. Cell Biol. 221 (6), e202108093. doi:10.1083/jcb.202108093
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu K., Jin H., Zhang S., Tang M., Meng X., Li Y., et al. (2023a). Intercellular genetic tracing of cardiac endothelium in the developing heart. Dev. Cell 58 (16), 1502–1512.e3. doi:10.1016/j.devcel.2023.05.021
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu N., Kawahira N., Nakashima Y., Nakano H., Iwase A., Uchijima Y., et al. (2023b). Notch and retinoic acid signals regulate macrophage formation from endocardium downstream of nkx2-5. Nat. Commun. 14 (1), 5398. doi:10.1038/s41467-023-41039-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Lyons I., Parsons L. M., Hartley L., Li R., Andrews J. E., Robb L., et al. (1995). Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene nkx2-5. Genes and Dev. 9 (13), 1654–1666. doi:10.1101/gad.9.13.1654
PubMed Abstract | CrossRef Full Text | Google Scholar
MacGrogan D., Luxán G., Driessen-Mol A., Bouten C., Baaijens F., Pompa J. L. (2014). How to make a heart valve: from embryonic development to bioengineering of living valve substitutes. Cold Spring Harb. Perspect. Med. 4 (11), a013912. doi:10.1101/cshperspect.a013912
PubMed Abstract | CrossRef Full Text | Google Scholar
Mandal L., Banerjee U., Hartenstein V. (2004). Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat. Genet. 36 (9), 1019–1023. doi:10.1038/ng1404
PubMed Abstract | CrossRef Full Text | Google Scholar
Masino A. M., Gallardo T. D., Wilcox C. A., Olson E. N., Williams R. S., Garry D. J. (2004). Transcriptional regulation of cardiac progenitor cell populations. Circulation Res. 95 (4), 389–397. doi:10.1161/01.RES.0000138302.02691.be
PubMed Abstract | CrossRef Full Text | Google Scholar
Moretti A., Caron L., Nakano A., Lam J. T., Bernshausen A., Chen Y., et al. (2006). Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127 (6), 1151–1165. doi:10.1016/j.cell.2006.10.029
PubMed Abstract | CrossRef Full Text | Google Scholar
Nakano A., Liu N. (2023). Response to matters arising: intercellular genetic tracing of cardiac endothelium in the developing heart. Dev. Cell 58 (16), 1513–1514. doi:10.1016/j.devcel.2023.05.020
PubMed Abstract | CrossRef Full Text | Google Scholar
Nakano A., Nakano H., Smith K. A., Palpant N. J. (2016). The developmental origins and lineage contributions of endocardial endothelium. Biochimica Biophysica Acta 1863 (7 Pt B), 1937–1947. doi:10.1016/j.bbamcr.2016.01.022
PubMed Abstract | CrossRef Full Text | Google Scholar
Nakano H., Liu X., Arshi A., Nakashima Y., van Handel B., Sasidharan R., et al. (2013). Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 4, 1564. doi:10.1038/ncomms2569
PubMed Abstract | CrossRef Full Text | Google Scholar
O’Donnell A., Yutzey K. E. (2020). Mechanisms of heart valve development and disease. Dev. Camb. Engl. 147 (13), dev183020. doi:10.1242/dev.183020
CrossRef Full Text | Google Scholar
Pestel J., Ramadass R., Gauvrit S., Helker C., Herzog W., Stainier D. Y. R. (2016). Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Dev. Camb. Engl. 143 (12), 2217–2227. doi:10.1242/dev.133272
PubMed Abstract | CrossRef Full Text | Google Scholar
Shigeta A., Huang V., Zuo J., Rana B., Nakashima Y., Lu Y., et al. (2019). Endocardially derived macrophages are essential for valvular remodeling. Dev. Cell 48 (5), 617–630. doi:10.1016/j.devcel.2019.01.021
PubMed Abstract | CrossRef Full Text | Google Scholar
Tanaka M., Chen Z., Bartunkova S., Yamasaki N., Izumo S. (1999). The cardiac homeobox gene csx/nkx2.5 lies genetically upstream of multiple genes essential for heart development. Dev. Camb. Engl. 126 (6), 1269–1280. doi:10.1242/dev.126.6.1269
PubMed Abstract | CrossRef Full Text | Google Scholar
Yzaguirre A. D., Speck N. A. (2016). Insights into blood cell formation from hemogenic endothelium in lesser-known anatomic sites. Dev. Dyn. An Official Publ. Am. Assoc. Anatomists 245 (10), 1011–1028. doi:10.1002/dvdy.24430
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang H., Lui K. O., Zhou B. (2018). Endocardial cell plasticity in cardiac development, diseases and regeneration. Circulation Res. 122 (5), 774–789. doi:10.1161/CIRCRESAHA.117.312136
留言 (0)