Diverticular disease, a prevalent gastrointestinal disorder, is particularly significant in Western countries, with prevalence rates as high as 30% in people in their 50s and more than 70% in people older than 80 (Everhart and Ruhl, 2009). In the United States, diverticular disease results in significant healthcare utilization, with more than 1.7 million outpatient visits and over 300,000 hospitalizations annually. Additionally, it leads to approximately 38,740 30-day readmissions and 4,780 deaths, contributing to healthcare expenditures of around $9 billion annually (Peery et al., 2022).Typical symptoms of the disease include abdominal pain, diarrhea, and bleeding. Depending on the severity, diverticular disease can be subdivided into simple diverticulosis (presence of symptoms but no evidence of inflammation), diverticulitis, and diverticular hemorrhage (Tursi et al., 2020). It is important to note that diverticulitis may occur in approximately 10% to 25% of patients with diverticular disease (Strate and Morris, 2019). Therefore, studying the pathogenesis of diverticular disease (especially intestinal diverticular disease) can provide valuable references for better prevention and treatment, holding significant clinical and social value.
The gut microbiota is a complex and diverse ecosystem composed of bacteria, fungi, viruses, and other microorganisms (Lozupone et al., 2012). These microbes co-evolved with the host and play crucial roles in regulating metabolism (Hacquard et al., 2015), immunity (Hooper et al., 2012), nervous system function (Cryan and Dinan, 2012), and maintaining the intestinal barrier (Adak and Khan, 2019), earning the title of the body’s “second genome”. However, when the gut microbiota is affected by internal and external factors, an imbalance may occur, leading to reduced microbial diversity or a disrupted balance between commensal and pathogenic bacteria (Ha et al., 2014). Studies have shown that such imbalance is closely associated with various diseases, particularly gastrointestinal disorders like intestinal diverticular disease (Canakis et al., 2020; Quaglio et al., 2022; Marasco et al., 2023). For instance, research indicates that a decrease in Clostridium cluster IV bacteria, known for their anti-inflammatory properties, is associated with the development of intestinal diverticular disease (Barbara et al., 2017). Another prospective study found that a decline in gut microbiota diversity, coupled with a reduction in commensal bacterial families and genera (such as Faecalibacterium and Ruminococcus) and an increase in potentially pathogenic bacteria (such as Fusobacteria), may be linked to a higher risk of diverticulitis (Mj et al., 2022). These commensal bacteria ferment undigested dietary fiber, producing metabolites such as short-chain fatty acids (SCFAs) that not only provide energy but also protect the gut by supporting the intestinal barrier and regulating the immune system (Arumugam et al., 2011; Vinolo et al., 2011). In contrast, pathogenic bacteria release enterotoxins that disrupt tight junctions in intestinal epithelial cells, weakening the barrier function (Hecht et al., 1992). These changes collectively may damage the structure and function of the intestinal wall, thereby increasing the risk of intestinal diverticular disease (Tursi, 2016). Although a few studies (Jones et al., 2018) have suggested that there may be no direct relationship between the composition of the gut microbiota and intestinal diverticular disease, the potential impact of microbiota alterations on this condition is increasingly recognized as more research emerges. Given the lack of conclusive causal evidence, further studies remain essential.
Mendelian randomization (MR) is widely recognized for its reliability as a statistical method. The core of the method is to utilize genetic variations as instrumental variables (IVs), which in turn provide an effective assessment of the causal relationship between exposure and outcome. In comparison to traditional observational studies, MR offers significant advantages in reducing bias, avoiding confounders, and guarding against reverse causality. Currently, MR has been applied in numerous fields, and has demonstrated its unique value in exploring the causal relationship between gut microbiota and various diseases, including appendicitis, anxiety disorders, and lymphoma (Wang et al., 2023; Liang et al., 2024; Li et al., 2024).
To date, however, few studies have employed MR to explore the causal relationship between gut microbiota and diverticular disease, particularly intestinal diverticular disease. To address this gap, our study leverages summary statistics from genome-wide association studies (GWAS) and applies a bidirectional two-sample MR approach. This methodology enables us to rigorously test the potential association between gut microbiota and intestinal diverticular disease, enhancing both the stability and reliability of our findings.
2 Materials and methods2.1 Study designIn the forward MR analysis, we investigated the potential influence of gut microbiota on the development of intestinal diverticular disease. In reverse MR, we examined the effect of intestinal diverticular disease on gut microbiota. In this process, each gut microbiota and intestinal diverticular disease-associated single-nucleotide polymorphisms (SNP) serve as IVs for inferring causal effects between them. To ensure the validity of the IVs, the study had to fulfill the three core assumptions of MR (Figure 1). These assumptions are as follows: 1. The “correlation” assumption, where IVs are strongly associated with exposure factors; 2. The “independence” assumption, IVs are not associated with confounders; 3. The “exclusivity” assumption, where IVs are not associated with the outcome.
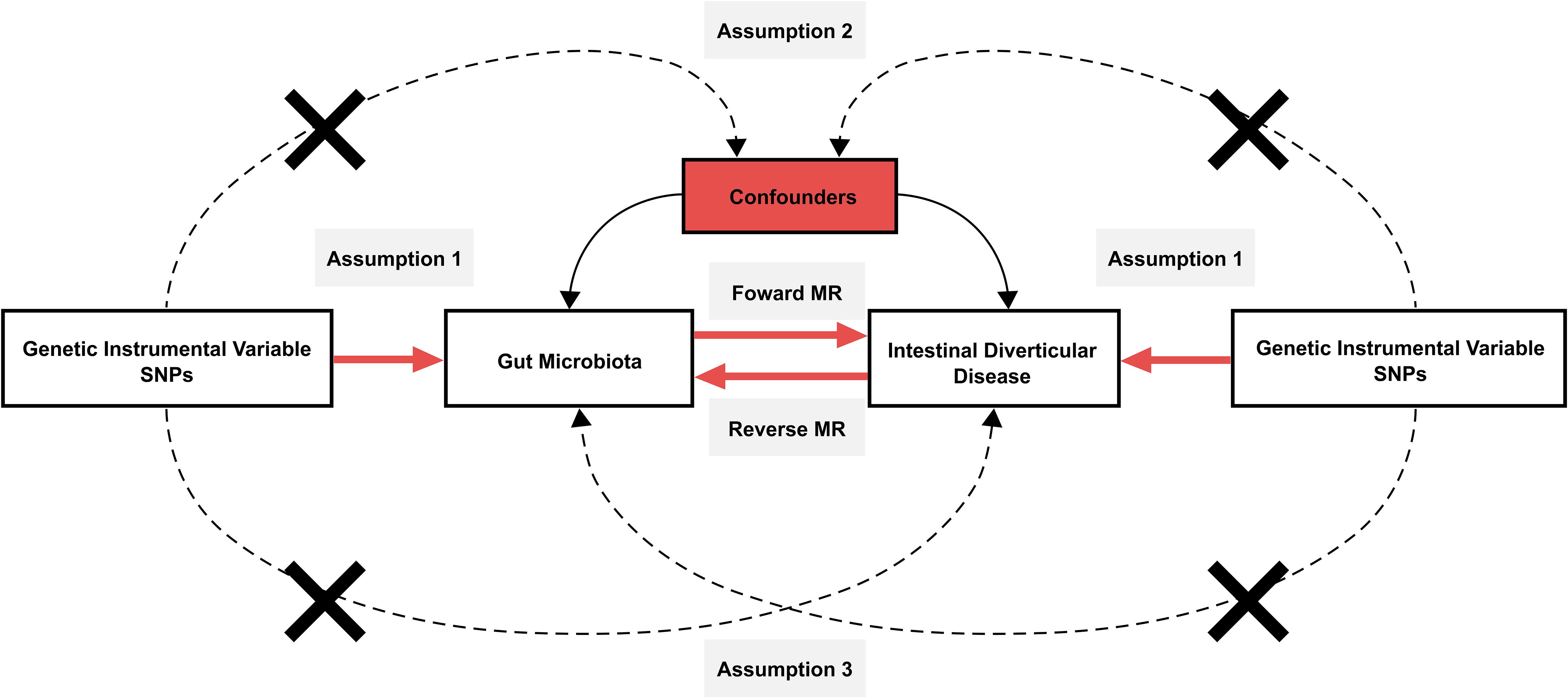
Figure 1. Schematic design of Mendelian randomization. Mendelian randomization requires valid genetic instrumental variables satisfying three assumptions.
2.2 Data sourcesThe GWAS data on gut microbiota were obtained from the FINRISK 2002 study in Finland, a large population-based prospective cohort study (Qin et al., 2022). The study conducted a comprehensive genetic analysis of fecal samples from 5,959 participants. This entailed an exhaustive sample collection and analysis process, which covered 2,801 microbial taxa and 7,967,866 human genetic variants. Following rigorous statistical analysis and screening, the study identified 471 gut microbiota taxonomic groups, including 11 phyla, 19 classes, 24 orders, 62 families, 146 genera, and 209 species. Following further data cleansing, taxonomic units that could not be accurately identified were eliminated, and 410 units were included as the main subjects of the study.
The GWAS data for intestinal diverticular disease were obtained from the IEU Open GWAS program “ukb-b-14796” (https://gwas.mrcieu.ac.uk/). The study covered 462,933 people, of which 5,193 were intestinal diverticular disease patients and 457,740 were controls. It is worth mentioning that the population participating in the above study was of European ancestry (Table 1).
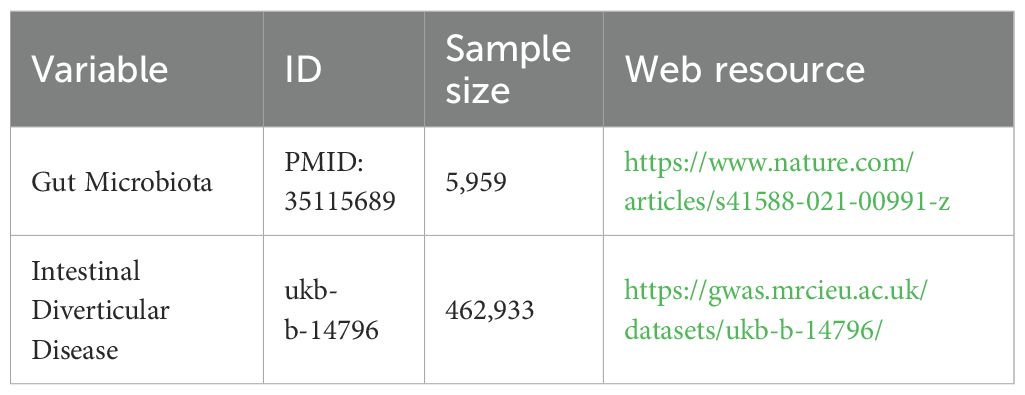
Table 1. Data sources and information used in this study.
2.3 Selection of IVsTo ensure the accuracy of the causal conclusion between gut microbiota and intestinal diverticular disease, we extracted SNPs that were significantly associated with exposure as IVs. The final screened SNPs were required to satisfy the following conditions: 1) Threshold p < 5x10-6; 2) Consistent with linkage disequilibrium (LD) with R2 < 0.001 and LD > 10,000; 3) F-statistic > 10. In addition, we utilized the LDlink to exclude SNPs that may be significantly associated with potential confounders (https://ldlink.nih.gov/?tab=home).
2.4 Data analysisIn assessing the causal relationship between exposure and outcome, we used a variety of MR methods, including inverse variance weighted (IVW), MR-Egger, weighted median, simple mode, weighted mode, leave-one-out sensitivity analysis, and MR-PRESSO. where IVW was used as the main analytical method to derive a combined causal estimate by combining the Wald ratios of all IVs based on the assumption that all IVs are valid variables (Burgess et al., 2023). At the same time, we corrected the p-values using the Bonferroni method by setting different significance p-values at different classification levels (phylum p< 4.545×10-03, class p< 2.632×10-03, order p< 2.083×10-03, family p< 8.065×10-04, genus p< 3.425×10-04, species p< 2.392×10-04) (Sedgwick, 2014). If the p-value is between the above significance p-value and 0.05, we consider that they have a potential causal relationship. MR-Egger and MR-PRESSO can be used to detect horizontal pleiotropy (p< 0.05). When the intercept of MR-Egger is not zero, it may imply the existence of horizontal pleiotropy, which may violate the basic assumptions of MR analysis. Leave-one-out sensitivity analysis was used to assess the degree of dependence of the results on a single IV by removing each SNP one by one and rerunning the MR analysis to observe the stability of the results. The Q-statistics of IVW and MR-Egger were used to assess the degree of heterogeneity among IVs. The presence of heterogeneity was indicated when the p-value of the heterogeneity test was less than 0.05. All analyses were based on “TwoSampleMR”, “MRPRESSO”, “ggplot2”, “foreach” and “foreach” software packages in R version 4.3.2.
3 Results3.1 Selection of IVsBased on the initially set criteria, we screened 410 gut microbiota taxonomic groups and intestinal diverticular disease for suitable SNPs as IVs, and the detailed results are shown in Supplementary Data Sheets 1–3.
3.2 MR results of the effect of gut microbiota on intestinal diverticular diseaseAccording to the results of the IVW analysis, there was a potential causal association between 11 microbial taxa and intestinal diverticular disease (p < 0.05) (Figure 2). Specifically, there was a positive correlation between increasing abundance of Caryophanales and diverticular disease risk at the order level (OR 1.031, 95%CI 1.029-1.049, p=0.001), whereas Chromatiales was negatively associated with diverticular disease risk (OR 0.991, 95%CI 0.983-1.000, and p=0.038). At the family level, Paenibacillaceae (OR 1.011, 95%CI 1.002-1.019, p=0.013) and Turicibacteraceae (OR 1.003, 95%CI 1.000-1.005, p=0.036) were positively associated with diverticular disease risk. At the genus level, Herbinix (OR 1.007, 95%CI 1.000-1.017, p=0.049) and Turicibacter (OR 1.002, 95%CI 1.000-1.004, p=0.046) were positively associated with the risk of diverticular disease, while Arcobacter (OR 0.994, 95%CI 0.990-0.999, p=0.222) and Herbidospora (OR 0.995, 95%CI 0.990-1.000, p=0.039) were negatively associated with diverticular disease risk. At the species level, Staphylococcus fleurettii (OR 1.002, 95%CI 1.000-1.004, p=0.035) was positively associated with the risk of diverticular disease, while Ligilactobacillus ruminis (OR 0.997, 95%CI 0.995-0.999, p= 0.006) and Megamonas funiformis (OR 0.997, 95%CI 0.995-0.999, p=0.012) were negatively associated with diverticular disease risk. Furthermore, the results of other MR methods are provided in the Supplementary Table 1, and the OR in these results are consistent with those of IVW. In order to more visually demonstrate the causal relationship between these microbial taxa and intestinal diverticular disease, we plotted a scatter plot (Figure 3).
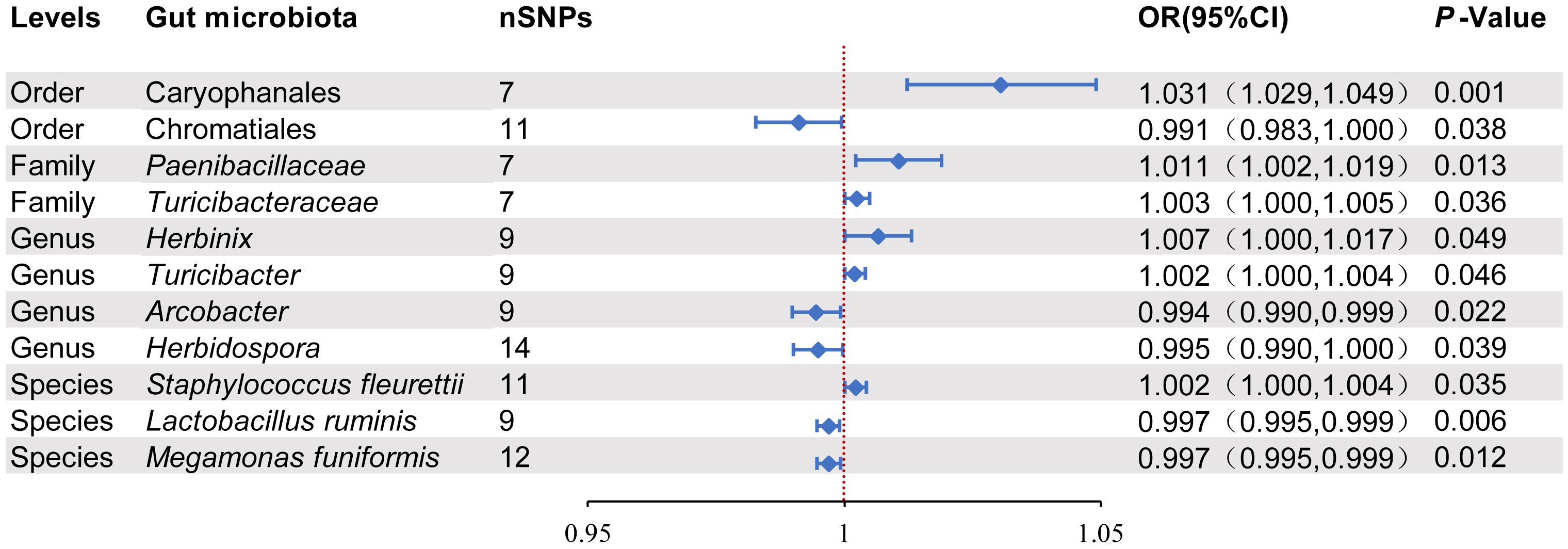
Figure 2. Positive MR results of causal links between gut microbiota on intestinal diverticular disease. SNP, Single-nucleotide polymorphism; OR, odds ratios; CI, Confidence interval.
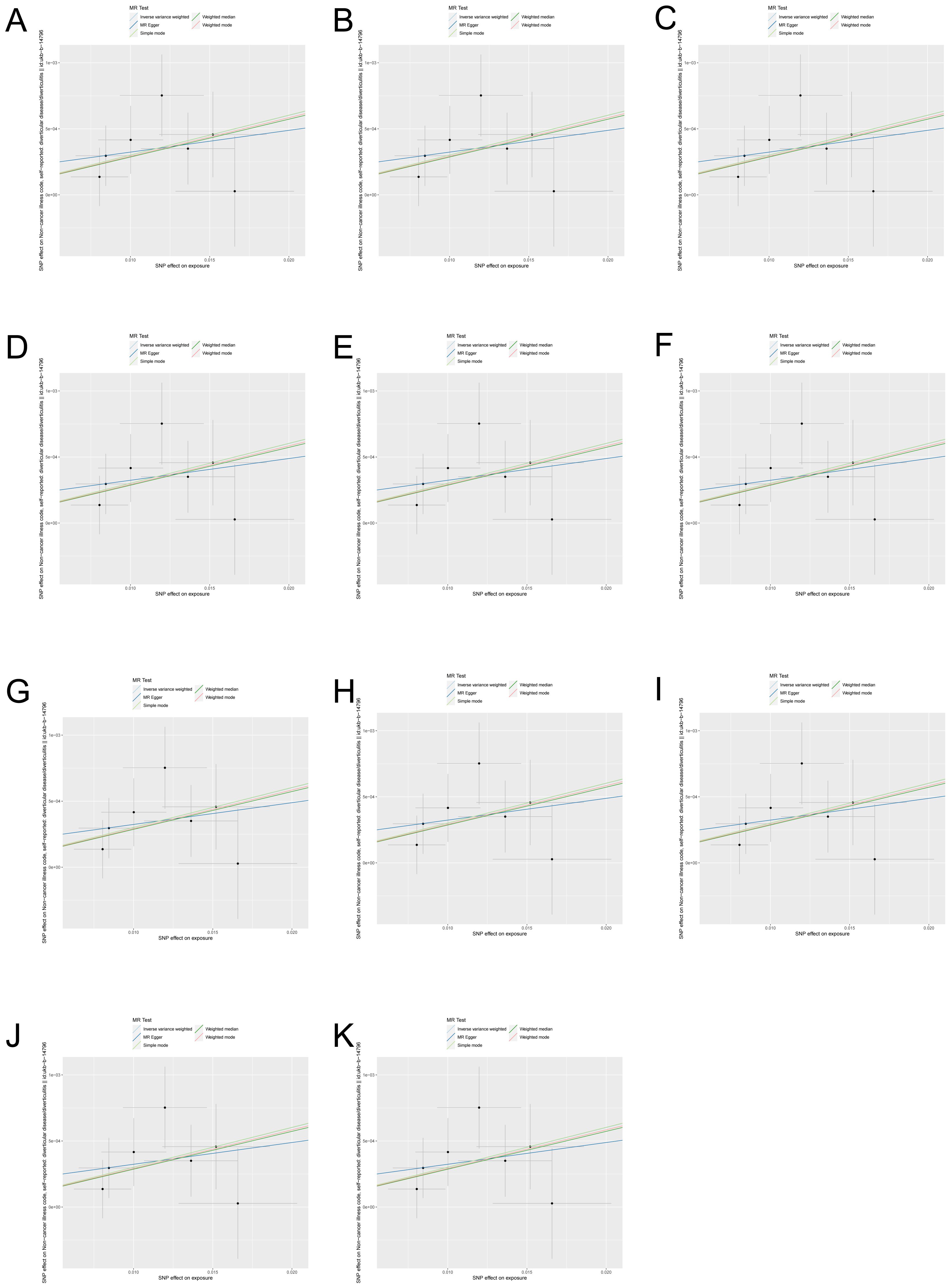
Figure 3. Scatter plots of gut microbiota with causal effects on intestinal diverticular disease. (A) Caryophanales; (B) Chromatiales; (C) Paenibacillaceae; (D) Turicibacteraceae; (E) Herbinix; (F) Turicibacter; (G) Arcobacter; (H) Herbidospora; (I) Staphylococcus fleurettii; (J) Ligilactobacillus ruminis; (K) Megamonas funiformis.
3.3 Heterogeneity and pleiotropy of IVsWe used MR-Egger intercept and MR-PRESSO to detect pleiotropy of all IVs, and the results showed nonexistent pleiotropy (p>0.05). Cochran’s Q test did not find significant heterogeneity of IVs (p>0.05) (Table 2). Finally, in order to present the results of the study visually, we used leave-one-out sensitivity analysis, forest plot and funnel plot to visualize the results (Supplementary Figures 1–3).
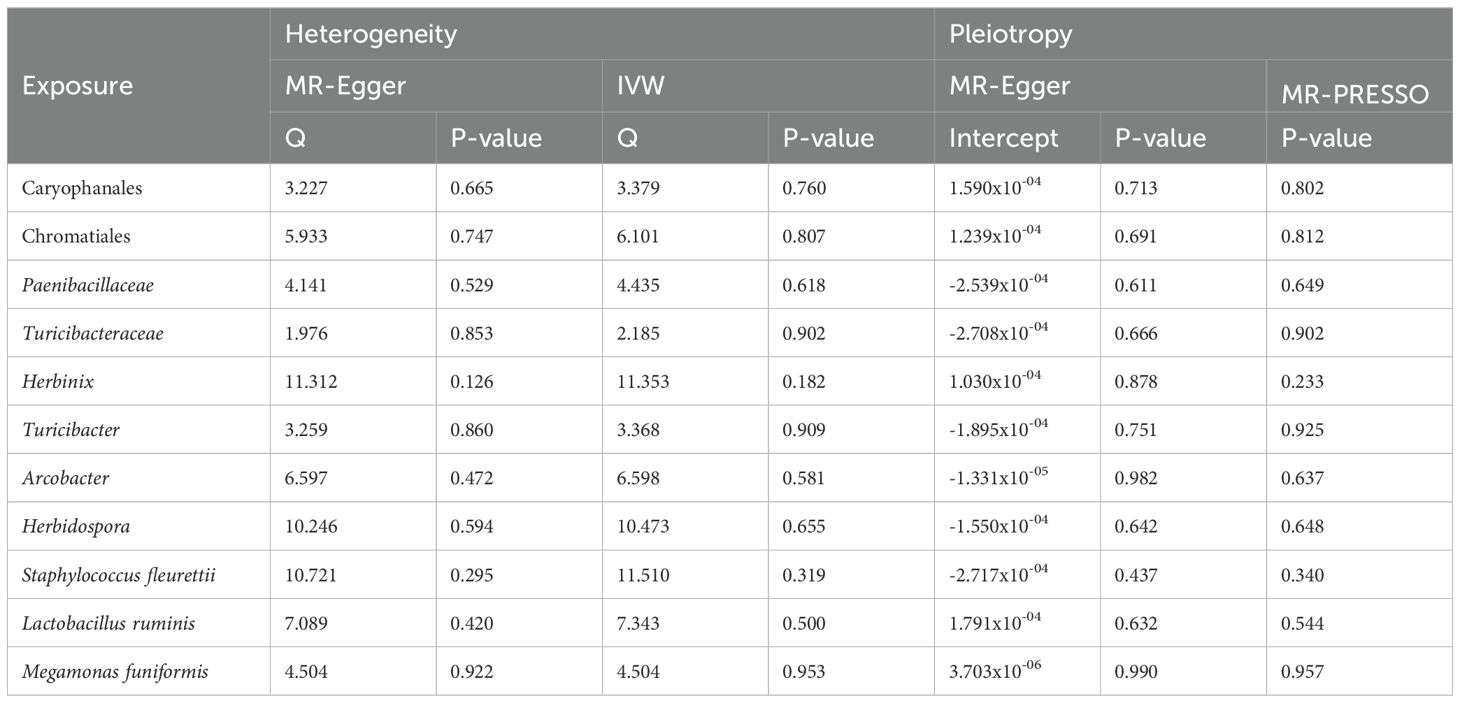
Table 2. Heterogeneity and horizontal pleiotropy of IVs.
3.4 Reverse MR resultsTo further explore whether there was a causal effect of intestinal diverticular disease on the above 11 microbial taxa, we further performed reverse MR analysis. Finally, they were not found to be statistically associated in the IVW method (Supplementary Table 2).
4 DiscussionThis study is the first of its kind to use MR to explore potential causal links between gut microbiota and intestinal diverticular disease. Based on genomic data from 5,959 individuals, we systematically analyzed the possible roles of 410 microbiota taxonomic groups in the pathogenesis of intestinal diverticular disease. Ultimately, our study revealed potential associations between changes in the abundance of 11 microbial taxa and intestinal diverticular disease. Specifically, an increased abundance of the microbial taxa Caryophanales, Paenibacillaceae, Herbinix, Turicibacter, Turicibacteraceae, and Staphylococcus fleurettii may promote the development of intestinal diverticular disease. In contrast, microbial taxa such as Chromatiales, Arcobacter, Herbidospora, Ligilactobacillus ruminis, and Megamonas funiformis demonstrated a potential protective effect against intestinal diverticular disease.
The gut microbiota is a vast assemblage of microorganisms that inhabit the human gut. It is comprised of hundreds of millions of microorganisms that work in concert to form a complex ecosystem (Sandler et al., 2002). They are mainly composed of the phylum Firmicutes and Bacteroidota, with a few belonging to the phylum Actinomycetota, Fusobacteria, and Pseudomonadota, among others (Eckburg et al., 2005). This microbiota composition is highly dynamic and influenced by factors such as age, diet, medication, lifestyle, and environmental exposures, which can all lead to shifts in microbial balance, often referred to as dysbiosis (Rinninella et al., 2019). Dysbiosis has been implicated in a range of systemic diseases, spanning neurological conditions like Parkinson’s disease (Wang et al., 2024), gastrointestinal disorders like inflammatory bowel disease (Lloyd-Price et al., 2019) and diverticular disease, and metabolic and immune-related diseases such as obesity (Le Chatelier et al., 2013) and autoimmune disorders (Alkader et al., 2023).
A study conducted in Italy observed a notable increase in the abundance of the phylum Firmicutes in patients diagnosed with intestinal diverticular disease, particularly within the family Ruminococcaceae, with levels exceeding twice those observed in the general population (Lopetuso et al., 2018). In our study, an increased abundance of five Firmicutes (Caryophanales, Paenibacillaceae, Herbinix, Turicibacter, and Staphylococcus fleurettii) was associated with an increased risk of intestinal diverticular disease, whereas two Pseudomonadota (Chromatiales and Arcobacter) demonstrated a protective effect against intestinal diverticular disease. Further, an analysis of fecal samples from 28 patients with diverticulosis revealed a link between diverticulitis and increased abundance of Pseudobutyrivibrio, Bifidobacterium, and Christensenellaceae (Kvasnovsky et al., 2018). This supports the observed pathogenic role of Herbinix in intestinal diverticular disease, given its familial association with Pseudobutyrivibrio (both belonging to Lachnospiraceae). Although the odds ratios of these microbiota are not particularly large, it is important to note that their cumulative effect, when acting synergistically, could still have a meaningful impact on the development of intestinal diverticular disease. Furthermore, the majority of microbiota positively associated with intestinal diverticular disease in this study belonged to the phylum Firmicutes, which aligns with previous studies linking Firmicutes bacteria to this disease. This consistency further reinforces the reliability of our findings.
Another descriptive, cross-sectional study showed a trend toward a decrease in the number of Clostridium cluster IX and Lactobacillaceae in symptomatic intestinal diverticular disease patients, which is consistent with the protective effect of Ligilactobacillus ruminis against intestinal diverticular disease found in our study (Barbara et al., 2017). This mechanism may be related to the ability of these bacteria to produce SCFAs (including acetic, propionic, and butyric acids). SCFAs play a multifaceted role in supporting intestinal health. They not only activate anti-inflammatory factors, such as IL-10, which help modulate immune responses, but also stimulate B cells to produce immunoglobulin A (IgA) (Hecht et al., 1992). This production of IgA is essential for reinforcing the gut’s immune defense, as IgA binds to pathogens and toxins, preventing them from penetrating the intestinal lining. In line with this, Megamonas funiformis, identified in our study as a protective bacterial group, demonstrates similar beneficial functions. Originally isolated from healthy human feces, Megamonas funiformis has recently been shown to alleviate fatty liver disease associated with metabolic dysfunction through its production of propionic acid (Sakon et al., 2008; Yang et al., 2023). This finding suggests it may exert comparable protective effects in intestinal diverticular disease by enhancing gut health and resilience. These insights reinforce the potential application of specific “probiotics” in treating or managing intestinal diverticular disease.
Staphylococcus fleurettii belongs to the genus Staphylococcus. Although there is a relative paucity of studies on its specific association with intestinal diverticular disease, Staphylococcus are widely recognized to be strongly associated with a variety of infections involving multiple sites such as the intestinal tract, urinary tract, and skin (Trautner and Darouiche, 2004; Raineri et al., 2022; Severn and Horswill, 2023). In addition, it is also worth noting that Staphylococcus fleurettii was initially isolated from goat cheese, a finding that further corroborates speculation in previous studies about a potential link between dietary patterns and intestinal diverticular disease (Lemes et al., 2021). Herbidospora has been isolated primarily from soil and plant samples. Although studies on its functionality in the gut are still insufficient, scientific studies have found that its subspecies, such as Herbidospora daliensis, have significant anti-inflammatory properties (Kudo et al., 1993; Chen et al., 2022). Based on this finding, it is reasonable to hypothesize that Herbidospora may have a protective effect against intestinal diverticular disease.
This study has significant advantages. First, this study is based on MR analysis of large-scale GWAS data, which effectively overcomes the limitations of insufficient sample size, confounding factor interference and reverse causation in observational studies. Second, by selecting a study population of European origin, this study effectively reduces the influence of ethnic differences on the study results.
However, there are several limitations to this study. First, the research focuses exclusively on a European cohort, which may limit the generalizability of the findings to other populations. Additionally, we acknowledge that the pathogenic mechanisms of intestinal diverticular disease may vary depending on its location and type. Due to data limitations, our study was unable to further differentiate between specific locations or types of intestinal diverticular disease. However, the majority of the data are based on patients with colonic intestinal diverticular disease, meaning that the results primarily reflect the relationship between gut microbiota and colonic intestinal diverticular disease. Finally, the lack of detailed information on age and lifestyle factors restricted further stratified analyses. Future research should aim to validate these findings, address these limitations, and explore in more detail the biological mechanisms underlying the relationship between gut microbiota and intestinal diverticular disease.
5 ConclusionThis study, employing a bidirectional two-sample MR approach, identifies potential causal relationships between 11 gut microbiota taxa and intestinal diverticular disease. The findings suggest that variations in microbiota abundance may influence the onset and progression of intestinal diverticular disease, with some taxa providing a protective effect and others increasing risk. Future research should validate these results across diverse ethnic groups and regions (such as Asia and Africa) to assess the impact of racial and geographic differences on the gut microbiota–disease relationship. Integrating functional genomics and experimental studies will be essential to further investigate the role of symbiotic and pathogenic bacteria in the pathogenesis of intestinal diverticular disease and gut ecological balance. Additionally, considering lifestyle factors, dietary habits, and their interactions with microbiota will be crucial in understanding their influence on disease risk. The effect of gut microbiota on intestinal diverticular disease in various sites and forms should also be explored.
In summary, this study expands our understanding of the gut microbiota–intestinal diverticular disease relationship, providing valuable insights for the development of personalized treatment and prevention strategies.
Data availability statementThe datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statementEthical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributionsBZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. DC: Data curation, Writing – review & editing. HZ: Data curation, Writing – review & editing. SL: Data curation, Writing – review & editing.
FundingThe author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
AcknowledgmentsWe would like to express our gratitude to Dr. Xiaojie Wang for his assistance in the revision of this manuscript.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1460504/full#supplementary-material
ReferencesAlkader, D. A. A., Asadi, N., Solangi, U., Singh, R., Rasuli, S. F., Farooq, M. J., et al. (2023). Exploring the role of gut microbiota in autoimmune thyroid disorders: a systematic review and meta-analysis. Front. Endocrinol. 14. doi: 10.3389/fendo.2023.1238146
PubMed Abstract | Crossref Full Text | Google Scholar
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
PubMed Abstract | Crossref Full Text | Google Scholar
Barbara, G., Scaioli, E., Barbaro, M. R., Biagi, E., Laghi, L., Cremon, C., et al. (2017). Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut 66, 1252–1261. doi: 10.1136/gutjnl-2016-312377
PubMed Abstract | Crossref Full Text | Google Scholar
Burgess, S., Davey Smith, G., Davies, N. M., Dudbridge, F., Gill, D., Glymour, M. M., et al. (2023). Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 4, 186. doi: 10.12688/wellcomeopenres.15555.3
PubMed Abstract | Crossref Full Text | Google Scholar
Chen, J. J., Lee, T. H., Cheng, M. J. (2022). Secondary metabolites with anti-inflammatory activities from an Actinobacteria herbidospora daliensis. Molecules (Basel Switzerland) 27, 1887. doi: 10.3390/molecules27061887
PubMed Abstract | Crossref Full Text | Google Scholar
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Sci. (New York N.Y.) 308, 1635–1638. doi: 10.1126/science.1110591
PubMed Abstract | Crossref Full Text | Google Scholar
Everhart, J. E., Ruhl, C. E. (2009). Burden of digestive diseases in the United States part II: lower gastrointestinal diseases. Gastroenterology 136, 741–754. doi: 10.1053/j.gastro.2009.01.015
PubMed Abstract | Crossref Full Text | Google Scholar
Ha, C. W., Lam, Y. Y., Holmes, A. J. (2014). Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J. Gastroenterol. 20, 16498–16517. doi: 10.3748/wjg.v20.i44.16498
PubMed Abstract | Crossref Full Text | Google Scholar
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
PubMed Abstract | Crossref Full Text | Google Scholar
Hecht, G., Koutsouris, A., Pothoulakis, C., LaMont, J. T., Madara, J. L. (1992). Clostridium difficile toxin B disrupts the barrier function of T84 monolayers. Gastroenterology 102, 416–423. doi: 10.1016/0016-5085(92)90085-d
PubMed Abstract | Crossref Full Text | Google Scholar
Hooper, L. V., Littman, D. R., Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Sci. (New York N.Y.) 336, 1268–1273. doi: 10.1126/science.1223490
PubMed Abstract | Crossref Full Text | Google Scholar
Jones, R. B., Fodor, A. A., Peery, A. F., Tsilimigras, M. C. B., Winglee, K., McCoy, A., et al. (2018). An aberrant microbiota is not strongly associated with incidental colonic diverticulosis. Sci. Rep. 8, 4951. doi: 10.1038/s41598-018-23023-z
PubMed Abstract | Crossref Full Text | Google Scholar
Kudo, T., Itoh, T., Miyadoh, S., Shomura, T., Seino, A. (1993). Herbidospora gen. nov., a new genus of the family Streptosporangiaceae Goodfellow et al. 1990. Int. J. systematic bacteriology 43, 319–328. doi: 10.1099/00207713-43-2-319
PubMed Abstract | Crossref Full Text | Google Scholar
Kvasnovsky, C. L., Leong, L. E. X., Choo, J. M., Abell, G. C. J., Papagrigoriadis, S., Bruce, K. D., et al. (2018). Clinical and symptom scores are significantly correlated with fecal microbiota features in patients with symptomatic uncomplicated diverticular disease: a pilot study. Eur. J. Gastroenterol. Hepatol. 30, 107–112. doi: 10.1097/MEG.0000000000000995
PubMed Abstract | Crossref Full Text | Google Scholar
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
PubMed Abstract | Crossref Full Text | Google Scholar
Lemes, V. B., Galdino, G. G., Romão, P., Reis, S. T. (2021). The relation between the diet and the diverticulitis pathophysiology: an integrative review. Arquivos gastroenterologia 58, 394–398. doi: 10.1590/S0004-2803.202100000-66
PubMed Abstract | Crossref Full Text | Google Scholar
Li, J., Fan, C., Wang, J., Tang, B., Cao, J., Hu, X., et al. (2024). Association between gut microbiota and anxiety disorders: a bidirectional two-sample mendelian randomization study. BMC Psychiatry 24, 398. doi: 10.1186/s12888-024-05824-x
PubMed Abstract | Crossref Full Text | Google Scholar
Liang, J., Liu, G., Wang, W., Xue, H. (2024). Causal relationships between gut microbiota and lymphoma: a bidirectional Mendelian randomization study. Front. Cell. infection Microbiol. 14. doi: 10.3389/fcimb.2024.1374775
PubMed Abstract | Crossref Full Text | Google Scholar
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
PubMed Abstract | Crossref Full Text | Google Scholar
Lopetuso, L. R., Petito, V., Graziani, C., Schiavoni, E., Paroni Sterbini, F., Poscia, A., et al. (2018). Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disorders. Digestive Dis. (Basel Switzerland) 36, 56–65. doi: 10.1159/000477205
PubMed Abstract | Crossref Full Text | Google Scholar
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
PubMed Abstract | Crossref Full Text | Google Scholar
Marasco, G., Buttitta, F., Cremon, C., Barbaro, M. R., Stanghellini, V., Barbara, G. (2023). The role of microbiota and its modulation in colonic diverticular disease. Neurogastroenterol. Motil. 35, e14615. doi: 10.1111/nmo.14615
PubMed Abstract | Crossref Full Text | Google Scholar
Mj, O., Turner, G. A., S., A., Frizelle, F. A., P., R. (2022). Distinct changes in the colonic microbiome associated with acute diverticulitis. Colorectal Dis. 24, 1591–1601. doi: 10.1111/codi.16271
PubMed Abstract | Crossref Full Text | Google Scholar
Peery, A. F., Crockett, S. D., Murphy, C. C., Jensen, E. T., Kim, H. P., Egberg, M. D., et al. (2022). Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2021. Gastroenterology 162, 621–644. doi: 10.1053/j.gastro.2021.10.017
PubMed Abstract | Crossref Full Text | Google Scholar
Qin, Y., Havulinna, A. S., Liu, Y., Jousilahti, P., Ritchie, S. C., Tokolyi, A., et al. (2022). Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 54, 134–142. doi: 10.1038/s41588-021-00991-z
PubMed Abstract | Crossref Full Text | Google Scholar
Quaglio, A. E. V., Grillo, T. G., De Oliveira, E. C. S., Di Stasi, L. C., Sassaki, L. Y. (2022). Gut microbiota, inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 28, 4053–4060. doi: 10.3748/wjg.v28.i30.4053
PubMed Abstract | Crossref Full Text | Google Scholar
Raineri, E. J. M., Altulea, D., van Dijl, J. M. (2022). Staphylococcal trafficking and infection-from 'nose to gut' and back. FEMS Microbiol. Rev. 46, fuab041. doi: 10.1093/femsre/fuab041
PubMed Abstract | Crossref Full Text | Google Scholar
Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G. A. D., Gasbarrini, A., et al. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7, 14. doi: 10.3390/microorganisms7010014
PubMed Abstract | Crossref Full Text | Google Scholar
Sakon, H., Nagai, F., Morotomi, M., Tanaka, R. (2008). Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. systematic evolutionary Microbiol. 58, 970–975. doi: 10.1099/ijs.0.65456-0
PubMed Abstract | Crossref Full Text | Google Scholar
Sandler, R. S., Everhart, J. E., Donowitz, M., Adams, E., Cronin, K., Goodman, C., et al. (2002). The burden of selected digestive diseases in the United States. Gastroenterology 122, 1500–1511. doi: 10.1053/gast.2002.32978
PubMed Abstract | Crossref Full Text | Google Scholar
Severn, M. M., Horswill, A. R. (2023). Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat. Rev. Microbiol. 21, 97–111. doi: 10.1038/s41579-022-00780-3
PubMed Abstract | Crossref Full Text | Google Scholar
Tursi, A., Scarpignato, C., Strate, L. L., Lanas, A., Kruis, W., Lahat, A., et al. (2020). Colonic diverticular disease. Nat. Rev. Dis. Primers 6, 20. doi: 10.1038/s41572-020-0153-5
留言 (0)