Neonatal calves (< 1-2 month old) are immunologically naïve and vulnerable to infectious diarrhea, a worldwide serious disease often caused by the protozoa Cryptosporidium parvum (C. parvum) with clinical symptoms including profuse watery stools, dehydration, acidosis, lethargy, and in rare cases, death (Lombardelli et al., 2019). C. parvum develops within a parasitophorous vacuole extracytoplasmatically but intracellularly in intestinal epithelial cells (IEC), primarily in the ileum, but also distal jejunum, and leads to profound changes in the IECs’ morphology, physiology, and transcription. C. parvum intestinal infection results in partial enlargement and swelling of lymph nodes, mild to moderate villus atrophy, detrimental effects on the mucosal and epithelial integrity, hindering normal absorption and secretion mechanisms, impairing the epithelial barrier function, and leading to an increase in intestinal permeability and leaky gut syndrome (Dumaine et al., 2021; Helmy and Hafez, 2022; Gamsjäger et al., 2023).
The intestinal barrier (IB) is primarily composed of the epithelium tightly sealed by tight junctions (TJ) and a mucus layer on top, which regulates nutrient absorption and functions as a physical barrier against harmful pathogens (Rios-Arce et al., 2017). The TJ are composed of several transmembrane proteins, including occludin (OCLN), claudins (CLDN), and immunoglobulin superfamily proteins, including junctional adhesion molecules (JAM), which interact with cytoskeletal linker proteins such as zonula occludens (ZO), forming a complex architecture that activates a multitude of cellular processes to maintain barrier integrity (Ahn et al., 2016; Usuda et al., 2021). There are other molecules and junctions located below TJ, such as adherens junctions (AJ), desmosomes, gap junctions (GJ), cell adhesion molecules (CAMs), and G protein-coupled receptors (GPCRs) superfamily, that are involved in cell-cell adhesion or binding to extracellular matrix (Schnell et al., 2013). Pathogens have evolved a variety of tactics to exploit junctional structures or destroy them and thereby providing a pathway into the underlying tissue, often provoking inflammatory cascades and diarrhea (Guttman and Finlay, 2009). A pathogen’s effects on intestinal permeability not only increase the risk of other pathogens’ invasion, but also influence absorption mechanisms, nutrient pools, and systemic metabolism (Aurora and Sanford, 2015; Di Vincenzo et al., 2024).
The major pathway for vectorial nutrient absorption across IECs is transcellular transport, which involves specific transporter proteins such as the ATP-binding cassette (ABC) transporter and the solute carriers (SLC) superfamilies (Klaassen and Aleksunes, 2010). In active transport, ABC transporters export substrates from cells in an energy-dependent manner, whereas in secondary active transport, members of the SLC family transport substances from the extracellular space (influx facilitated transporters) through electrochemical potential differences or ion gradients generated by primary active transporters (Haberkorn et al., 2021). SLC and ABC transporters can transport a wide variety of molecules, ranging from simple ions, sugars and amino acids to complex substrates such as lipids, vitamins, proteins, and xenobiotics, with a broad range of specificity from low to high even within a family. Transporters are essential for maintaining metabolic homeostasis, but also play a key physiological role in many cellular functions. Thus, deficits in transporters, even in one, depending on their degree of specification, can cause serious health problems (Lin et al., 2015). In addition to transporters, channels are also important players in the transport system, facilitating transport of water and other small solutes across biological membranes (Hodges and Gill, 2010). However, this area has been largely overlooked in the pathophysiology of infectious diarrhea in neonatal calves.
The recent development of high-throughput RNA-Sequencing (RNA-Seq) technologies has provided hypothesis-neutral information about the composition and abundance of transcriptomes. Combined with bioinformatics analysis, this could provide unique insight into a pathophysiological condition that is usually hard to model. Developing management and treatment for cryptosporidiosis can be greatly influenced by a deep understanding of the epithelial barriers, junctions, and transport mechanisms. The objective of the present study was therefore to investigate different aspects of intestinal cell junctions as well as substrate translocation in neonatal calves infected with C. parvum using next-generation sequencing (NGS).
2 Material and methods2.1 Experimental design and sample collectionThis study was framed under a recent comprehensive project described in detail by (Dengler et al., 2023) in accordance with the German legislation on the protection of animals, and licensed by the Landesdirektion Leipzig as TVV 19/20. In brief, ten healthy male neonatal calves (Bos taurus, Holstein-Friesian) were selected and transported to the University of Leipzig within their first 24 h of life. At day 1 of life, calves were infected by oral application of 2 × 107C. parvum oocysts (infected, n=5) or received pure water (CTRL, n = 5). Infection was confirmed through regular clinical monitoring of calves, measurement and scoring of fecal consistency, and examination of fecal shedding of C. parvum oocysts using the immunofluorescence assay kit MERIFLUOR® Cryptosporidium/Giardia (Meridian Bioscience, Inc., Cincinnati, USA). In addition, a snap test (BoDia, Fassisi, Göttingen, Germany) was used to test the presence of E. coli K99, rotavirus, coronavirus, and C. parvum in fecal samples before and 7 days post infection. After slaughtering the calves on day 7 post infection, the jejunum epithelium was manually stripped off from the underlying muscle and stored at -80°C.
2.2 RNA extraction, quantification, library construction, and sequencingThe RNA extraction, library preparation, and NGS analysis was previously described in detail (Veshkini et al., 2024). In brief, individually extracted RNA (NucleoSpin RNA kit, Macherey-Nagel, Düren, Germany) from each sample was assessed for RNA integrity number and quantified using a Bioanalyzer (Agilent Genomics, Waldbronn, Germany) and Qbit (Fisher Scientific), respectively. mRNA transcriptome libraries were synthesized using Illumina TruSeq Stranded mRNA library preparation kits (Illumina, San Diego, USA) and 1 microgram of total RNA as an input, and sequenced on an HiSeq2500 (Illumina, San Diego, USA) with 2 x 100 bp cycles.
2.3 Data processing, read mapping, and statistical analysisAfter demultiplexing, the obtained reads were analyzed for quality control, trimming, alignment and expression counts at gene and transcript level using the nf-core RNAseq v3.4 pipeline (https://nf-co.re/rnaseq/3.12.0) with default settings. For read alignment, the ARS-UCD1.2_Btau5.0.1Y assembly run 9 (Hayes and Daetwyler, 2019) without unplaced NKLS contigs served as backbone. The Bos taurus Ensembl genome annotation v105 provided gene and transcript coordinates. Gene expression counts were calculated via Salmon (v. 1.5.2) within the nf-core RNAseq pipeline.
Differentially expressed genes (DEG) were assessed by setting up an infection versus control group model in the DESeq2 1.26.0 package (Love et al., 2014) and limited to the transcripts having a transcript per million (TPM) value > 1 in at least 4 samples. Calculated P-values were adjusted for multiple testing via Benjamini-Hochberg. Only genes with a false discovery rate (FDR)BH < 0.05 were accepted as statistically significant.
2.4 Statistical and bioinformatic analysisThe bioinformatic analysis was limited to a subset of DEG related to junctions (TJ, AJ, GJ), CAM, GPCRs, transporters, and ion channels (termed “barriers and transporters-DEG”, Supplementary Table S1) due to the large number of identified DEG. The immune-related DEG were discussed elsewhere (Veshkini et al., 2024). In order to select candidate genes, we searched our database against public databases such as STRING (Version 12), DAVID (https://davidbioinformatics.nih.gov/, version 2023 q1), and TransportDB (version 2.0). Gene ontology (GO) annotation and functional enrichment analysis including biological process and cellular component was executed by ClueGo (v2.5.9) in Cytoscape software according to the following criteria: FDR < 0.05 and having at least two identified DEG within each pathway (Bindea et al., 2009, 2013). The network nodes and edges data were retrieved from the ClueGo plugin and plotted using yFiles radial layout in Cytoscape software (v3.9.1).
3 Results3.1 RNA sequencingIn the mucosal cells, 12,908 expressed genes at TPM >1 in at least four samples were identified, including 11,844 known genes and 1,064 genes with yet unknown function (ENSBTAG) (Supplementary Table S2). Principal component analysis (PCA) was used to compare gene expression patterns in mucosal cells collected from infected and non-infected calves (Figure 1). Despite unsupervised analysis, the two groups stand as two well-separated clusters, which indicates large differences in gene expression between the two groups related to the infection. The PC1 and PC2 together explain nearly 70% of the total variation.
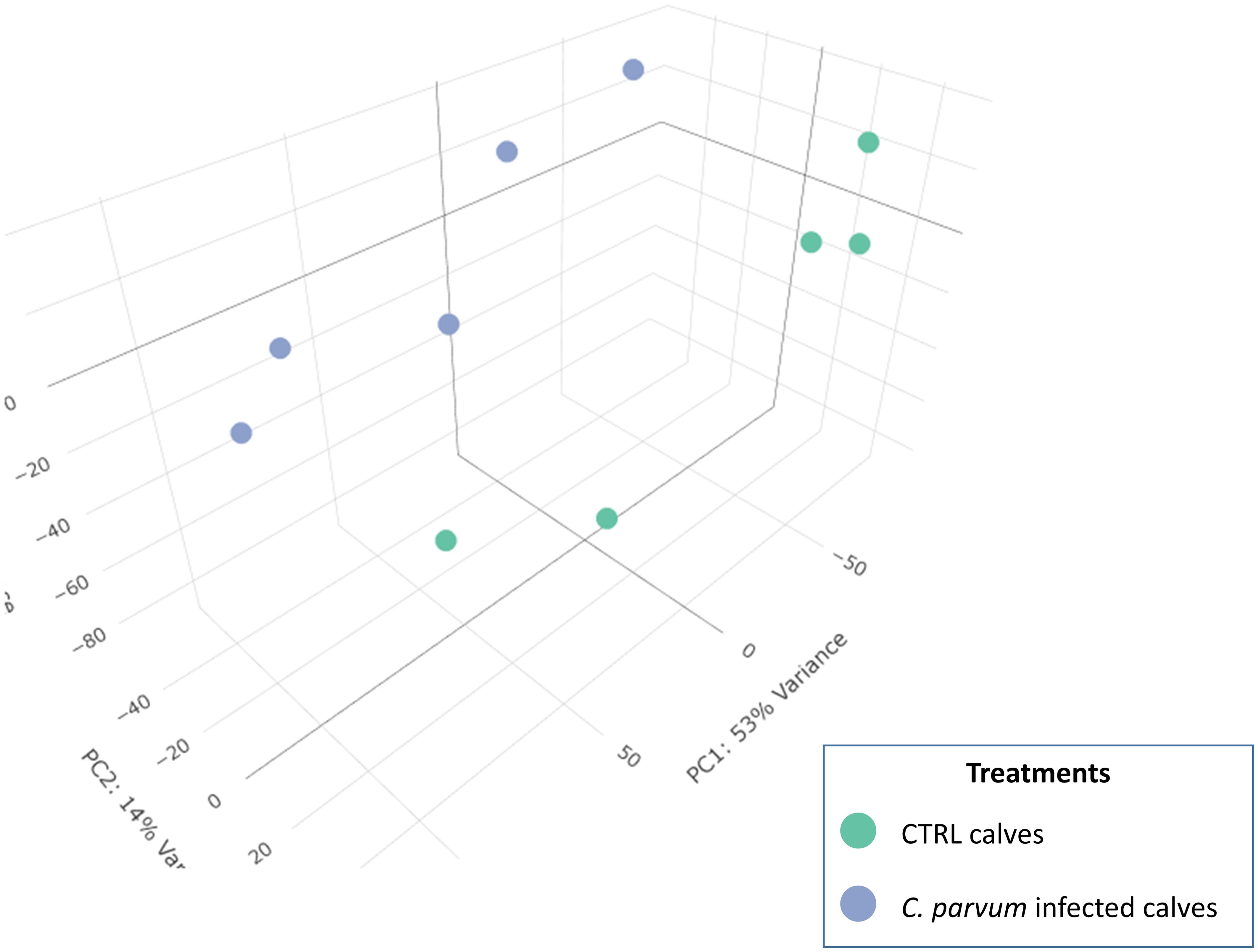
Figure 1. 3D Principal component analysis (PCA) of the normalized RNAseq data in calves infected or not with Cryptosporidium parvum. A PCA is an unsupervised machine learning model that identifies individuals with similar characteristics to bring out strong patterns from large and complex datasets. The dots represent individual calves and are grouped into two recognizable clusters based on treatment groups (infected or not with C. parvum). This observation emphasizes that the treatment effect is the primary factor affecting the gene expression results in the current dataset. There is a value associated with each component (PC), which indicates its variance percentage explained by the model.
3.2 Differentially expressed gene analysis between infected and non-infected calves and their associated biological pathwaysThere were 562 lower and 405 higher expressed genes at a FDR < 0.05, log2-fold change range: ≈ -6 to +6 (Supplementary Table S3). As a complement to our latest study focused on immune markers, this study focused on barriers and transporters-DEG including transcripts of 460 gene encoding TJ, AJ, GJ, CAM, ATPase transporters, ABC transporters, SLC transporters, and channels (Supplementary Table S1). Among barrier- and transporter-DEGs, there were 59 genes with higher expression and 109 genes with lower expression in infected calves as highlighted in the Figure 2 (the full list is provided in Supplementary Table S4).
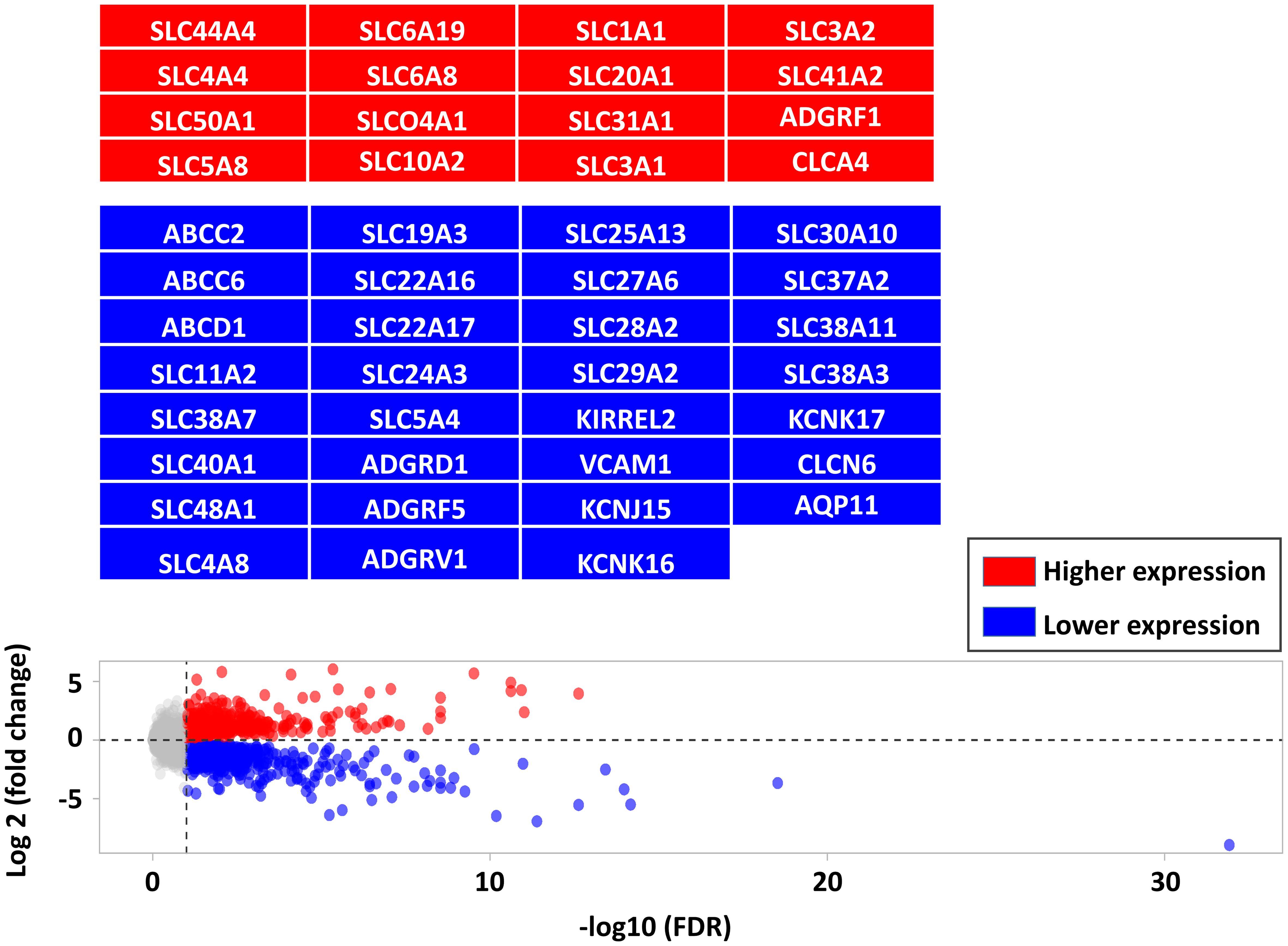
Figure 2. Comparing the differentially expressed genes (DEG, FDR < 0.05) between CTRL and infected calves. DEG are shown in red (higher expressed in infected calves) and blue (lower expressed in infected calves) and highlighted in corresponding boxes.
C. parvum infection significantly impacted the transcript expression of AJ, CAM, GPCR, transporters, and channels but not TJ or GJ. Gene ontology analysis was performed to predict up- and downregulated genes’ biological processes and their directions in a holistic manner and highlighted the key genes involved in the network (Figure 3). Among the pathways annotated to the DEG are those involved in the transport and metabolism of organic acids, carbohydrate derivate, lipids and fatty acids, amino acids, and ions (cations and anions) (Supplementary Table S5). The network shows deregulated pathways involved in transmembrane transport of ions, carbohydrates, and other organic compounds, affecting the cellular homeostasis.
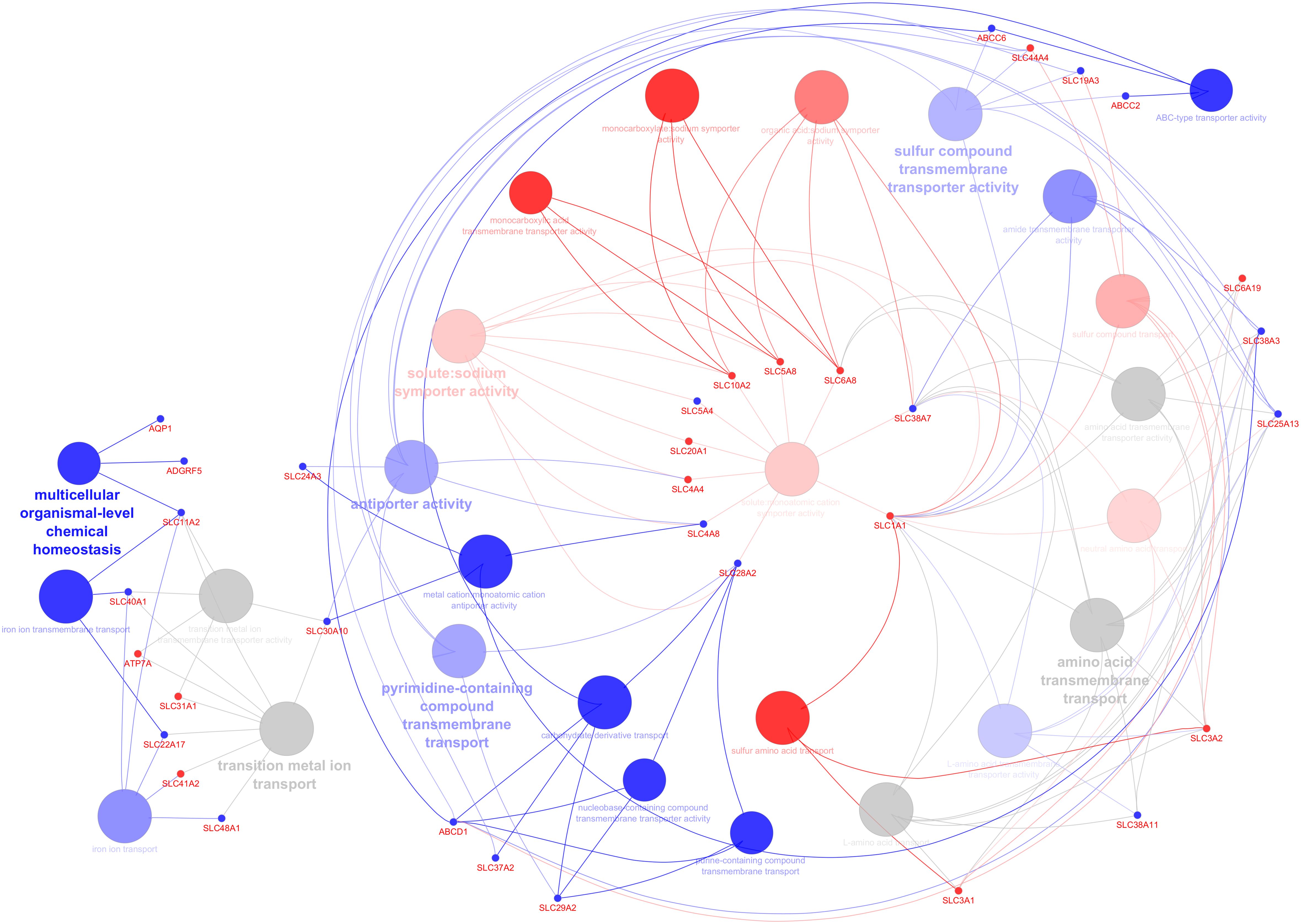
Figure 3. Predicting the biological processes’ direction associated with differentially expressed genes in the infected calves. The network shows interconnections among different biological pathways. Biological pathways are colored according to their predicted upregulation (red), downregulation (blue), or neutral (gray, the direction is not distinguishable). The hue intensity corresponds to false discovery rate. The highlighted genes are those that were initially or centrally involved in pathway enrichment, with red and blue dots indicating genes with higher and lower expression, respectively.
At cellular component level, DEGs were annotated to various cell compartments including the morphological structure of the cellular membrane such as basolateral, and brush border as well as intracellular organelles such as endosome and endoplasmic reticulum (Figure 4; Supplementary Table S6). The number of differentially expressed cell membrane-associated genes are higher in the apical part of the cell than in the basolateral part.
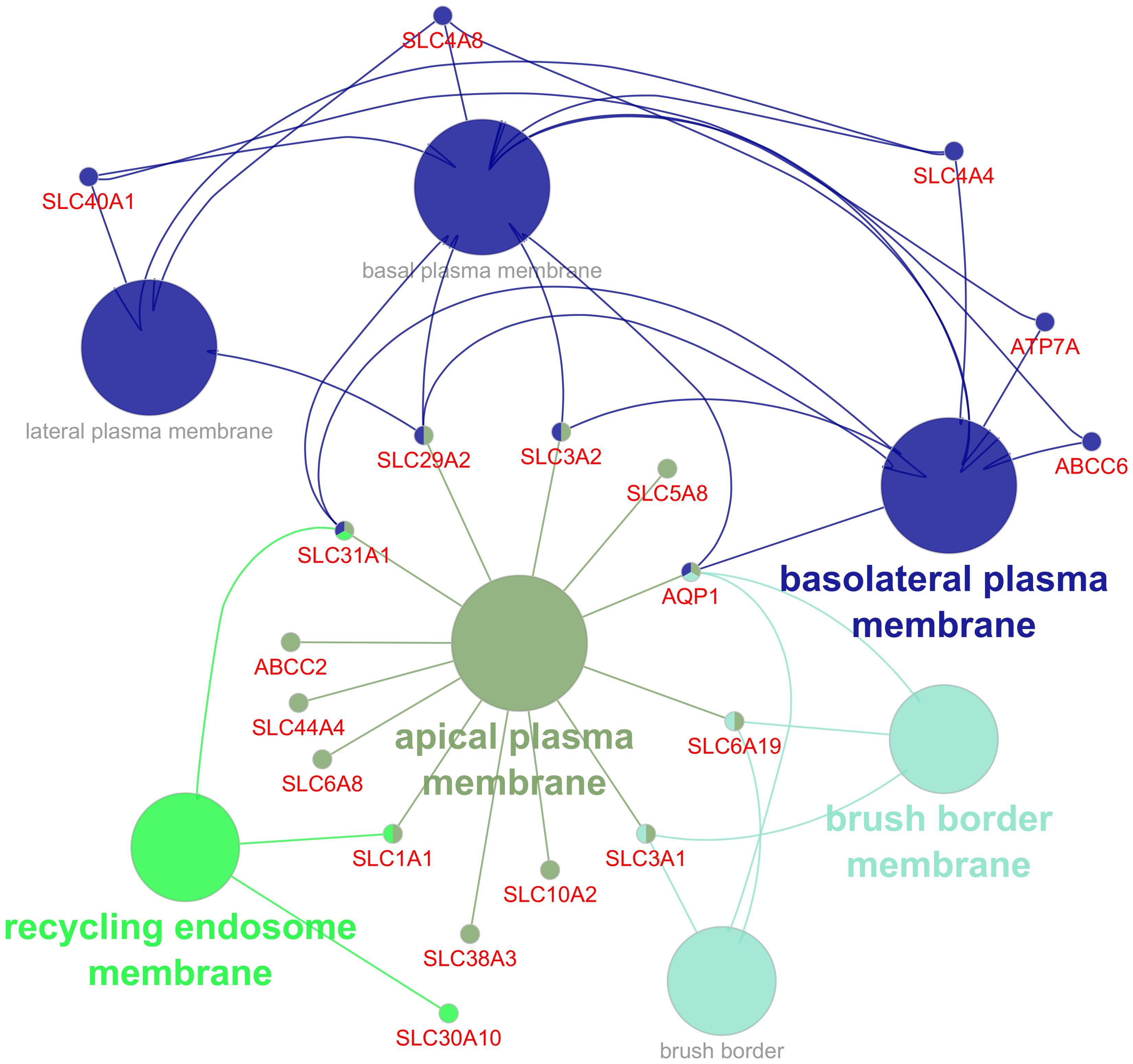
Figure 4. Gene ontology (GO) functional enrichment analysis of cellular component (CC) annotated to differentially expressed genes in the infected group. Different colors indicate clusters automatically assigned by the ClueGo, with circles representing pathways and dots representing genes.
4 DiscussionThere has been evidence that C. parvum infection is associated with the disruption of the host’s intestinal epithelial barrier integrity and increased permeability; however, the underlying molecular mechanisms in dairy calves have not been completely elucidated. We have previously reported the impact of C. parvum on calves’ clinical health status, zootechnical performance, and immunohematological parameters (Dengler et al., 2023; Veshkini et al., 2024). In brief, our histopathological results showed that C. parvum-infected calves had villus atrophy and reduced villus-crypt ratios, suggesting tissue damage (Supplementary Figure S7). The possibility of cellular damage by C. parvum may also be supported by the enrichment of oxidative stress, apoptosis, and programmed cell death signaling pathways in the jejunum mucosa of the infected calves (Supplementary Table S8). Additionally, C. parvum infection was confirmed at the jejunum tissue by comparing the abundance of oxidative stress, apoptosis, and programmed cell death related proteins extracted from our proteomics analysis (Supplementary Figures S9, S10).
The infected calves had lower glucose and higher urea plasma concentrations, but body weight, internal body temperature, and immunohematology markers were not significantly changed. Regarding the immune system, we have found that inflammatory signaling pathways such as Toll-like receptors (TLRs) and the nuclear factor-kappa B (NF-κB) are induced following infection, but the adaptive immune response is not yet fully responsive (Veshkini et al., 2024). This study complements those findings and mainly focused on aspects of calf intestinal nutrient transport during C. parvum infection. There are distinct pathways used by the intestinal epithelium to transport substances from the gut lumen into the circulation or vice versa, depending on their size, hydrophobicity, and physicochemical characteristics. To better comprehend the effect of C. parvum on intestinal permeability and pathophysiology of diarrhea, the following sections will discuss the molecular aspects of each pathway separately.
4.1 Paracellular transport and intestinal permeability regulationParacellular transport involves passive but selective transport of substances across an epithelium through cell junctions, including TJ, AJ, GJ, desmosomes, and hemidesmosomes, which are arranged into at least two distinct pathways: high-capacity pores (mainly regulated by CLDN family) and low-capacity leaks (regulated by OCLN and ZO family) (Zheng et al., 2021). During stimulation or pathological conditions, disruption of these junctions’ integrity can increase paracellular permeability, allowing pathogens to enter the systemic circulation and affect systemic immune function (Chelakkot et al., 2018).
C. parvum infection influenced the expression of calcium-dependent adhesion (cadherins superfamily) through reduction of cadherin related family member 1 (CDHR1) and cadherin 13 (CDH13), and induction of CDH26 and protocadherin 1 (PCDH1). The cadherin family is required for the formation of AJ (George and Beeching, 2006) and consequently TJ (McCole, 2014), thus their deregulation may reflect on barrier functions. In accordance, studies in mice and humans have shown that C. parvum infection downregulates components of TJs and AJs to translocate through the intestinal mucosa and invade their host (Kumar et al., 2018; Lamisere et al., 2022). There is evidence that the physical damage to the epithelial cells by C. parvum contributes to increased intestinal permeability or leaky gut syndrome in calf and mice models (De Sablet et al., 2016; Gamsjäger et al., 2023), in which dysregulation of AJs might be one of the reasons. In contrast to TJs, which seal paracellular gaps between adjacent epithelial cells and maintain electrochemical gradients necessary for efficient transcellular ion transport (Tsukita et al., 2001), AJs provide mechanical linkages (cell–cell contacts) between adjacent epithelial cells (Yap et al., 1997). This evidence suggests that C. parvum infection may impair junctions integrity, resulting in increased intestinal permeability, and in turn facilitating secondary infections.
Other adhesion molecules, such as those belonging to the CAM and GPCR families, were also affected by infection, including a lower expression of vascular cell adhesion molecules 1 (VCAM1), Kirre-like nephrin family adhesion molecule 2 (KIRREL2), adhesion G protein-coupled receptors (ADGR)-D1, ADGRF5, and ADGRV1, and a higher expression of ADGRF1. Unfortunately, the role of CAM and GPCR families in the pathophysiology of diarrhea in ruminants is poorly understood. Gut permeability and development also depend heavily on inflammatory and immune responses (Jridi et al., 2020). As a part of interactions between hosts and pathogens, tumor necrosis factor (TNF)-α and interleukin (IL)-1β produced by inflammatory monocytes aid C. parvum in changing the intestinal barrier and permeability (De Sablet et al., 2016). A previous mouse model experiment revealed that C. parvum infection altered immunohistochemical localization of Wnt pathway components as well as adherens junction ultrastructure in ileocecal epithelium (Benamrouz et al., 2014). TNFα signaling induces VCAM-1 expression, which regulates inflammation-related vascular adhesion and leukocyte migration across the endothelial barrier and entry into sites of immune complex deposition (Kong et al., 2018). Also, the ADGR family appears to transmit external signals, such as stimulants, to induce physiological responses that regulate mucosal immunity and maintain intestinal barrier function (Sun et al., 2017). At one side, physical damage caused by C. parvum infection affects TJ and AJ’s regulation and localization; however, inflammation and immune responses may also be their regulators.
4.2 Transmembrane transport4.2.1 ATP-binding cassette and ATPases transportersThe primary active transporters are ATP-dependent, including the family of ABC transporters and ATPases (ion pumps) that transport ions, lipids, carbohydrates, and xenobiotics against a concentration gradient (Schumann et al., 2020). ABC transporters play a crucial role in the pathophysiology of infectious diarrhea for two reasons: they facilitate secretion of various ions such as chloride and modulate xenobiotic absorption, distribution, metabolism, and secretion, but they can also interact with pathogens, potentially contributing to host protection (Mercado-Lubo and McCormick, 2010).
A number of ABC transporter genes were expressed in our data set (Supplementary Table 2), but only ABCC2, ABCC6, and ABCD1 expressions were reduced by infection. Nies and Keppler (2007) previously reviewed the substrate specificity of human and rat ABCC2. In the gut epithelium, ABCC2 localizes exclusively in the apical brush border membrane of villi, serving to efflux anions such as chloride and excrete endogenous and xenobiotic substances, especially in conjunction with glutathione, glucuronate, or sulfate into the intestinal lumen (Nies and Keppler, 2007). During intestinal infection (by Salmonella typhimurium), ABCC2 was shown to be upregulated (Pazos et al., 2008), probably to facilitate the apical efflux of the neutrophil chemoattractant hepoxilin A3 (HXA3) (Mrsny et al., 2004). Even though C. parvum induces intestinal inflammation, we have previously shown that neutrophil chemotaxis and differentiation associated pathways were downregulated (Veshkini et al., 2024), suggesting a minor role of neutrophils in neonatal calves right after the peak of infection. ABC transporter expression is under the control of transcription factors such as the retinoid X receptor (RXR), pleomorphic adenoma gene (PLAG), and zinc finger transcription factor specificity protein 1 (SP1) which imprints a tissue-specific expression (Abruzzese et al., 2022). In contrast to ABCC2, the ABCC6 transporter is located at the basolateral membrane and serves multiple functions, however the precise nature of its substrate(s) is unknown (Vanakker et al., 2013). It can be postulated that nutrient drainage by C. parvum interferes with the ABCC2 and ABCC6 function and expression, thereby disrupting the secretion and homeostasis of one or more (yet unknown) substrates. Furthermore, ABCD1, which is located at the peroxisome membrane, facilitates the import of coenzyme A-activated saturated very long-chain fatty acids (VLCFA; >C22:0) into peroxisomes for break down by β-oxidation, a crucial step in activating innate immunity, especially pro-inflammatory signaling in monocytes and macrophages (Zierfuss et al., 2022). Since fatty acids are required for both glycolysis and lipogenesis to cover the rapid energy demands and produce pro-inflammatory mediators, a downregulation of ABCD1 appears to interfere with the immune response and lead to the accumulation of VLCFA in plasma and tissues.
In addition, C. parvum infection affected members of ATPase transporters, resulting in higher expression of ATPase Ca2+ transporting 2 (ATP2A2) and ATPase Copper Transporting Alpha (ATP7A). ATP2A2 is an intracellular pump primarily located in the sarcoplasmic reticula and endoplasmic reticula of muscle tissues (Dhitavat et al., 2003). It functions as a regulator of the contraction/relaxation cycle by hydrolyzing ATP and translocating calcium from the cytosol into the sarcoplasmic reticulum lumen, thus probably playing a role in the intestinal motility to compensate diminished absorption. This claim is, however, unsupported by any evidence, and further research is required to approve it.
4.2.2 SLC transportersSolute carrier proteins (SLC) are a large and diverse group of membrane proteins which mediate passive and secondary active transport of ions, nucleotides, and sugars across biological membranes. In this study 240 solute carrier proteins (SLC) were identified in our data set (Supplementary Table 2), of which 18 genes had higher and 14 genes had lower expression levels (Figure 2). Bioinformatics analysis suggested that the transport and homeostasis of cation, anion, and metal ion, carboxylic acid, sulfur, pyrimidine containing compounds, nucleobase containing compounds, purine containing compounds, and carbohydrate derivate were disturbed due to the lower expression levels of SLC48A1, SLC22A17, SLC40A1, SLC11A2, SLC30A10, SLC37A2, SLC29A2, SLC24A3, SLC4A8, SLC5A4, SLC19A3 and the higher expression levels of SLC3A2, SLC20A1, SLC6A19, SLC31A1, SLC4A4, SLC5A8, SLC10A2, SLC6A8 (Figure 3; Supplementary Table S5).
In the pathophysiology of diarrhea, dysregulation of intestinal ion transport is a critical factor (Das et al., 2018). Some of the differentially expressed SLC transporters are involved in the ion transport system across (in or out) cell membranes. In particular, influx divalent metal transporter 1 (DMT1), also known as SLC11A2, is the solute transporter responsible for transport of ferrous iron across the brush border membrane of intestinal epithelial cells (Kayaaltı et al., 2015). Afterwards, the basolateral efflux channel ferroportin (FPN, SLC40A1) mediates the intracellular translocation and export of iron into the circulation in conjunction with multicopper ferroxidase, including hephaestin (Heph) and/or ceruloplasmin (Cp), and hepcidin (Dlouhy et al., 2019). These two SLC transporters are exclusively iron transporters, and their downregulation in our study indicates diminished iron turnover. Iron availability is crucial not only to support erythropoiesis, immune cell differentiation and function, metabolic processes, and cellular respiration but also essential for pathogens’ proliferation and pathogenicity (Grander et al., 2022). There is evidence indicating a close relationship between DMT1 and FPN transporters and inflammation. Previous work reported that intestinal inflammation interferes with iron metabolism and DMT1 expression, affecting erythropoietic (Urrutia et al., 2013) and macrophage function (Grander et al., 2022). Activated Toll-like receptors (TLRs) and inflammatory factors such as IL-6 are known to induce the production of hepcidin, which consequently reduces iron availability by suppressing the expression of FPN1 through the hepcidin-FPN1 axis (Cai et al., 2021). TNF exposure reduced DMT1 expression in human intestinal cells (in vitro) (Johnson et al., 2004) and reduced duodenal iron transport in mice (in vivo) (Laftah et al., 2006). In accordance, we reported earlier that TNF-α is induced in response to C. parvum infection but neither erythrocytes and immune cell count nor hemoglobin and hematocrit concentrations were significantly affected (Veshkini et al., 2024).
The other group of differentially expressed SLC transporters in our study are involved into transport of more complex compounds such as sugars, amino acids, vitamins, nucleotides, carbohydrates, oligopeptides, and drugs. In the small intestine, thiamine (Vitamin B1) is transported across cell membranes by SLC19A3, where it is phosphorylated to its active form, thiamine pyrophosphate (TPP), and acts as a cofactor of transketolase. TPP is later transported by SLC25A19 to mitochondria, where it is a cofactor of pyruvate dehydrogenase complexes, branched chain ketothiolase dehydrogenases, and alpha-ketoglutarate dehydrogenases (Plecko and Steinfeld, 2017). SLC19A3 downregulation while SLC25A19 remains unchanged may affect the pyruvate oxidation in TCA cycle as well as energy and amino acids metabolism. SLC25A13, encoding a citrin protein, is an important mitochondrial solute transporter involved in the urea metabolism by swapping mitochondrial aspartate for cytosolic glutamate, where it can be used later for nucleotide synthesis pathways (Lin et al., 2011). Thus, it can be postulated that downregulation of SLC25A13 may affect the malate-aspartate shuttle, gluconeogenesis, amino acid homeostasis, TCA cycle, and protein turnover.
The higher expression levels of SLC transporters appears to be due to feedback mechanisms initiated by inflammation as compensation for vital compounds and also to regulate the activity of immune cells, but less information is available in cattle. ASBT is an active sodium-dependent bile acid transporter (encoded by SLC10A2) responsible for ileal bile acid (BA) reabsorption and has also been linked to intestinal disorders like diarrhea (Jung et al., 2004). Moreover, it plays an important role in regulating lipid and cholesterol homeostasis as well as determining the size of the BAs pool (Han et al., 2015). ASBT gene expression is primarily controlled by transcription factors such as caudal-type homeobox-1 (CDX1) and -2 (CDX2) and hepatocyte nuclear factor 1-α (HNF1-α), nuclear receptors such as retinoid X receptor (RXR) and farnesoid x receptor (FXR), and intestinal flora, which plays a critical role in both pathological and physiological processes as reviewed by (Yang et al., 2020). The reasons for the SLC10A2 higher expression is not clear but it seems to be a compensatory mechanism to reabsorb BA and decrease the excretion of fecal BAs and keep the liver cholesterol homeostasis in check.
Solute carrier family 1, member 1 (SLC1A1; also known as EAAT3 or EAAC1) is a major epithelial transporter of L-glutamate and D/L-aspartate especially in the intestines (Bailey et al., 2011; Sheng et al., 2022). The sodium bicarbonate cotransporter (NBCe1) encoded by SLC4A4 plays a role in pH regulation and homeostasis by transporting the bicarbonate from the blood to the lumen (Cappellesso et al., 2022). Positive correlations were found between SLC1A1 expression and levels of infiltrating CD8+T cells and dendritic cells, and between SLC4A4 expression and CD8+T cell infiltration levels (Zhou et al., 2020). A number of organic anion transporting polypeptides (OATPs) substrates are transported by SLCO4A1, including steroid hormone conjugates, pro-inflammatory prostaglandin PGE2, thyroid hormones, and xenobiotics. SLCO4A1 expression is under control of inflammation-associated pathways such as NF-κB, and TNF-receptor 2 signaling cascades (Koller et al., 2022). As immune cells depend on SLCs to induce rapid and robust metabolic reprogramming, thereby controlling their expression through major immune associated nuclear transcription factors, this seems to be a consequence of an activated immune response.
4.2.3 Channels and membrane transporters with channel like propertiesIn our study, a subset of genes was identified to function as ion channels, notably chloride channels, potassium channels (KCN family), magnesium transporters, copper transporters, and heme transporters (Supplementary Table 2). Among those, the infected calves had lower expression of KCNJ15, KCNK16, and KCNK17, as well as chloride voltage-gated channel 6 (CLCA6), whereas calcium-sensitive chloride transporting channel 4 (CLCA4) had higher expression.
During infectious diarrhea, disruption of ion channel function may lead to alterations in electrolyte, nutrient, and fluid transport, potentially causing acid-base imbalances. In this regard, a good example would be the complex relationship between ion channels and lactose absorption in the intestinal epithelium under diarrheal conditions (Misselwitz et al., 2019). C. parvum-induced diarrhea leads to reduced lactase activity at the brush border, lower absorption of lactose metabolites and sodium via sodium/glucose cotransporter 1 (SGLT1), and increased passage of lactose to the distal small intestine and colon (Gomez et al., 2022). Due to the coupled transport of sodium and potassium with glucose and galactose, reduced lactose absorption means less glucose is available for SGLT1, potentially decreasing sodium absorption.
The function of potassium channels is closely connected with the (re)absorption of Na+, Cl−, and water as well as secretion of K+, HCO3− in the basolateral and luminal membranes of the epithelial cells (Heitzmann and Warth, 2008). Chloride channels regulate intestinal Cl− secretion and have emerged as potential targets in the diarrhea treatment (Ko et al., 2014; Thiagarajah et al., 2015). At the crypt base, cells predominantly secrete fluid containing Cl− and HCO3– but the surface epithelial cells’ function is to reabsorb Na+ and to secrete HCO3− and K+ to preserve the ion gradient at the crypt-villus axis (Heitzmann and Warth, 2008). Therefore, ion channel deregulation, leading to acid-base imbalances, as a part of physiologic and metabolic pathways leading to bacterial D-lactate production, could partly explain diarrheic calf metabolic (D-lactic) acidosis.
The absorption of chloride is essential for fluid absorption, and when dysregulated by infection, it affects the absorption of water and other solutes. In this regard, another cluster of DEG was associated with the transportation of solutes such as water and cholesterol (Supplementary Table 4). In response to infection, aquaporin (AQP11) expression was reduced, whereas ATPase transporters (ATP2A2, ATP7A) expression were significantly elevated. AQPs are water channel proteins that facilitate the absorption of fluid (primarily water) in many tissues including the small intestine and colon and are responsive to osmotic gradients (Zhu et al., 2016; Tomita et al., 2017; Zhang et al., 2019). The osmoregulation and mucosal fluid fluxes associated with ion and aquaporin channels make them a potential target for diarrhea treatment.
5 ConclusionA thorough understanding of the pathophysiology of infectious diarrhea, including the absorption and excretion mechanisms, is the critical step for developing antidiarrheal agents. Using NGS and bioinformatics analysis, this study investigated how host-pathogen interactions may affect intestinal epithelial permeability and cellular transport mechanisms. The study highlighted critical DEGs encoding junctions, adhesions molecules, transporters, and channels associated with the pathophysiology of infectious diarrhea and provided an insight into their complex interactions, expressions, and regulations after infection. Among, infection-induced differential expression of adhesion molecules such as the cadherin family may affect intestinal permeability and paracellular absorption. Meanwhile, differential expressions of SLC and ABC transporters, along with ion and water channels, not only affect nutrient transmembrane transport, but also affect acid-base balance, electrochemical gradients, and osmotic homeostasis, which may partly contribute to metabolic acidosis in neonatal calves. Further studies and validation with additional methods should be conducted to determine which of these transporters of channels are potent candidate for designing new treatment strategies, whether these changes are related to pathogen impact or perhaps as a results of immune response preventing pathogens from obtaining host nutrients.
Data availability statementThe datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statementThe animal study was approved by Landesdirektion Leipzig as TVV 19/20. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributionsAV: Data curation, Formal analysis, Visualization, Writing – original draft. CK: Data curation, Methodology, Resources, Writing – review & editing. FD: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review & editing. LB: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. WL: Investigation, Methodology, Project administration, Writing – review & editing. CH: Investigation, Methodology, Writing – review & editing. RU: Investigation, Methodology, Project administration, Writing – review & editing. CD: Investigation, Methodology, Resources, Writing – review & editing. HH: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by a grant from the Leipzig veterinary junior scientist support program financed by the “Freundeskreis Tiermedizin”, the Faculty of Veterinary Medicine, and by Ceva Santé Animale. Core budget of the FBN.
AcknowledgmentsThe authors express their appreciation to Claudia Kmiecik, Birgit Wego, Hilke Brandt, Kristine Gürtler for tissue sampling and to Simone Wöhl, Manuela Lötzke and Sebastian Gaedecke for RNA isolation, library preparation and NGS sequencing. We are most grateful to Britta Beck, Maxi Berberich, Thomas Grochow, Manuela Kirchner, Clara Kiesewetter, Anna Kraft, Lea-Christina Murnik, Beate Schneidewind, Andreas Schaller, Melanie Stölzle and René Schumacher for their support with the animal experiments.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1495309/full#supplementary-material.
ReferencesAbruzzese, V., Sukowati, C. H. C., Tiribelli, C., Matera, I., Ostuni, A., Bisaccia, F. (2022). The expression level of ABCC6 transporter in colon cancer cells correlates with the activation of different intracellular signaling pathways. Pathophysiology 29, 173–186. doi: 10.3390/pathophysiology29020015
PubMed Abstract | Crossref Full Text | Google Scholar
Ahn, C., Shin, D. H., Lee, D., Kang, S. M., Seok, J. H., Kang, H. Y., et al. (2016). Expression of claudins, occludin, junction adhesion molecule A and zona occludens 1 in canine organs. Mol. Med. Rep. 14, 3697–3703. doi: 10.3892/mmr.2016.5725
PubMed Abstract | Crossref Full Text | Google Scholar
Aurora, R., Sanford, T. (2015). Host microbiota contributes to health and response to disease. Mo Med. 112, 317–322.
Bailey, C. G., Ryan, R. M., Thoeng, A. D., Ng, C., King, K., Vanslambrouck, J. M., et al. (2011). Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J. Clin. Invest. 121, 446–453. doi: 10.1172/jci44474
PubMed Abstract | Crossref Full Text | Google Scholar
Benamrouz, S., Conseil, V., Chabé, M., Praet, M., Audebert, C., Blervaque, R., et al. (2014). Cryptosporidium parvum-induced ileo-caecal adenocarcinoma and Wnt signaling in a mouse model. Dis. Model. Mech. 7, 693–700. doi: 10.1242/dmm.013292
PubMed Abstract | Crossref Full Text | Google Scholar
Bindea, G., Galon, J., Mlecnik, B. (2013). CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663. doi: 10.1093/bioinformatics/btt019
PubMed Abstract | Crossref Full Text | Google Scholar
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
PubMed Abstract | Crossref Full Text | Google Scholar
Cai, C., Zeng, D., Gao, Q., Ma, L., Zeng, B., Zhou, Y., et al. (2021). Decreased ferroportin in hepatocytes promotes macrophages polarize towards an M2-like phenotype and liver fibrosis. Sci. Rep. 11, 13386. doi: 10.1038/s41598-021-92839-z
PubMed Abstract | Crossref Full Text | Google Scholar
Cappellesso, F., Orban, M.-P., Shirgaonkar, N., Berardi, E., Serneels, J., Neveu, M.-A., et al. (2022). Targeting the bicarbonate transporter SLC4A4 overcomes immunosuppression and immunotherapy resistance in pancreatic cancer. Nat. Cancer 3, 1464–1483. doi: 10.1038/s43018-022-00470-2
PubMed Abstract | Crossref Full Text | Google Scholar
Chelakkot, C., Ghim, J., Ryu, S. H. (2018). Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 1–9. doi: 10.1038/s12276-018-0126-x
PubMed Abstract | Crossref Full Text | Google Scholar
Das, S., Jayaratne, R., Barrett, K. E. (2018). The role of ion transporters in the pathophysiology of infectious diarrhea. Cell Mol. Gastroenterol. Hepatol. 6, 33–45. doi: 10.1016/j.jcmgh.2018.02.009
PubMed Abstract | Crossref Full Text | Google Scholar
Dengler, F., Hammon, H. M., Liermann, W., Görs, S., Bachmann, L., Helm, C., et al. (2023). Cryptosporidium parvum competes with the intestinal epithelial cells for glucose and impairs systemic glucose supply in neonatal calves. Veterinary Res. 54, 40. doi: 10.1186/s13567-023-01172-y
PubMed Abstract | Crossref Full Text | Google Scholar
De Sablet, T., Potiron, L., Marquis, M., Bussière, F. I., Lacroix-Lamandé, S., Laurent, F. (2016). Cryptosporidium parvum increases intestinal permeability through interaction with epithelial cells and IL-1β and TNFα released by inflammatory monocytes. Cell Microbiol. 18, 1871–1880. doi: 10.1111/cmi.12632
PubMed Abstract | Crossref Full Text | Google Scholar
Dhitavat, J., Dode, L., Leslie, N., Sakuntabhai, A., Lorette, G., Hovnanian, A. (2003). Mutations in the sarcoplasmic/endoplasmic reticulum ca2+ ATPase isoform cause darier’s disease. J. Invest. Dermatol. 121, 486–489. doi: 10.1046/j.1523-1747.2003.12410.x
PubMed Abstract | Crossref Full Text | Google Scholar
Di Vincenzo, F., Del Gaudio, A., Petito, V., Lopetuso, L. R., Scaldaferri, F. (2024). Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Internal Emergency Med. 19, 275–293. doi: 10.1007/s11739-023-03374-w
PubMed Abstract | Crossref Full Text | Google Scholar
Dlouhy, A. C., Bailey, D. K., Steimle, B. L., Parker, H. V., Kosman, D. J. (2019). Fluorescence resonance energy transfer links membrane ferroportin, hephaestin but not ferroportin, amyloid precursor protein complex with iron efflux. J. Biol. Chem. 294, 4202–4214. doi: 10.1074/jbc.RA118.005142
PubMed Abstract | Crossref Full Text | Google Scholar
Dumaine, J. E., Sateriale, A., Gibson, A. R., Reddy, A. G., Gullicksrud, J. A., Hunter, E. N., et al. (2021). The enteric pathogen Cryptosporidium parvum exports proteins into the cytosol of the infected host cell. eLife 10, e70451. doi: 10.7554/eLife.70451
PubMed Abstract | Crossref Full Text | Google Scholar
Gamsjäger, L., Cirone, K. M., Schluessel, S., Campsall, M., Herik, A., Lahiri, P., et al. (2023). Host innate immune responses and microbiome profile of neonatal calves challenged with Cryptosporidium parvum and the effect of bovine colostrum supplementation. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1165312
PubMed Abstract | Crossref Full Text | Google Scholar
George, S. J., Beeching, C. A. (2006). Cadherin:catenin complex: A novel regulator of vascular smooth muscle cell behaviour. Atherosclerosis 188, 1–11. doi: 10.1016/j.atherosclerosis.2005.12.017
留言 (0)