Colorectal cancer (CRC) stands as a prevailing malignancy impacting the digestive tract, presenting a substantial threat to global health. In 2020, the global incidence of CRC accounted for approximately 1.93 million cases (10%),[1] rendering it the third most prevalent cancer type, following breast cancer and lung cancer. Moreover, CRC significantly impacted cancer-related mortality, with approximately 920,000 deaths (9.2%), ranking second among all cancer-related mortalities.[2] In China, the incidence of CRC accounted for approximately 550,000 cases (12.2%), positioning as the second most prevalent cancer type.[3] Correspondingly, CRC-related mortalities reached approximately 280,000 deaths (9.5%), securing its place as the fifth leading cause of cancer-related deaths in the country.[4] The incidence and mortality rates of CRC in China exhibit a gradual elevation compared with the global average, signifying a rapid increase in the burden imposed by its malignancy.[2,4] Contrary to developed countries experiencing a gradual decrease in CRC incidence and mortality, China continues to report a progressive rise in cases.[5] Furthermore, CRC cases have been frequently reported among the younger Chinese population, with higher burden in males, urban areas, and the eastern regions of China than in females, rural areas, and the western regions, respectively.[6]
Despite the effectiveness of early surgery and post-operative chemotherapy, CRC remains challenging due to its high recurrence rate, metastasis, and drug resistance.[7,8] The presence of heterogeneity and poor prognosis necessitates identifying new targets for developing novel anti-tumor drugs.[9,10] One potential target is ribosomal protein L22-like 1 (RPL22L1), a protein-coding gene that regulates ribosome composition through RPL22 and governs morphogenesis by modulating pre-messenger RNA (mRNA) splicing.[11,12] In addition, prior studies have linked RPL22L1 to the promotion of ovarian cancer (OV) metastasis through epithelial-to-mesenchymal transition,[13] and its involvement has been correlated with unfavorable prognostic outcomes and 5-fluorouracil resistance in CRC.[14] Moreover, investigations have suggested its promising potential as a diagnostic and prognostic marker in prostate cancer progression.[15,16] However, the precise function and underlying mechanisms of RPL22L1 concerning CRC growth are still unknown.
The key objective of this study is to use bioinformatics analysis and molecular biology studies to thoroughly investigate RPL22L1 expression and its prognostic implications in CRC. It also aims to elucidate the underlying molecular mechanism responsible for its cancer-promoting role, emphasizing its potential as a promising anti-tumor target.
MATERIAL AND METHODS EthicsAll experimental procedures adhered to the Standard Operating Procedures for the Laboratory Animal and were approved by the Biological and Medical Ethics Committee, School of Basic Medical Sciences of Jiamusi University (JDJCYXY2023041). This study does not involve patients, therefore informed consent from patients is not applicable.
Bioinformatics analysisRNA expression profiles and clinical information for colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) were retrieved from the Cancer Genome Atlas (TCGA) through the University of California Santa Cruz (UCSC) Xena website (https://xenabrowser.net/datapages/). The RPL22L1 expression data underwent log2-transformation for intergroup comparison. Moreover, datasets from various sources, including GSE9348, GSE23878, GSE73360, GSE81582, GSE20916, GSE8671, GSE39582, and GSE17536, were obtained from the publicly available Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The R (4.2.1) software was used for analyzing and visualizing the expression alterations of RPL22L1 between different cancer tissues and corresponding normal tissues, employing the “ggplot2” package. The online tool The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) (https://ualcan.path.uab.edu/analysis-prot.html) was employed to assess RPL22L1 protein levels,[17] whereas TISCH2 (http://tisch.comp-genomics.org/home/) was used to examine the expression of RPL22L1 in various cells within the CRC tumor microenvironment (TME).[18]
Subsequently, the prognostic value of RPL22L1 expression in CRC was evaluated. The “surv_cutpoint” function of R package “survminer” was used to calculate the optimal cutoff value of RPL22L1 expression, and the “survfit” function of R package “survival” was used to analyze the difference in prognosis between the two groups.
Then, Gene Set Enrichment Analysis (GSEA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were conducted using the “clusterProfiler” package.[19]
Finally, the correlation between RPL22L1 expression and CRC TME index was analyzed. “ESTIMATE” (1.0.13) package, a method for inferring the degree of infiltration of stromal and immune cells in tumors, was used to calculate the stromalScore, immuneScore, and ESTIMATEScore of each tumor patient. Pearson’s correlation coefficient was calculated between RPL22L1 expression and each index of TME using the “corr.test” function of the R package “psych” (2.1.6).
Cell culture and groupingNormal colonic epithelial cells (NCM460, BNCC339288), CRC cell lines SW480 (BNCC100604), LOVO (BNCC338601), RKO (BNCC100173), and HCT116 (BNCC287750) were purchased from BeiNa Culture Collection (Xinyang, China, https://www.bncc.com/) and grown at 37°C in 5% carbon dioxide (CO2) in high-glucose Dulbecco’s Modified Eagle Medium (11965092, ThermoFisher Scientific, Waltham, MA) with L-glutamine (A2916801, ThermoFisher Scientific), 10% fetal bovine serum (FBS, 10099158, ThermoFisher Scientific), and 1% penicillin/Streptomycin solution (10 μL/mL, C0222, Beyotime, Shanghai, China). All cells were identified through Short Tandem Repeat (STR) profiling and tested negative of mycoplasma contamination.
Cells were allocated to the groups of control short hairpin RNA (shCtrl) and shRPL22L1 for our subsequent analysis on the effects of RPL22L1 on CRC growth. In addition to these groups, cells were further allocated to the groups of Rapamycin (CRC cells were treated with 10 nM rapamycin (S1842, Beyotime) for 48 h) and shRPL22L1+MHY1485 (CRC cells were subjected to the RNA interference and treated with 5 μM MHY1485 [A426126, Sangon Biotech, Shanghai, China] for 48 h) to delve into the effects of RPL22L1 on mechanistic target of rapamycin complex 1 (mTORC1) in CRC.
RNA interferenceIn the transient interference experiment, 5.0 × 105 cells were treated with 4 μL Lipo8000™ (C0533, Beyotime) and 80 pmol small interfering RNA (siRNA) at the specific target locations (GenePharma, Shanghai, China). They are labeled as the groups of siRNA-52, siRNA-141, and siRNA-325, and those treated with the negative control siRNA were allocated to the group of NC. The cells were lysed in TriZol reagent (15596026, ThermoFisher Scientific) for subsequent investigations after 72 h. Supplementary Table 1 contains a list of the siRNA sequences employed.
CRC cells were infected with LV10N-Puro-RPL22L1-Homo-52 (GenePharma) at an Multiplicity of Infection (MOI) of 10 and allocated to the group of shRPL22L1 to achieve RPL22L1 knockdown for stable interference, and those infected with the control lentivirus were named as the group of shCtrl.
Cell growth assayIn this procedure, six-well plates were injected with 5.0 ×104 RKO and/or HCT116 cells from the various groups. The cells were stained using crystal violet solution (C0121, Beyotime), photographed in a digital camera (D500, Nikon, Tokyo, Japan), and counted after 72 h to determine the relative cell number. In addition, the DepMap portal (https://depmap. org/portal/) was used to analyze the impact of RPL22L1 on CRC cell growth.[20]
Wound healing assayIn six-well plates, 80% confluent monolayers of RKO and HCT116 cells were cultured. Sterilized 200 μL pipette tips were used to create the wound, and the debris was gently removed by washing with the medium. After culturing for 24 h in serum-free medium, the narrowing of the wound area was photographed on the inverted microscope (IX71, Olympus, Tokyo, Japan) and assessed using ImageJ software (National Institutes of Health, Bethesda, MD). The wound healing rate was calculated as ([0 h width - 36 h width]/0 h width) × 100%.
Transwell assayIn this experiment, 2 × 104 cells were suspended in 200 μL of serum-free medium in the upper chamber, whereas 600 μL of culture medium containing 10% FBS was added to the lower chamber of a Transwell plate (3422, Corning Inc., Corning, NY). The cells in the upper chamber were removed using a cotton swab after 36 h of incubation. Subsequently, the remaining cells were fixed and stained. An inverted microscope (IX71, Olympus) was then applied to observe and record the cells. The relative cell number was calculated as (number of cells in random fields/average number of three random fields in control group).
RNA-seq assayA total of 5 × 106 cells per sample were collected and sent to GENEWIZ Biotechnology Co., Ltd. for RNA-sequencing (RNAseq), with three replicates per group. For enrichment analysis, KEGG and GSEA were also employed.
Real-time reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)Total RNA isolation was carried out using Trizol (15596026, ThermoFisher Scientific), following the recommended guidelines of the manufacturer. For complementary DNA synthesis and real-time PCR, the BeyoFast™ SYBR Green One-Step qRT-PCR Kit (D7268S, Beyotime) was employed following the provided protocols. The relative level of RPL22L1 mRNA was determined using the 2-ΔΔCt method.[21] The primers applied in this research were synthesized by GENEWIZ (Suzhou, China) and are listed in Supplementary Table 2.
Western blotCell samples were lysed using radioimmunoprecipitation assay buffer (P0013B, Beyotime). Subsequently, equal amounts of the lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrohoresis (SDS-PAGE) gel electrophoresis and transferred onto Polyvinylidene Fluoride Membrane (PVDF) membranes (IPVH00010, Millipore, St Louis, MO). Following the 1 h blocking step with 5% non-fat milk dissolved in Tris buffer containing 1% Tween-20, the PVDF membranes were incubated with primary antibodies of RPL22L1 (1:1000, DF9870, Affinity Biosciences, Cincinnati, OH), marker of proliferation Ki-67 (MKI67, 1:1000, AG2646, Beyotime), Phospho-P70S6K (Thr389) (1:1000, #9205, Cell Signaling Technology, Danvers, MA), S6KB (1:1000, #34475, Cell Signaling Technology), minichromosomal maintenance complex component 3 (MCM3, 1:1000, AG2608, Beyotime), MCM7 (1:1000, AF7434, Beyotime), WEE1 G2 checkpoint kinase (WEE1, 1:1000, 29474-1-AP, Proteintech, Rosemont, IL), cleaved caspase 3 (1:1000, AF0081, Beyotime), E-cadherin (1:1000, 20874-1-AP, Proteintech), N-cadherin (1:1000, 22018-1-AP, Proteintech), growth arrest and DNA damage-inducible beta (GADD45B, 1:1000, ab205252, Abcam, Cambridge, UK), and Beta Actin (ACTB) (1:2000, AF5003, Beyotime) at 4℃ overnight. Subsequently, the membranes underwent incubation with the secondary antibody (A0208, Beyotime) at room temperature for 1 h. Visualization of the bands was accomplished using an efficient chemiluminescence (ECL) kit (P0018S, Beyotime), and the intensities of the bands were quantified using ImageJ software (National Institutes of Health).
Apoptosis and cell cycle detection using flow cytometryFor the cell cycle and apoptosis assay, the CRC cells were prepared according to the guidelines of the cell cycle and apoptosis analysis kit (C1052, Beyotime). Detection was conducted using a flow cytometer (Sysmex CyFlow Cube8, Norderstedt, Germany), and the data analysis was carried out using FCS Express V3 software (De Novo Software, Glendale, CA) and FlowJo v10.8.1 (FlowJo LLC., Ashland, OR).
Animal experimentsA total of 14 male BALB/c nude mice, aged 8–12 weeks, were acquired from Yangzhou University (Yangzhou, China) and routinely fed in the specific pathogen-free environment with the sterile rodent chow and tap water. For the xenograft assay, all mice were allocated to the shCtrl and shRPL22L1 groups (n = 7 for each group). Subsequently, each mouse received an injection of 7.0 × 106 CRC cells in 100 mL under the axillary arm. The mice were euthanized after 28 days using overdose of CO2, and the tumors were removed, captured on camera, and measured using a Vernier caliper.
Statistics analysisStatistical analyses were conducted using R software (4.2.1) and GraphPad Prism 9.5.1 (GraphPad Software Inc., La Jolla, CA). Unpaired Student’s t-test and paired t-test were used to compare differences between unpaired and paired samples, respectively. Survival curves were generated using the Kaplan–Meier method, and the significance of these differences was assessed using the log-rank test. Pearson’s test was conducted for correlation analysis. Furthermore, the criterion for determining statistical significance was set to P < 0.05.
RESULTS High expression of RPL22L1 in various cancers, including CRCThe Cancer Genome Atlas (TCGA) data analysis revealed that 7609 differentially expressed genes (DEGs) in colon adenocarcinoma (COAD) [Supplementary Figure 1a], 3612 Differently Expressed Genes (DEGs) in rectum adenocarcinoma (READ) [Supplementary Figure 1b], and 2882 DEGs were common in both CRC types [Supplementary Figure 1c]. The detailed information is shown in Supplementary Table 3. Following an extensive literature review, RPL22L1 was selected as the target gene for further research. A pan-cancer analysis was conducted to assess RPL22L1 expression in various tumors. In the context of unpaired samples, RPL22L1 expression exhibited a substantial upregulation in 26 cancer types (P < 0.05), encompassing COAD and READ, while displaying a significant downregulation in kidney chromophobe (KICH) and ovarian cancer (OV) (P < 0.01) [Supplementary Figure 1d]. Similarly, RPL22L1 expression exhibited significant upregulation in 15 cancer types (P < 0.05), including COAD and READ, while exhibiting a substantial downregulation in KICH (P < 0.01) [Supplementary Figure 1e]. Finally, pan-cancer analysis based on proteomics confirmed that the RPL22L1 protein was significantly upregulated in five tumors (P < 0.001), including COAD [Supplementary Figure 1f].
Moreover, comprehensive analysis of multiple datasets consistently exhibited a considerable upregulation of RPL22L1 expression in CRC tissues compared with normal tissues across various datasets, including GSE9348, GSE23878, GSE73360, GSE81582, GSE20916, GSE8671, and TCGA (P < 0.05) [Figure 1a-h]. In addition, RPL22L1 expression exhibited significantly higher levels in each pathological stage compared with normal tissues (P < 0.001) [Figure 1i], and the microsatellite instability-high (MSI-H) group exhibited significantly elevated RPL22L1 expression levels compared with the microsatellite stable (MSS) group (P < 0.001) [Figure 1j].
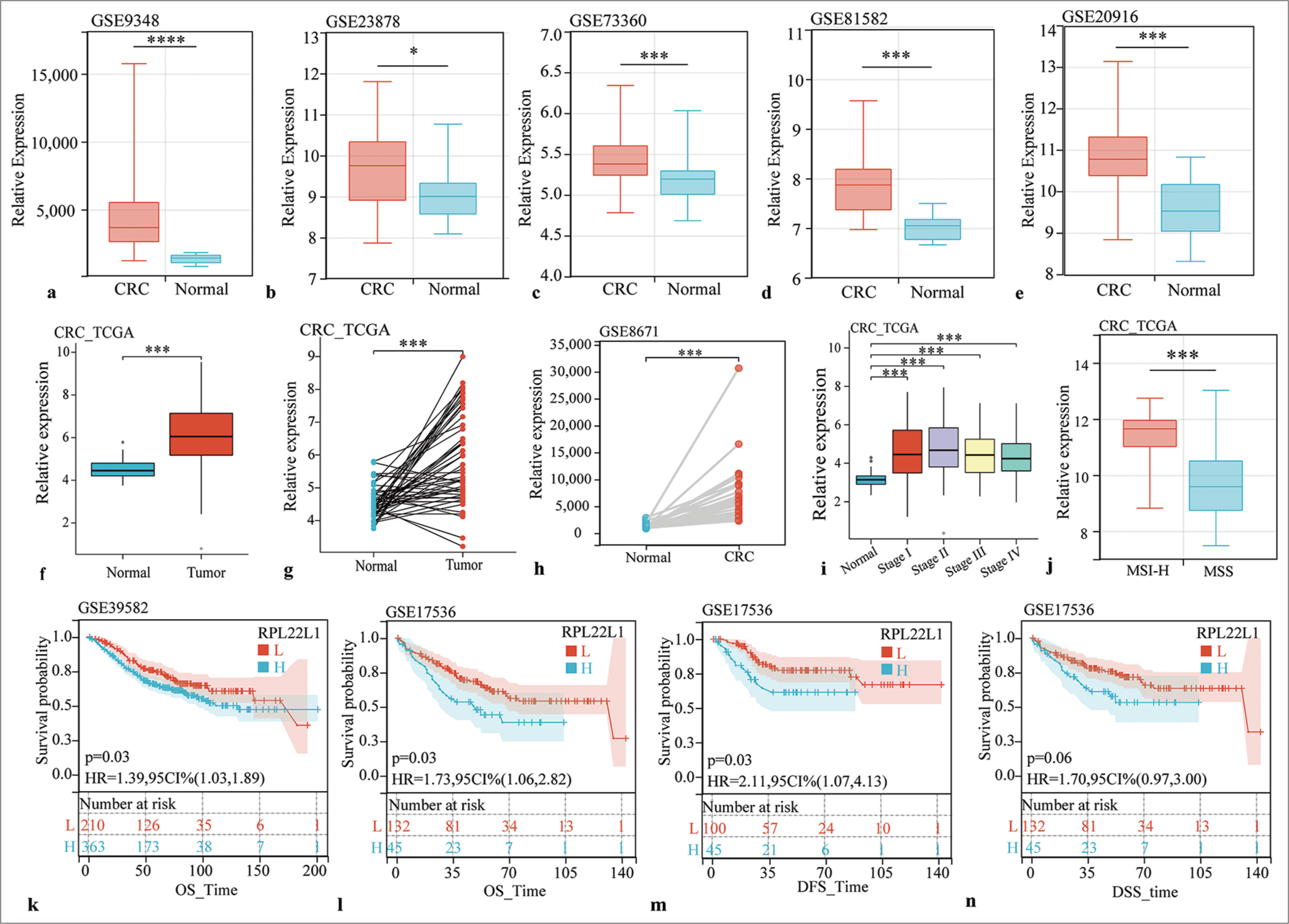
Export to PPT
Prognostic significance of RPL22L1Prognostic analyses in various datasets were conducted to explore the clinical relevance of high RPL22L1 expression in CRC. In GSE39582, a substantial association was observed between high RPL22L1 expression and poor overall survival (OS) (P = 0.03) [Figure 1k]. Furthermore, in GSE17536, a negative association was found between high RPL22L1 expression with OS (P = 0.03) [Figure 1l], disease-free survival (P = 0.03) [Figure 1m], and disease-specific survival (P = 0.06) outcomes [Figure 1n]. These findings suggest that the high expression of RPL22L1 can promote CRC, and targeting RPL22L1 could offer potential anticancer benefits.
GSEA analysis based on TCGA data suggested that RPL22L1 may be involved in the cell cycle, G2M checkpoint, mTORC1 signaling, Hypoxia pathway , and apoptosis pathways in COAD [Supplementary Figure 2a and b]. In addition, it may be involved in the cell cycle, E2F targets, G2M checkpoint, and mTORC1 signaling pathways in READ [Supplementary Figure 2c and d].
RPL22L1 expression is not correlated with the CRC TME indexThe top 20 DEGs in COAD and READ are additionally presented in Supplementary Figure 3a and b. RPL22L1 is expressed in various stromal and immunological cells in addition to tumor cells, according to the TISCH2 database study [Supplementary Figure 3c and d]. Furthermore, according to Pearson’s correlation coefficients, the expression of RPL22L1 was not substantially associated with the StromalScore, ImmunoScore, or ESTIMATEScore [Supplementary Figure 4ac], suggesting that RPL22L1 may not exhibit a remarkable impact on CRC TME regulation.
Knockdown of RPL22L1 inhibits proliferation and migration of CRC cell linesThe transient interference experiment revealed that all three pairs of siRNAs presented a significant reduction in RPL22L1 mRNA and protein levels (P < 0.001) [Supplementary Figure 5a-c]. Cellular experiment showed that the RPL22L1 protein level was increased in CRC cell lines compared with NCM460 cells (P < 0.05) [Figure 2a]. Moreover, the stably transfected cell lines exhibited a significant decrease in RPL22L1 mRNA and protein levels (P < 0.001, P < 0.001) in RKO [Figure 2b and c] and HCT116 cells [Figure 2d and e]. The knockdown of RPL22L1 resulted in significant inhibition of the proliferation in both RKO (P < 0.001) [Figure 2f] and HCT116 cells (P < 0.001) [Figure 2g]. Analysis of the DepMap database also confirmed that knocking out and/or knocking down the RPL22L1 gene inhibited the growth of various CRC cell lines, such as RKO and HCT116 [Figure 2h and i]. In addition, wound healing experiments revealed that knocking down RPL22L1 significantly inhibited the migration of RKO (P < 0.05) [Figure 2j] and HCT116 cells (P < 0.01) [Figure 2k], which was also confirmed by the Transwell experiments (P < 0.001) [Figure 2l].
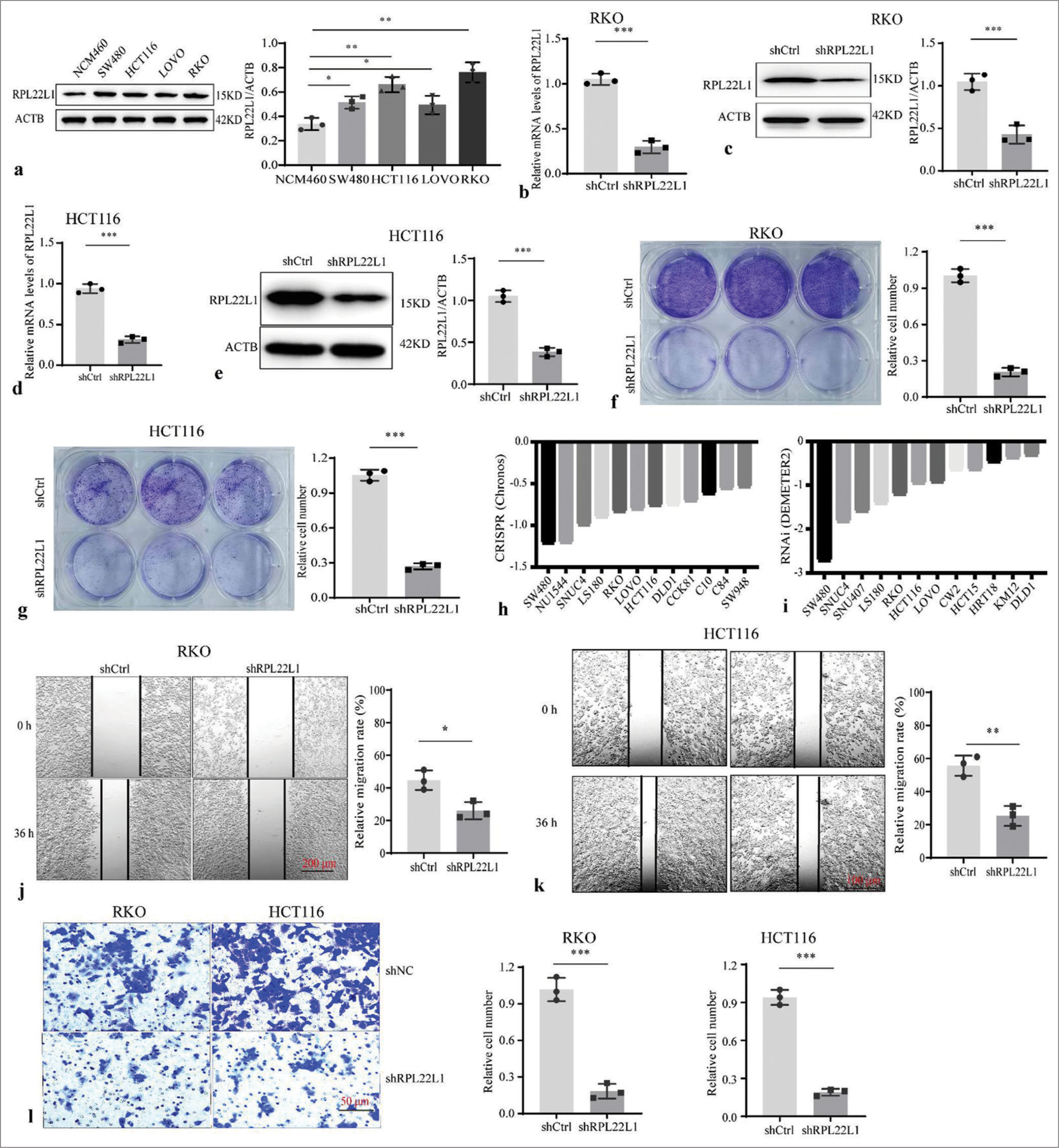
Export to PPT
Knocking down RPL22L1 arrests the cell cycle and promotes apoptosisFlow cytometry analysis was conducted to evaluate the influence on cell cycle and apoptosis, determining the underlying inhibitory effect of RPL22L1 knockdown on cell growth. Knocking down of RPL22L1 resulted in a significant arrest of the G1 and S phases of the cell cycle (P < 0.01, P < 0.05), along with a decrease in the percentage of cells in the G2 phase (P < 0.001) [Figure 3a and b]. Moreover, a remarkable increase in the proportion of apoptotic cells was observed on RPL22L1 knockdown (P < 0.001) [Figure 3c and d].
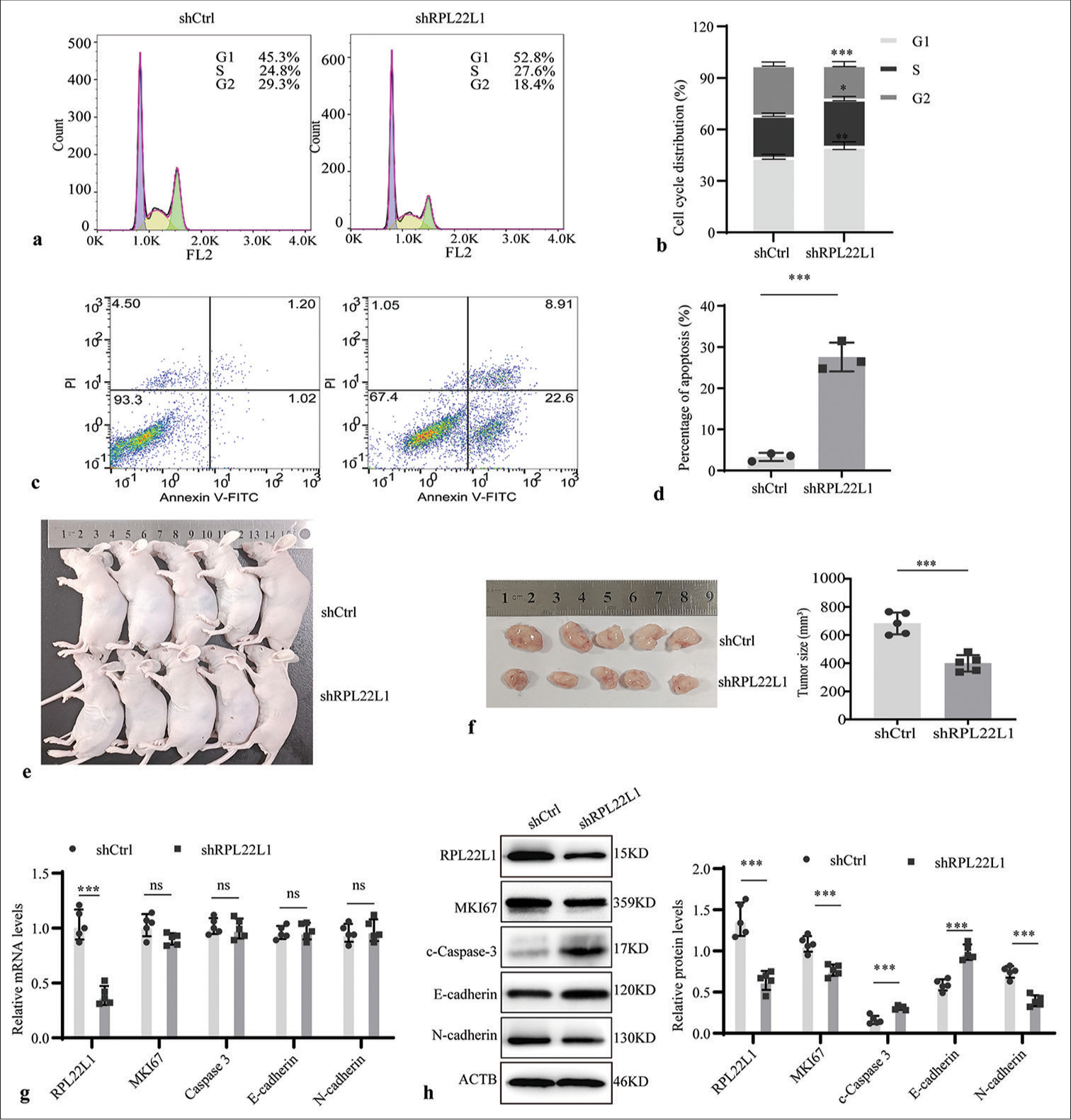
Export to PPT
Knocking down RPL22L1 reduced tumor volume in nude miceIn tumor-bearing mice, the knockdown of RPL22L1 remarkably reduced the tumor volume in vivo (P < 0.001) [Figure 3e and f]. In addition, the qRT-PCR results suggested that RPL22L1 silencing only diminished its mRNA level, exhibiting no evident effect on the levels of MKI67, Caspase-3, E-cadherin, and N-cadherin (P > 0.05) [Figure 3g]. Furthermore, Western blot analyses validated a substantial decrease in RPL22L1, MKI67, and N-cadherin and upregulation of Caspase 3 and E-cadherin in protein levels in the shRPL22L1 group when compared with the shCtrl group (P < 0.001) [Figure 3h].
mTOR pathway mediates the inhibitory effect of low levels of RPL22L1 on cell cycleAn RNAseq assay was carried out to understand the molecular mechanism underlying the tumor-suppressive effects of RPL22L1 knockdown [Supplementary Table 4]. The analysis unveiled 1672 upregulated genes and 946 downregulated genes after RPL22L1 knockdown [Figure 4a]. GSEA revealed that knocking down RPL22L1 promoted cell apoptosis and inhibited DNA replication and cell cycle [Figure 4b]. A heat map was used to illustrate the differential gene expression related to apoptosis, cell cycle, and mTORC1 signaling [Figure 4c]. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis highlighted the primary involvement of downregulated genes in E2F targets, G2M checkpoint, mTORC1 signaling, and HYPOXIA pathways [Figure 4d]. Finally, the knockdown of RPL22L1 substantially decreased the mRNA levels of MCM3, MCM7, GADD45B, and WEE1 (P < 0.001), whereas the levels of MKI67 and S6KB were evidently unaffected (P > 0.05) [Figure 4e]. The relevant results from western blot also manifested that the RPL22L1 knockdown reduced the protein level of MCM3, MCM7, GADD45B, WEE1, MKI67, and P70S6K (p < 0.05), whereas the SK6B level was unaffected evidently (P > 0.05) [Figure 4f and g]. Furthermore, the findings of the recovery experiment highlighted that the mTOR pathway may mediate the suppressive impact of reduced RPL22L1 expression on MCM3, MCM7, and WEE1 expressions, with the exception of GADD45B [Figure 4h].
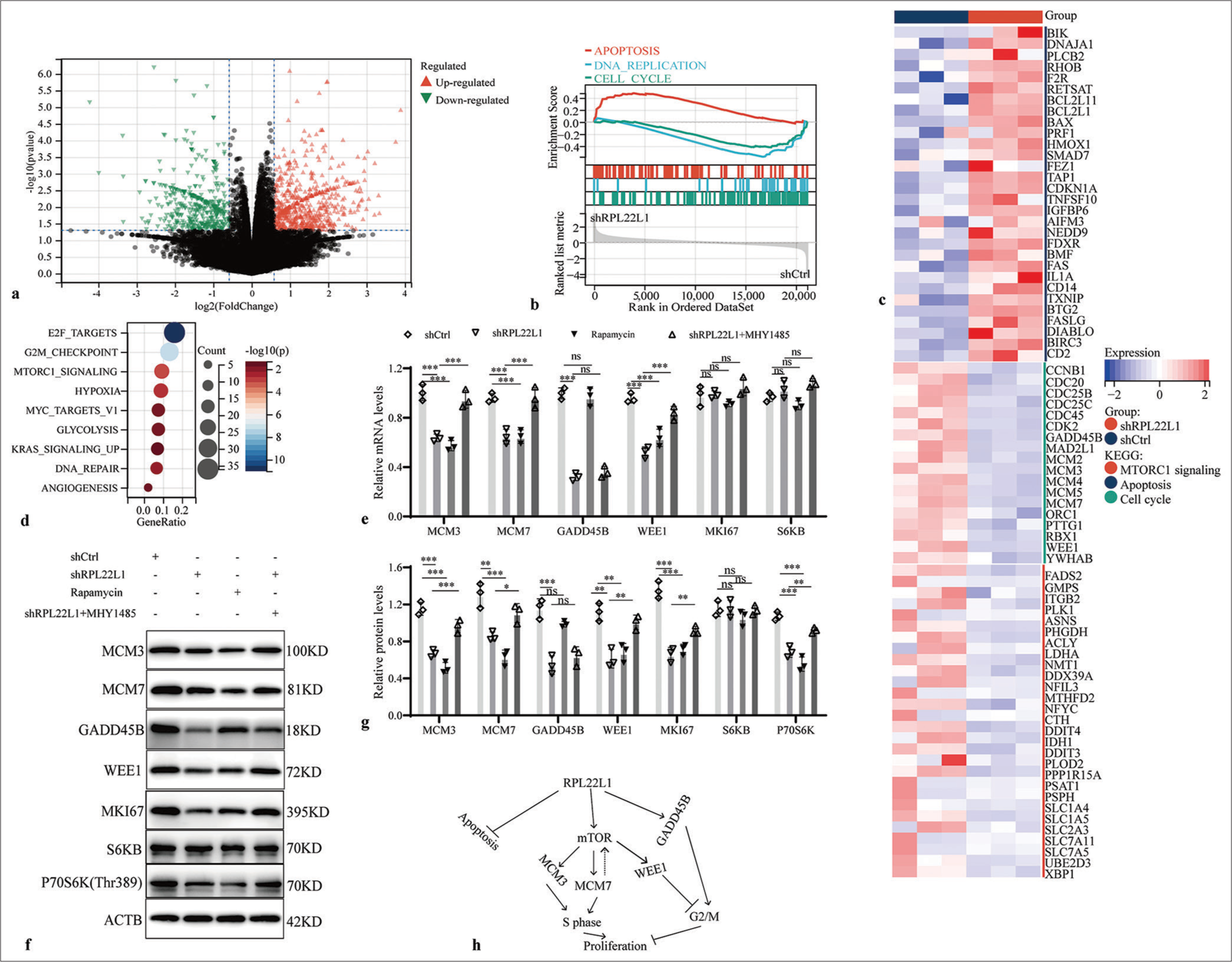
Export to PPT
DISCUSSIONCRC ranks as the third most prevalent cancer globally, and its incidence continuously increases. Patients with CRC present varied prognoses and respond differently to the same treatment plan.[22] The heterogeneity and drug resistance of CRC highlight the vital need to continuously develop new targets in anti-tumor research.[10,23]
To screen for novel anti-CRC targets, the expression profile data of COAD and READ were obtained from TCGA. Subsequent screening for DEGs yielded 2882 DEGs following the intersection. Finally, RPL22L1 gene was selected as the subject of investigation, considering the expression abundance of each gene and the existing literature. The pan-cancer analysis revealed a substantial elevation in the RPL22L1 mRNA and protein levels across various cancers, including CRC. Moreover, multiple GEO datasets consistently confirmed its upregulated expression in CRC. Notably, the MSI-H group exhibited significantly higher RPL22L1 expression levels than the microsatellite stable (MSS) group, highlighting the key function of the RPL22L1 gene in the survival of microsatellite instability-high (MSI-H) CRC cells. Furthermore, the expression of the RPL22L1 gene was detected in tumor cells, stromal cells, and immune cells. High RPL22L1 expression showed a significant association with unfavorable prognosis in CRC patients, indicating its potential pro-cancer effect. GSEA based on TCGA data revealed that RPL22L1 may participate in the cell cycle, G2M checkpoint, mTORC1 signaling, HYPOXIA, and E2F targets, further indicating that RPL22L1 acts as an oncogene in CRC.
Tumor microenvironment (TME), an important regulator of tumorigenesis, development, metastasis, and drug resistance, comprises endothelial cells, immune cells, stromal cells, proteins, and metabolites.[24,25] Analysis of the CRC TME revealed no significant correlation between RPL22L1 and TME index, suggesting that RPL22L1 may not exert its cancer-promoting function through TME regulation. Moreover, cytological function experiments, following RPL22L1 knockdown, exhibited a remarkable inhibition of proliferation and migration of CRC cell lines RKO and HCT116. The analysis of genome-wide knockdown and/or knockdown screening library data corroborated the crucial role of RPL22L1 as a dependent gene for the growth of various CRC cells, such as RKO and HCT116.[20]In vivo experiments further validated that RPL22L1 knockdown led to substantial inhibition of tumor growth. Flow cytometry also showed that the knockdown of RPL22L1 not only blocked the G1 and S phases of the cell cycle but also promoted apoptosis. Thus, these findings confirm that RPL22L1 has oncogenic potential in CRC.
An RNAseq assay was performed to unravel the molecular mechanism behind the cell cycle arrest and apoptosis promotion caused by RPL22L1 knockdown. A total of 1672 upregulated and 946 downregulated genes were identified as a result of RPL22L1 knockdown. Furthermore, GSEA suggested that the RPL22L1 knockdown activated the apoptosis pathway while inhibiting DNA replication and the cell cycle pathway. KEGG analysis further revealed that the genes with low expression after RPL22L1 knockdown were mainly involved in E2F targets, G2M checkpoint, mtorc1 signaling, and HYPOXIA. Moreover, knocking down the RPL22L1 gene significantly reduced the mRNA or protein levels of various factors, such as MCM3, MCM7, GADD45B, WEE1, MKI67, and Phospho-P70S6K. MCM3 and MCM7 have been reported as diagnostic and prognostic markers for various tumors, respectively.[26-29] They are also involved in the formation of the MCM2-7 complex (MCM complex), a helicase necessary for initiating and extending DNA replication in eukaryotic cells and unwinding DNA.[30,31] The decrease in MCM3 and MCM7 expressions inhibited the formation of the MCM2-7 complex (MCM complex) and delayed DNA synthesis.
Most tumor cells lack the G1 checkpoint function, rendering the G2 checkpoint critical for tumor survival. The transition from G2 to the M phase is modulated by the cyclin-dependent kinase 1 (CDK1) and Cyclin B complex, modulated by diverse feedback mechanisms.[32,33] GADD45B, a member of the growth arrest DNA damage-inducible gene family, is crucial in DNA damage repair, cell growth, apoptosis, and metastasis.[33-35] As a member of the G2M checkpoint, the activation of GADD45B can block the G2 phase and inhibit cell proliferation.[32,36] In addition, Wee1-like protein kinase (WEE1), a negative regulator of the G2M checkpoint, blocks G2 to M transition through the inhibitory phosphorylation of CDK1 Tyr15.[33,36] At present, various small molecule inhibitors targeting WEE1 have been developed to promote G2/M transition and subsequently induce cancer cell death.[37,38] In this study, knocking down RPL22L1 significantly downregulated the expression levels of GADD45B and WEE1. This finding suggests a potential reduction in the G2/M checkpoint function, leading to faster G2 to M transition.
Moreover, findings from the recovery experiments demonstrated that the mTOR pathway may mediate the suppressive effects of reduced RPL22L1 levels on the expression of MCM3, MCM7, and WEE1, with the exception of GADD45B. The mTOR pathway has been previously reported to regulate protein synthesis, cell growth, and proliferation.[39,40] For example, the silencing or overexpression of MCM7 has been exhibited to promote autophagy and apoptosis in cutaneous melanoma cells or cell proliferation and migration in esophageal squamous cell carcinoma by regulating the protein kinase B 1/Mechanistic target of rapamycin (AKT1/mTOR) pathway.[41,42] This finding suggests a possible positive feedback loop between MCM7 and the mTOR pathway. In addition, the targeting on mTOR pathway has been observed to overcome the resistance to WEE1 inhibition in small-cell lung cancer cells.[43] This finding is consistent with the results of this investigation, which demonstrated that mTOR promotes the expression of WEE1. Consequently, inhibiting the WEE1 function may activate negative feedback mechanisms that enable the mTOR pathway.
SUMMARYThis study identifies RPL22L1 as a highly expressed marker with unfavorable prognostic implications for CRC patients. Moreover, the knockdown of RPL22L1 exhibited significant inhibition of CRC growth and migration. For the first time, this study revealed the mechanism through which the mTOR pathway mediates the cell cycle blockade effect of RPL22L1. However, the limitation is that the molecular mechanism by which RPL22L1 regulates the mTOR pathway and apoptosis has not been thoroughly revealed. Thus, the protein level of RPL22l should be determined. Future studies will focus on elucidating the molecular mechanism through which RPL22L1 promotes the mTOR pathway and inhibits apoptosis, providing a theoretical basis for developing anti-CRC drug targeting RPL22L1.
AVAILABILITY OF DATA AND MATERIALSAll datasets generated for this study are available on reasonable request.
ABBREVIATIONSCRC – Colorectal cancer
RPL22L1 – Ribosomal Protein L22-Like 1
TCGA – The cancer genome atlas
GEO – Gene expression omnibus
RNAseq – RNA sequencing
RT-qPCR – Real-time reverse transcriptase-polymerase chain reaction
TME – Tumor microenvironment
OV – Ovarian cancer
COAD – Colon adenocarcinoma
READ – Rectum adenocarcinoma
UCSC – University of California Santa Cruz
GSEA – Gene set enrichment analysis
KEGG – Kyoto encyclopedia of genes and genomes
DEGs – Differentially expressed genes
KICH – Kidney chromophobe
MSI-H – Microsatellite instability-high
MSS – Microsatellite stable
OS – Overall survival
DFS – Disease-free survival
DSS – Disease-specific survival
shCtrl – Control short hairpin RNA
siRPL22L1 – RPL22L1-specific short hairpin RNA
MCM complex – MCM2–7 complex
CDK1 – Cyclin-dependent kinase 1
AUTHOR CONTRIBUTIONSCML and XND: Designed the study; HZ and SL: Downloaded, processed, and analyzed the data; CML and XND: Performed experiments: and CML: Wrote the manuscript; CML, XND, HZ, and SL: Revised the manuscript. All authors reviewed and approved the final manuscript.
留言 (0)