Streptococcus suis (S. suis, SS) is a multi-animal commensal pathogen that colonises the upper respiratory tract of pigs, particularly the tonsils and nasal passages, and some serotypes of SS are pathogenic, mainly through wound infection (Hatrongjit et al., 2024). They can cause a wide range of diseases in pigs, including acute septicaemia, meningitis, arthritis, endocarditis and pneumonia, posing a serious threat to the development of pig farming and public health (Jiang et al., 2024). SS has been subdivided into 35 serotypes, including types 1 to 34 and 1/2, according to its capsule antigen (CPS) (Gao et al., 2024). Streptococcus suis type 1 (SS1), Streptococcus suis type 2 (SS2), Streptococcus suis type 7 (SS7) and Streptococcus suis type 9 (SS9) are the causative agents in pigs, but SS2, SS7 and SS9 are the most pathogenic (Dong et al., 2023; Hatrongjit et al., 2023; Cao et al., 2024; de et al., 2024). Among them, SS2 is the most serious threat, classified as a zoonotic infectious disease, and is the predominant causative agent both in the population and in pig herds, with the highest pathogenicity and prevalence, and is the primary target of detection for outbreak surveillance and pathogen identification, e.g (Liu et al., 2024; Yin et al., 2024). 436 cases of SS infection were detected in Thailand from January to September 2023, of which 9 died (Tungwongjulaniam et al., 2024). In addition, with the development of intensive farming, the prevalence of SS infections in china cannot be ignored, but due to the numerous serotypes of SS, the complexity of virulence factors and the interaction and linkage of virulence factors and pathogenic mechanism are still not clear, and there is a lack of effective means used to prevent and control the disease (Scherrer et al., 2024). Therefore, the establishment of rapid and effective diagnostic methods and the accumulation of epidemiological data are of great importance for the development of vaccines and the prevention and control of the disease.
Currently, bacterial isolation and culture with remains the gold standard for the diagnosis of SS (Okuhama-Yoshida et al., 2023; Uruén et al., 2023), but the method is time-consuming, with low sensitivity and high operational requirements, which is not conducive to promotion and outbreak monitoring, and the serological method cannot differentiate between podless and self-coagulating strains (Thu et al., 2021; Dong et al., 2023). With the development of molecular biology, single or multiplex PCR (Kerdsin et al., 2014), MLST (Xia et al., 2024), single fluorescent quantitative PCR (Wang et al., 2024) and other methods have been established according to the universal gene of SS or different serotype-specific podocardial antigenic genes, which have realised the identification and typing of SS at the gene level, compensated for the inadequacy of serological typing and improved the accuracy and sensitivity of detection (Wang et al., 2024). However, these methods do not allow multiple tests to be performed simultaneously, which increases the workload and cost of epidemic surveillance. In order to interrupt the spread of epidemics in a timely manner, rapid, sensitive and accurate early detection is essential for the implementation of stringent hygiene and biosecurity measures. Therefore, this study has established a rapid, sensitive and specific quadruplex TaqMan fluorescence quantitative PCR method for the simultaneous detection of SS2, SS7, SS9 and other serotypes, which are currently the most harmful and widespread.
2 Materials and methods2.1 Strains and clinical samplesStreptococcus suis type 1 (SS1), Streptococcus suis type 2 (SS2), Streptococcus suis type 7 (SS7), Streptococcus suis type 9 (SS9), Streptococcus suis type 14 (SS14), Escherichia coli (E. coli), Pasteurella multocida (P. multocida), Staphylococcus aureus (S. aureus), Streptococcus agalactiae (S. agalactiae), Streptococcus pneumoniae (S. pneumoniae), Actinobacillus pleuropneumoniae (A. pleuropneumoniae), Mycoplasma hyopneumoniae (M. hyopneumoniae), Enterococcus faecalis (E. faecalis), Streptococcus pyogenes (S. pyogenes), Glaesserella parasuis (G. parasuis), Swine Influenza Virus (SIV), Porcine Pseudorabies Virus (PRV), Porcine Circovirus Type 2 (PCV2), Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) are maintained in this laboratory. 156 pig swabs and pig tissue samples (joint fluid, tonsils, lungs) were collected from March 2023-June 2024 from selected pig farms in Heilongjiang Province.
2.2 Design of primers and probesBased on the genes gdh (GenBank ID: AY853916.1), cps2J(GenBank ID: AM946016.1), cps7H (GenBank ID: BR001004), cps9J (GenBank ID: KC537370), we designed specific primers and probes using Primer Express 3.0.1 to select the conserved regions (Table 1). The primers were synthesised by Invitrogen (Shanghai). The 5′end of the probe was labelled with FAM, NED, ROX and Cy5 fluorescence reporter groups, and the 3′end was labelled with the corresponding MGB fluorescence quenching group.
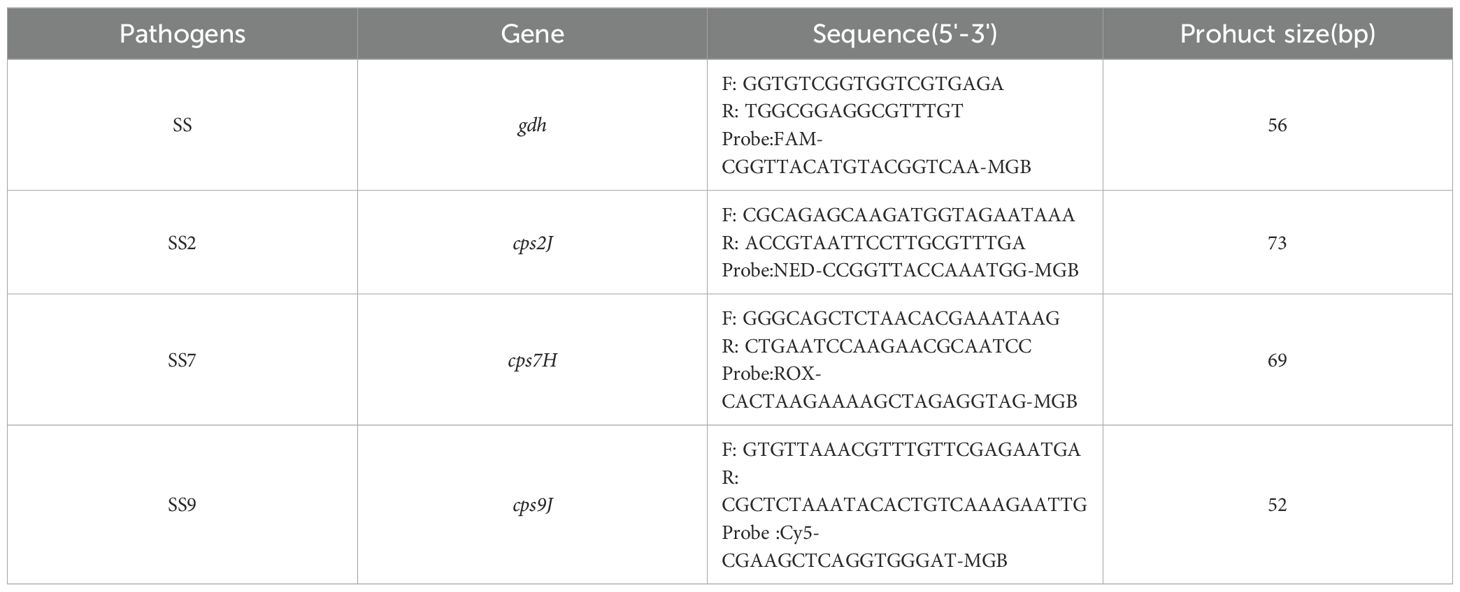
Table 1. Fluorescence quantitative PCR primers and probes.
2.3 Optimisation of reaction conditionsIn the 25 μL reaction system, 0.2-1.0 μL each of the upstream and downstream primers (10 μmol/L) and 0.2-1.0 μL of the probe (10 μmol/L) were added, and the annealing temperatures used were 50, 52, 54, 56, 58, 60 and 62 °C, and the number of cycles was 35, 40 and 45, respectively. The lowest cycle threshold (Ct value) and the highest ΔRn were used as indicators to obtain the optimal reaction system and procedure for quadruplex TaqMan fluorescence quantitative PCR.
2.4 Standard curve establishment and sensitivity testThe primers in Table 2 were used to amplify the target genes gdh, cps2J, cps7H and cps9J, respectively, and their products were recovered and purified and ligated into the pMD-18T vector and the recombinant plasmids of SS, SS2, SS7 and SS9 were transhumanised into the DH5α receptor cells and the culture was expanded and extracted with the kits after correct identification of bacteriophage PCR and sequencing. Recombinant plasmids (recombinant plasmid standards were designated P-SS, P-SS2, P-SS7 and P-SS9). The concentration of the plasmid was determined using a UV spectrophotometer, and the plasmid was diluted to a final concentration in the range of 4×100~4×1010 copies/uL and mixed in equal volumes so that the final concentrations were all in the range of 1×100 copies/μl~1×1010 copies/μl (Each gradient is repeated three times), which was then detected using the optimised quadruplex TaqMan fluorescence quantitative PCR assay.
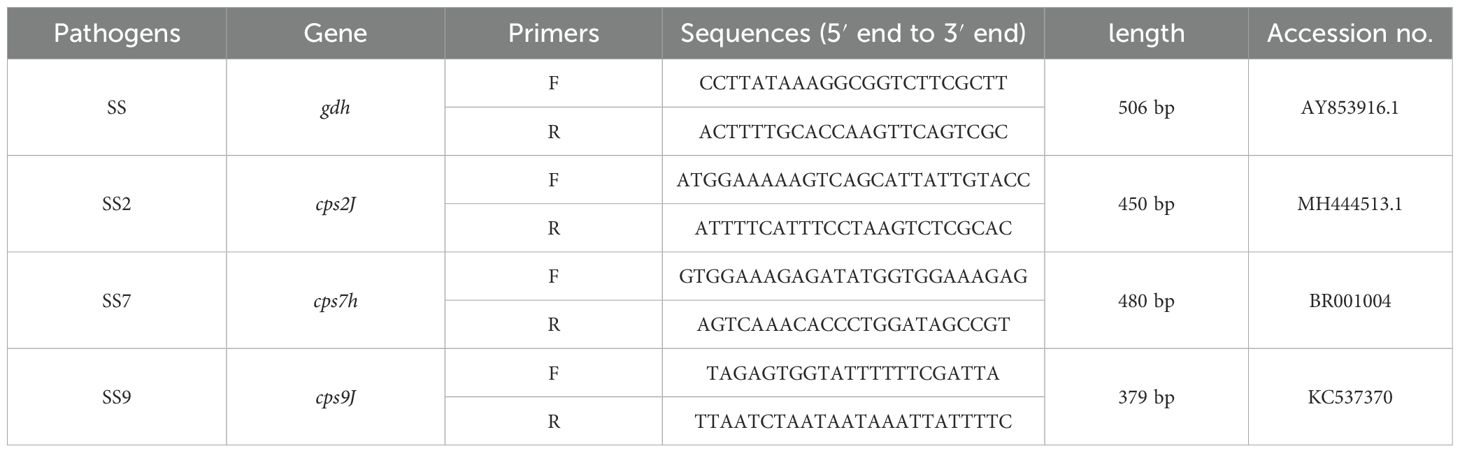
Table 2. Four sets of primers used to prepare plasmid standards.
2.5 Test for specificitySS1, SS2, SS7, SS9, SS14, E. coli, P. multocida, S. aureus, S. agalactiae, S. pneumoniae, A. pleuropneumoniae, M. hyopneumoniae, E. faecalis, S. pyogenes, G. parasuis, SIV, PRV, PCV2, PRRSV DNA/RNA as templates, recombinant plasmid standards P-SS, P-SS2, P-SS7 and P-SS9 were used as positive controls and sterile water as a blank control using an established quadruplex TaqMan fluorescence quantitative PCR.
2.6 Repeatability testingThe recombinant plasmids of SS, SS2, SS7 and SS9 with concentrations of 107 copies/µL, 105 copies/µL and 103 copies/µL were used as templates for the inter- and intra-batch experiments and the standard deviation of the Ct value and the coefficient of variation were calculated.
2.7 Clinical sample testingFrom March 2023 to June 2024, 156 pig swabs and tissue samples (joint fluid, tonsils, lungs) were collected from pigs in some pig farms in Heilongjiang Province who exhibited symptoms such as elevated body temperature, persistent fever, depression, decreased appetite, and difficulty breathing. Nucleic acids were extracted from the above samples using the corresponding commercially available kits and then detected using the established quadruplex TaqMan fluorescence quantitative PCR assay, while the extracted nucleic acids were detected using the reported PCR assay to compare the compliance rates (Wongsawan et al., 2015; Xin et al., 2023).
3 Results and analyses3.1 Results of optimisation of reaction conditionsAfter optimising the primers, probes, annealing temperature and cycle number of quadruplex TaqMan fluorescence quantitative PCR, it was found that the optimal annealing temperature was 60°C, the final concentrations of the primers were 0.08μmol/L, 0.14μmol/L, 0. 1μmol/L, and 0.12μmol/L for SS, SS2, SS7, and SS9, respectively, and the final concentrations of the probe were 0.08μmol/L, 0.08μmol/L, 0.12μmol/L, and 0.12μmol/L, respectively, and the optimal number of cycles was 40, which resulted in the lowest Ct value and the highest fluorescence signal. Final reaction system for quantitative PCR with fourfold fluorescence (Table 3) and reaction conditions (Table 4).
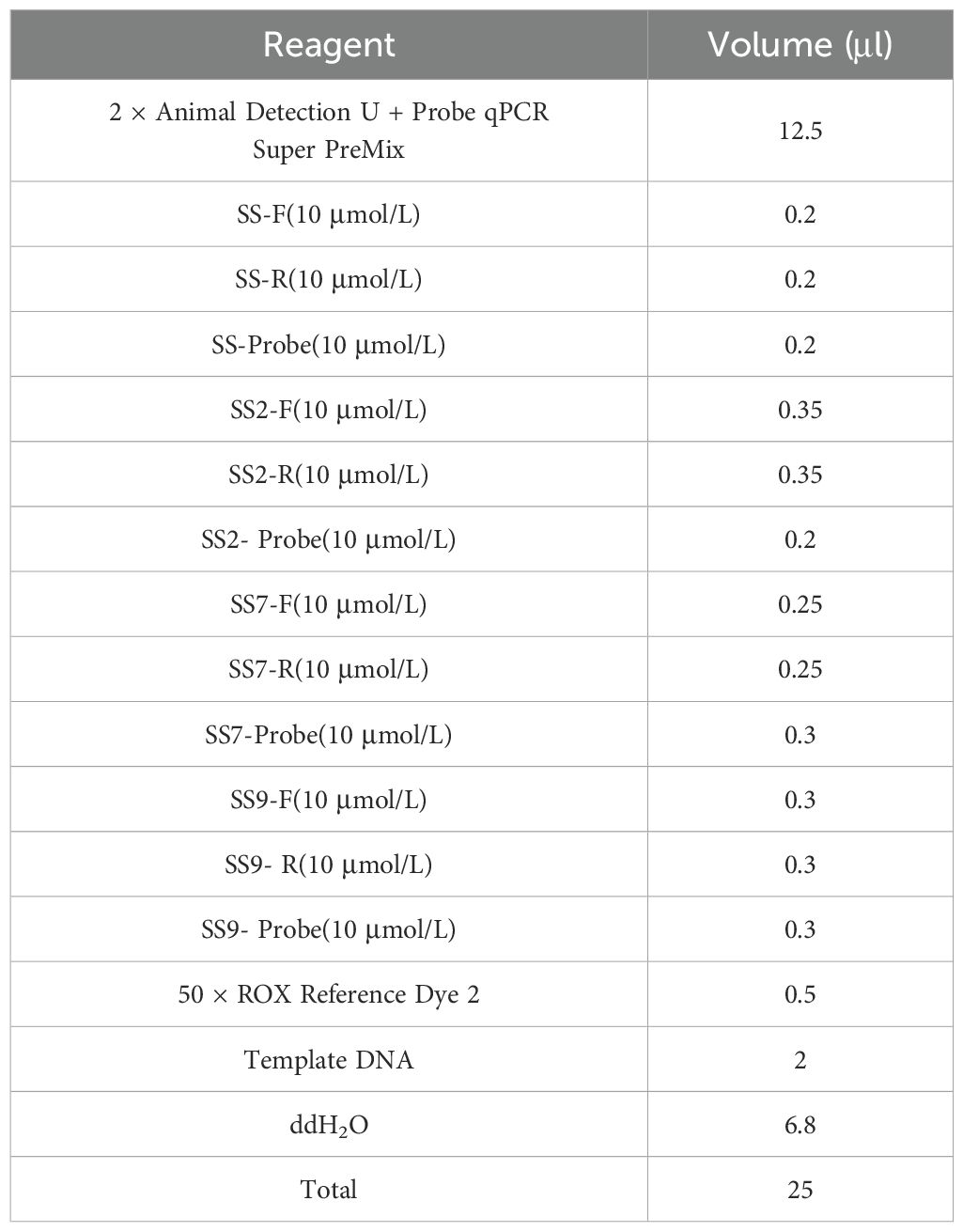
Table 3. Quadruplex TaqMan fluorescence quantitative PCR reaction system.
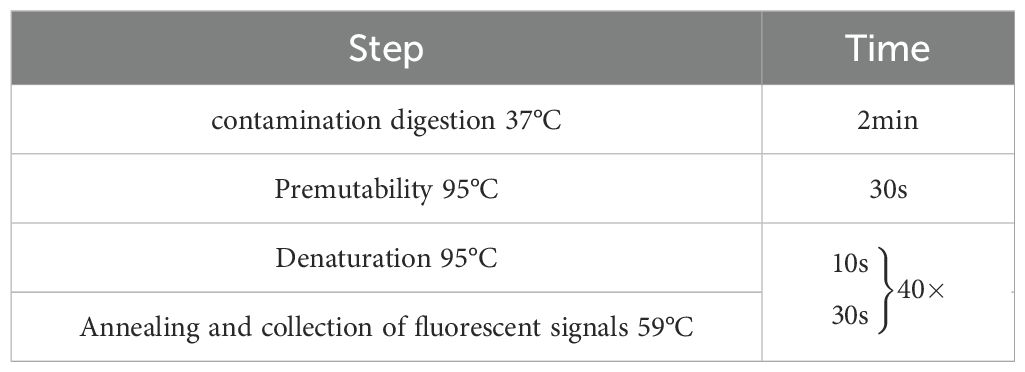
Table 4. Quadruplex fluorescence quantitative PCR reaction procedure.
3.2 Standard curves and minimum detection limitsP-SS, P-SS2, P-SS7 and P-SS9 recombinant plasmid standards 1×1010copies/μl- 1×104copies/μl were selected to plot standard curves. As shown in Figure 1, four standard curves were plotted and the amplification efficiencies were 95.192%, 105.956%, 87.917%, 100.231% with R2 of 0.999, 0.999, 0.997, 0.998, respectively. The established quadruplex TaqMan fluorescence quantitative PCR assay effectively detected a minimum concentration of 10 copies for the P-SS and P-SS9 recombinant plasmid standards, while the minimum concentration for the P-SS2 and P-SS7 recombinant plasmid standards was 100 copies (Figure 2). SS, SS2, SS7, and SS9 positive controls (FAM, NED, ROX, and Cy5) all had typical S-shaped amplification curves, and negative controls (FAM, NED, ROX, and Cy5) all had no amplification curves and a Ct value ≥40 or no value. The test is valid if this condition is met. If the Ct value of the test sample is <35 and a typical amplification curve appears, it is judged as positive; when 35≤Ct value <40, it is judged as suspicious and doubled for re-testing; when the Ct value is ≥40 or no value and no typical amplification curve, it is judged as negative.
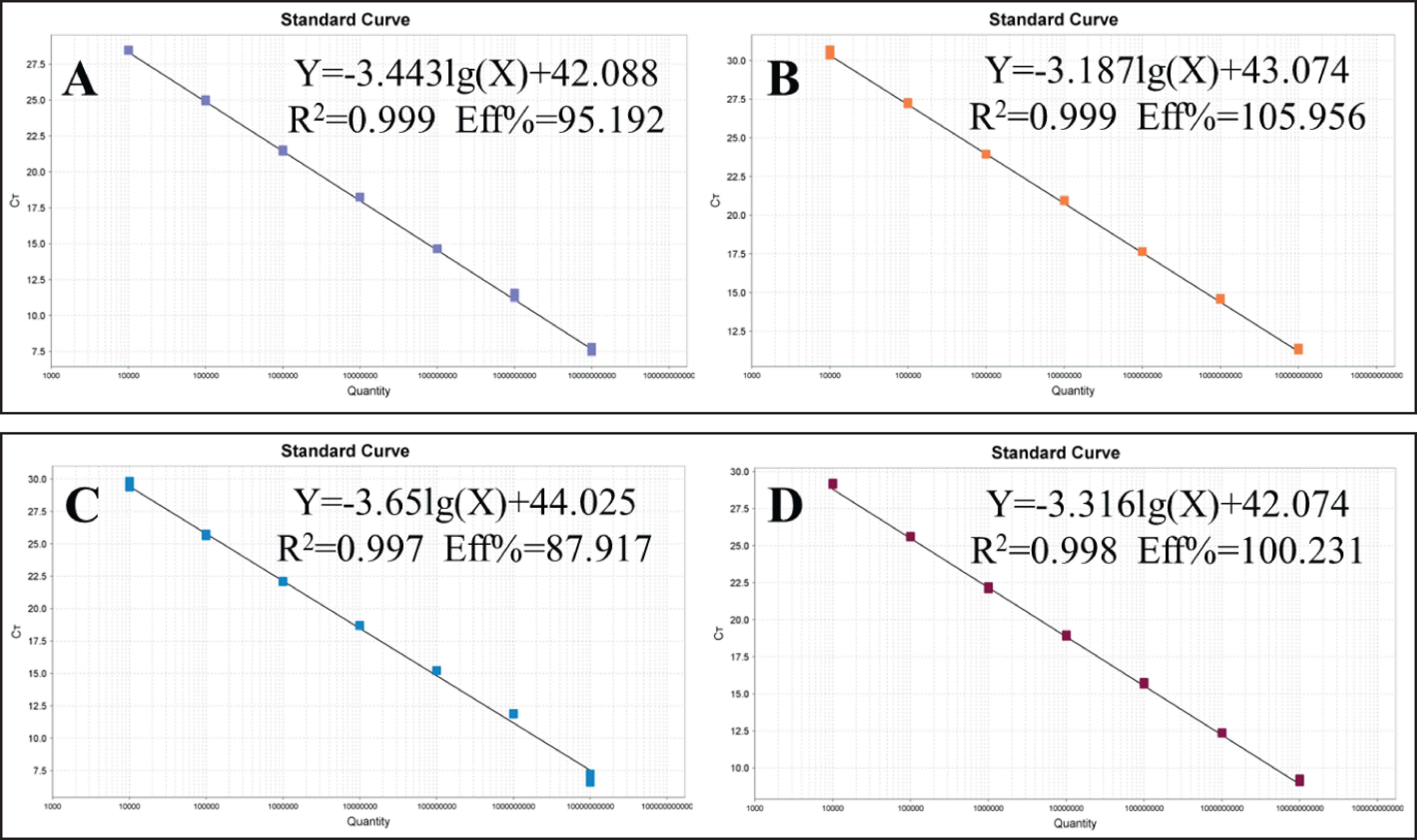
Figure 1. Standard curve of quadruplex TaqMan fluorescence quantitative PCR. (A) Streptococcus suis generalis; (B) Streptococcus suis type 2; (C) Streptococcus suis type 7; (D) Streptococcus suis type 9.
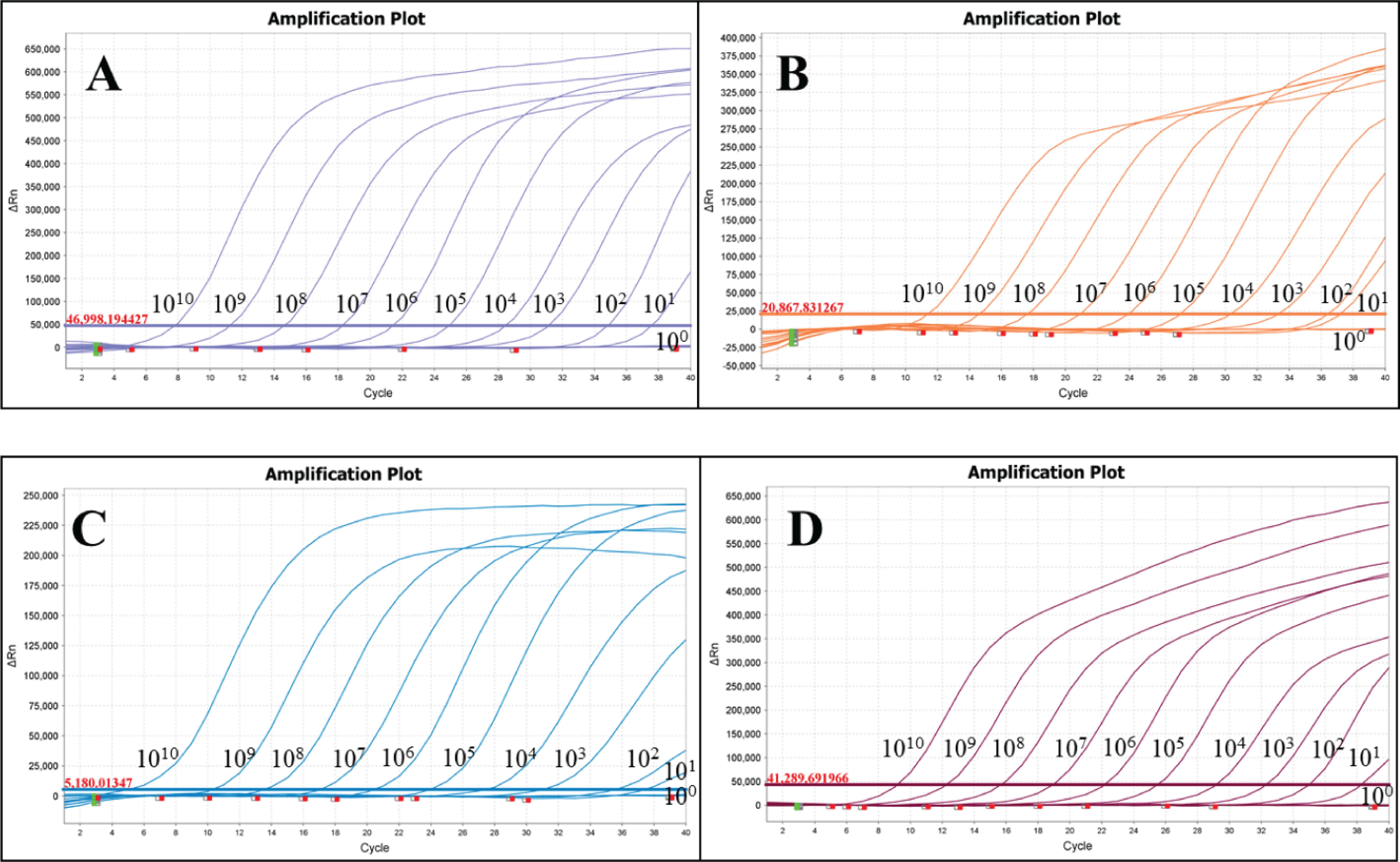
Figure 2. Sensitivity test of quadruplex TaqMan fluorescence quantitative PCR. (A) Streptococcus suis generalis; (B) Streptococcus suis type 2; (C) Streptococcus suis type 7; (D) Streptococcus suis type 9.
3.3 Specificity verification resultsSS1, SS2, SS7, SS9, SS14, E. coli, P. multocida, S. aureus, S. agalactiae, S. pneumoniae, A. pleuropneumoniae, M. hyopneumoniae, E. faecalis, S. pyogenes, G. parasuis, SIV, PRV, PCV2, PRRSV were used as templates for the detection of DNA/RNA, and recombinant plasmid standards P-SS, P-SS2, P-SS7 and P-SS9 were used as positive controls. As shown in Figure 3, the quadruplex TaqMan fluorescence quantitative PCR assay showed typical amplification curves for SS1, SS2, SS7, SS9, SS14, and the positive control, and there were no amplification curves and Ct values for the nucleic acids of the sterile water and the non-target pathogens, indicating that the method had high specificity. The evaluation results are described in Table 5.
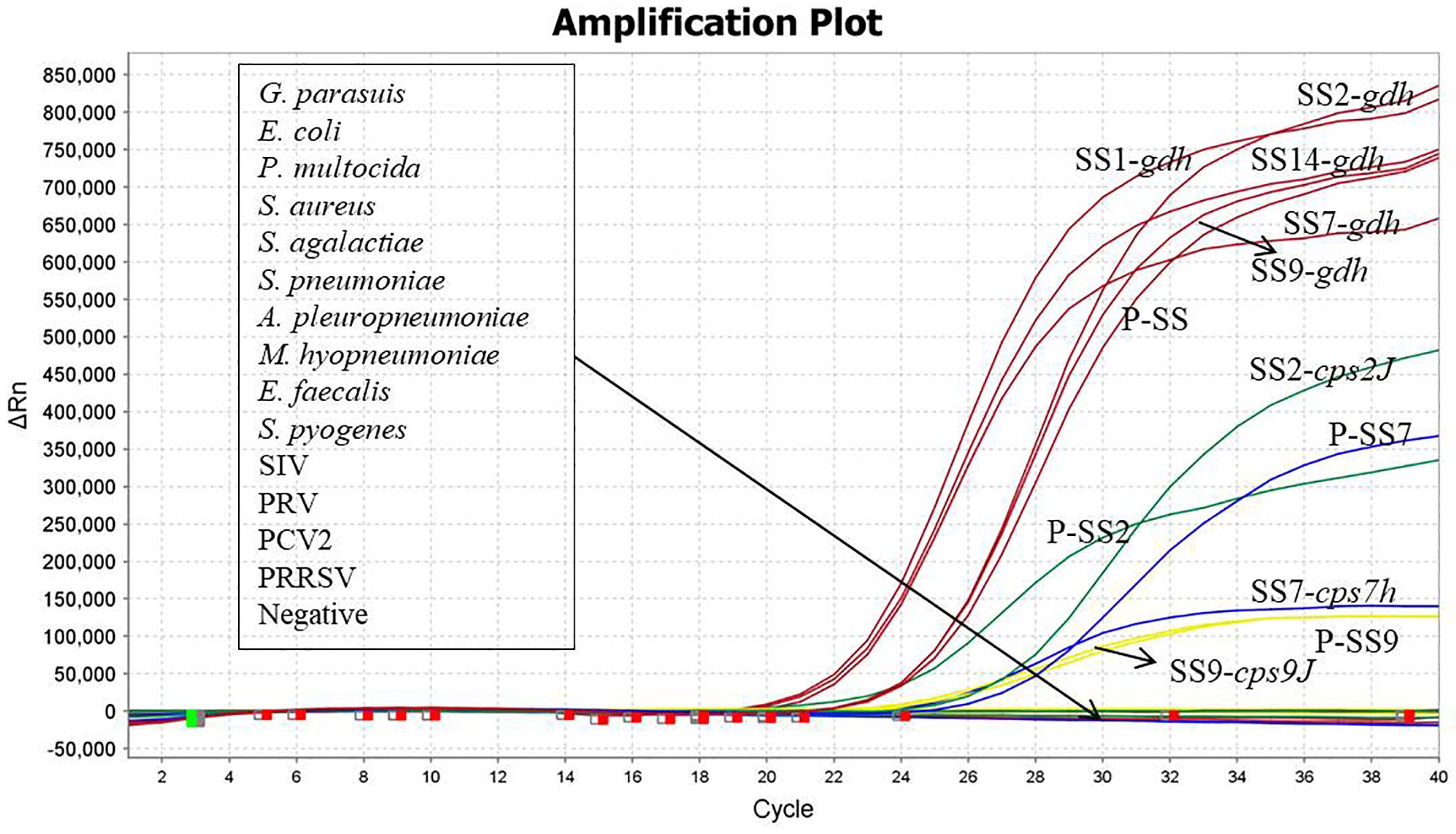
Figure 3. Specificity verification.

Table 5. Results and description of quadruplex TaqMan fluorescence quantitative PCR assay.
3.4 Repeatability test resultsRecombinant plasmids of SS, SS2, SS7 and SS9 were selected at a high concentration of 107, 105 copies/µL and a low concentration of 103 copies/µL for quadruplex TaqMan fluorescence quantitative PCR amplification, which was statistically analysed by observing the changes in the thresholds of the amplification curves and for the Ct values. The results showed that the coefficients of variation for the Ct values of SS, SS2, SS7 and SS9 were 0.21% ~ 1.10% (Table 6), indicating good reproducibility of this experiment.
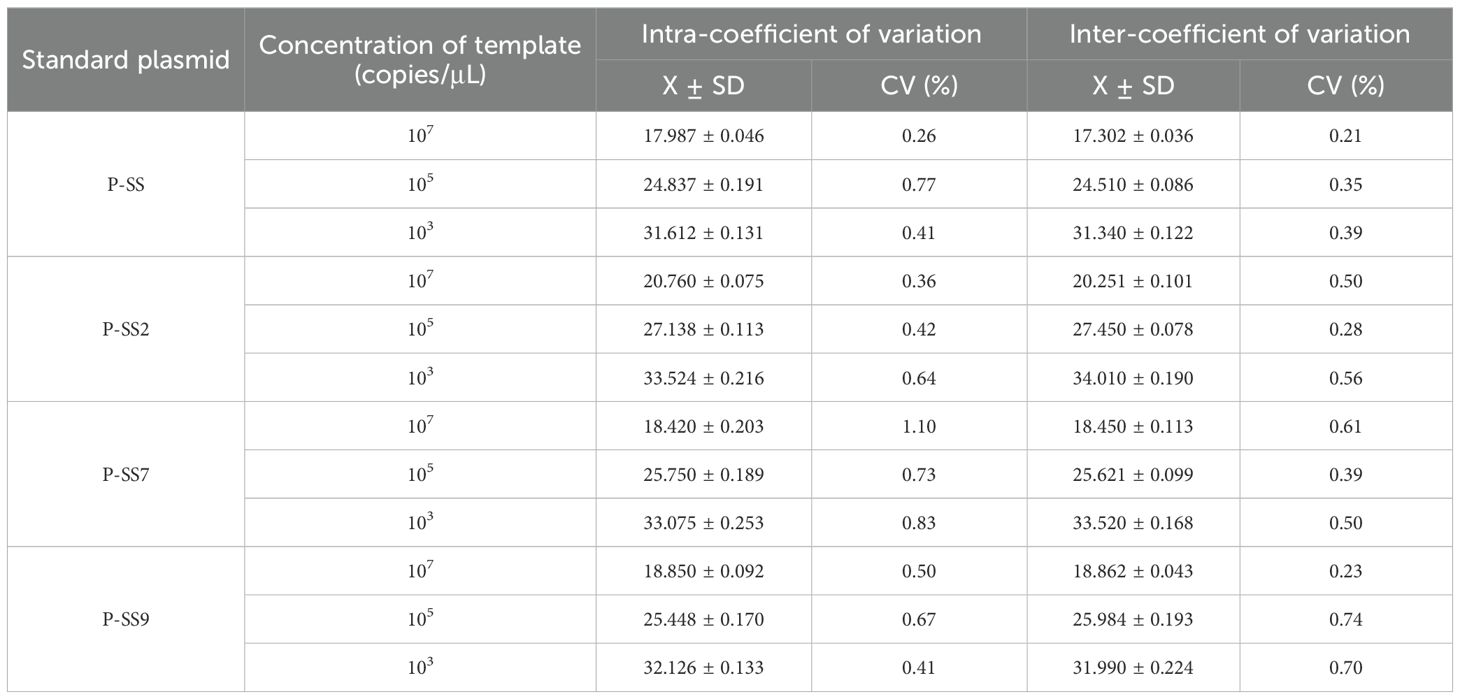
Table 6. Reproducibility of the quadruplex TaqMan fluorescence quantitative PCR method.
3.5 Clinical sample test resultsA total of 156 clinical samples collected were tested simultaneously using the quadruplex TaqMan fluorescence quantitative PCR assay established in this study and the reported assays. The results showed 68 positive and 88 negative samples (Table 7). The SS positivity rate was 5.77% (9/156), SS2 positivity rate was 20.51% (32/156), SS7 positivity rate was 8.33% (13/156) and SS9 positivity rate was 9.6% (15/156). No mixed infections were detected in clinical sample testing.
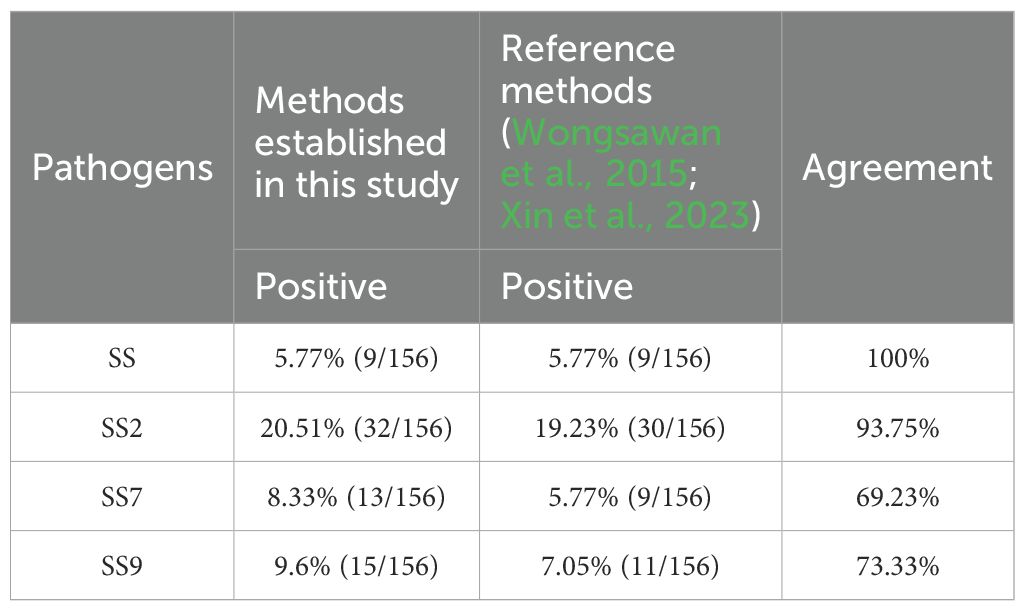
Table 7. Test results of clinical samples.
4 DiscussionIn this study, we developed a sensitive, specific and reproducible quadruplex TaqMan fluorescent quantitative PCR typing method based on the gdh, cps2J, cps7J and cps9J genes, which can differentiate between SS2, SS7, SS9 and other serotypes. The method does not cross-react with any of the non-target viruses or bacteria associated with infected pigs and accurately differentiates between SS2, SS7, SS9 and other serotypes of SS; The minimum detection limit for the recombinant plasmid standards P-SS and P-SS9 was 10 copies, and the minimum detection limit for P-SS2 and P-SS7 was 100 copies, which is comparable to the sensitivity of the quadruplex TaqMan fluorescence quantitative PCR assay for detecting SS and SS2 established by Xin L et al (Xin et al., 2023), whose method was only able to differentiate between SS2 and the other serotypes of SS. Coefficients of variation for both inter- and intra-batch replicates ranged from 0.21% to 1.10%. As there is no quadruplex TaqMan fluorescence quantitative PCR method that can differentiate between SS2, SS7, SS9 and other serotypes of SS, the establishment of this method is of great importance for rapid clinical typing and for guiding the precise use of medication and the timely adoption of appropriate measures.
Glutamate dehydrogenase (GDH), an important virulence-associated factor of SS, is expressed on the cell surface with NAD(P)H-dependent enzymatic activity and is a key enzyme linking carbon and nitrogen metabolism, which is important for bacterial pathogenicity (Chittick and Okwumabua, 2024). The study showed that the sequence of the gdh gene encoding glutamate dehydrogenase is highly conserved and that the gene is closely related to the reported gdh gene of SS (Silva et al., 2006). The nucleic acid sequences showed 97.2% to 98.4% homology with SS2, SS7 and SS9, of which 97.2% to 97.3% with SS2 strains, 98.4% with SS7 strains and 97.6% with SS9 strains (Smith et al., 1999; Okwumabua et al., 2001; Xu et al., 2021; Zhou et al., 2024). The amino acid sequences they encode were even more homologous, with more than 98.9% homology with sequences in GenBank (Okwumabua et al., 2001). Therefore, the gdh gene is a good target for the diagnosis of SS. CPS is the basis for distinguishing serotypes of SS (Goyette-Desjardins et al., 2020). Among them, cps2J, cps7H and cps9J are genes encoding specific podoplanet polysaccharide antigens of SS2, SS7 and SS9 respectively (Dekker et al., 2016; Jiang et al., 2022), which are closely related to the virulence and pathogenicity of SS and play an important role in bacterial survival and host infection. In addition, these genes are also used in molecular biology research for detection and analysis by PCR and other techniques to help understand the species of SS (Dekker et al., 2016; Wang et al., 2024), virulence factors and how they interact with the host, which is of great scientific importance and practical value in the prevention and control of porcine streptococcal infections.
The prevalence of Streptococcus suis in pigs is characterised by a wide spatial distribution that varies between regions and over time (Petrocchi Rilo et al., 2024; Uruén et al., 2024). According to the survey, 19 strains of SS were isolated from nine districts in Hubei Province in 2021-2023, including 10 serotypes, of which SS9 was the most prevalent, accounting for 21.05% of the total (Xia et al., 2024). Although SS2 was the most commonly reported strain infecting both humans and pigs, Wang M et al., showed that serotype 14 accounted for 71.1% and serotype 2 for 28.19% of ST-type SS isolates from Guangxi Zhuang Autonomous Region, China, during 2007-2018 (Wang et al., 2019). However, overseas, for example, testing of live pigs and pork at the Chiang Mai market in Thailand showed a positive rate of 84% for SS, with 34% positive for SS2, which remains the predominant serotype (Guntala et al., 2024). The most common serotypes of SS isolated from sick pigs in western Canada were SS2 (9.3%) and SS7 (7.8%) (de et al., 2024). Another study showed that the global prevalence of SS2 isolated from pigs was 13.6%, with approximately 10% in healthy pigs and 16% in sick pigs. The prevalence of SS2 did not change significantly over time. This suggests that SS2 continues to be a problem in the pig industry and a threat to human health (Keonam et al., 2024). In this study, the highest positivity rate was for SS2 (20.51%), followed bySS9 (9.6%) and SS7 (8.33%), while other serotypes of SS were also detected in the clinical samples tested (5.77%). In addition, the comparison with existing detection methods showed that the detection rate of the quadruplex TaqMan fluorescence quantitative PCR assay established in this study was higher than that of the conventional PCR method, and the concordance rate with the existing quadruplex TaqMan fluorescence quantitative PCR assay was higher. On the other hand, some of the samples were swabs, and although swabs are the easiest to collect, the nucleic acid content of the target pathogens was low after nucleic acid extraction, coupled with the limited sensitivity of conventional PCR, so there was a discrepancy in the detection rate of compliance between the two methods.
In conclusion, this study has established a sensitive, specific, rapid and efficient quadruplex TaqMan fluorescence quantitative PCR assay that can simultaneously differentiate between SS2, SS7, SS9 and other serotypes of SS, with the aim of providing technical support for rapid typing of SS and accurate prevention and treatment.
Data availability statementThe original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributionsHW: Conceptualization, Writing – original draft, Writing – review & editing. JC: Conceptualization, Writing – original draft. YS: Conceptualization, Writing – original draft. TA: Writing – review & editing, Data curation. YW: Investigation, Writing – original draft, Writing – review & editing. HC: Writing – original draft, Investigation. CY: Conceptualization, Writing – original draft. CX: Conceptualization, Writing – original draft. HZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by grants from the National Key R&D Program of China (2023YFF0724604); National Key R&D Program Young Scientist Project (2021YFF0703100); Natural Science Foundation of China (32072898); Cultivation, Quality Control and Detection technology of high grade agricultural experimental animal Pig (GZ20210010); Research on Improving the quality of Breeding and testing of experimental animal Resources (1610302022018); Basic research on quality control and genetic resistance of experimental pigs (SKLVBP202120); Basic research on quality control and genetic resistance of experimental pigs (SKLVBP202101).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesCao, X., Jia, K., Liu, Q., Yin, H., Yu, X., Hu, X., et al. (2024). The critical role of NLRP3 inflammasome activation in Streptococcus suis-induced blood-brain barrier disruption. Vet. Microbiol. 295, 110161. doi: 10.1016/j.vetmic.2024.110161
PubMed Abstract | Crossref Full Text | Google Scholar
Chittick, L., Okwumabua, O. (2024). Loss of expression of the glutamate dehydrogenase (gdh) of Streptococcus suis serotype 2 compromises growth and pathogenicity. Microb. Pathog. 188, 106565. doi: 10.1016/j.micpath.2024.106565
PubMed Abstract | Crossref Full Text | Google Scholar
Dekker, N., Daemen, I., Verstappen, K., de Greeff, A., Smith, H., Duim, B. (2016). Simultaneous quantification and differentiation of streptococcus suis serotypes 2 and 9 by quantitative real-time PCR, evaluated in tonsillar and nasal samples of pigs. Pathogens. 5. doi: 10.3390/pathogens5030046
PubMed Abstract | Crossref Full Text | Google Scholar
de, O. C. M., Gamage, R., Christensen, J. (2024). Molecular profile and epidemiological traits of Streptococcus suis isolated from diseased pigs in western Canada reveal multiple-serotype infection: Implications for disease control. Can. Vet. J. 65, 429–436.
PubMed Abstract | Google Scholar
Dong, N., Wang, Z., Sun, Q., Chen, X., Zhang, H., Zheng, J., et al. (2023). Establishment and application of an indirect ELISA for the detection of antibodies to porcine streptococcus suis based on a recombinant GMD protein. Anim. (Basel) 13. doi: 10.3390/ani13040719
PubMed Abstract | Crossref Full Text | Google Scholar
Gao, S., Mao, C., Yuan, S., Quan, Y., Jin, W., Shen, Y., et al. (2024). AI-2 quorum sensing-induced galactose metabolism activation in Streptococcus suis enhances capsular polysaccharide-associated virulence. Vet. Res. 55, 80. doi: 10.1186/s13567-024-01335-5
PubMed Abstract | Crossref Full Text | Google Scholar
Goyette-Desjardins, G., Auger, J. P., Dolbec, D., Vinogradov, E., Okura, M., Takamatsu, D., et al. (2020). Comparative study of immunogenic properties of purified capsular polysaccharides from streptococcus suis serotypes 3, 7, 8, and 9: the serotype 3 polysaccharide induces an opsonizing igG response. Infect. Immun. 88(10), e00377–20. doi: 10.1128/IAI.00377-20
PubMed Abstract | Crossref Full Text | Google Scholar
Guntala, R., Khamai, L., Srisai, N., Ounjaijean, S., Khamduang, W., Hongjaisee, S. (2024). Contamination of Streptococcus suis and S. suis Serotype 2 in Raw Pork and Edible Pig Organs: A Public Health Concern in Chiang Mai, Thailand. Foods. 13 (13), 2119. doi: 10.3390/foods13132119
PubMed Abstract | Crossref Full Text | Google Scholar
Hatrongjit, R., Fittipaldi, N., Gottschalk, M., Kerdsin, A. (2024). Genomic epidemiology in Streptococcus suis: Moving beyond traditional typing techniques. Heliyon. 10, e27818. doi: 10.1016/j.heliyon.2024.e27818
PubMed Abstract | Crossref Full Text | Google Scholar
Hatrongjit, R., Fittipaldi, N., Jenjaroenpun, P., Wongsurawat, T., Visetnan, S., Zheng, H., et al. (2023). Genomic comparison of two Streptococcus suis serotype 1 strains recovered from porcine and human disease cases. Sci. Rep. 13, 5380. doi: 10.1038/s41598-023-32724-z
PubMed Abstract | Crossref Full Text | Google Scholar
Jiang, X., Zhu, L., Zhan, D. (2022). Development of a recombinase polymerase amplification assay for rapid detection of Streptococcus suis type 2 in nasopharyngeal swab samples. Diagn. Microbiol. Infect. Dis. 102, 115594. doi: 10.1016/j.diagmicrobio.2021.115594
PubMed Abstract | Crossref Full Text | Google Scholar
Jiang, Z. J., Hong, J. C., Tang, Q. X., Lin, B. W., Zhang, W. Q., Xia, H., et al. (2024). Streptococcus suis meningoencephalitis diagnosed with metagenomic next-generation sequencing: A case report with literature review. J. Infect. Chemother. 30, 544–547. doi: 10.1016/j.jiac.2023.11.017
PubMed Abstract | Crossref Full Text | Google Scholar
Keonam, K., Nam, N. H., Saksangawong, C., Sringam, P., Saipan, P., Kongpechr, S., et al. (2024). Prevalence of Streptococcus suis serotype 2 isolated from pigs: A systematic review and meta-analysis. Vet. World. 17, 233–244. doi: 10.14202/vetworld.
PubMed Abstract | Crossref Full Text | Google Scholar
Kerdsin, A., Akeda, Y., Hatrongjit, R., Detchawna, U., Sekizaki, T., Hamada, S., et al. (2014). Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 63, 824–830. doi: 10.1099/jmm.0.069757-0
PubMed Abstract | Crossref Full Text | Google Scholar
Okuhama-Yoshida, E., Nakayama, M., Hattori, M., Takamatsu, D., Okura, M. (2023). Improvement of the mismatch amplification mutation assay-PCR for discrimination between Streptococcus suis serotypes 2 and 1/2. J. Microbiol. Methods 214, 106828. doi: 10.1016/j.mimet.2023.106828
PubMed Abstract | Crossref Full Text | Google Scholar
Okwumabua, O., Persaud, J. S., Reddy, P. G. (2001). Cloning and characterization of the gene encoding the glutamate dehydrogenase of Streptococcus suis serotype 2. Clin. Diagn. Lab. Immunol. 8, 251–257. doi: 10.1128/CDLI.8.2.251-257.2001
PubMed Abstract | Crossref Full Text | Google Scholar
Petrocchi Rilo, M., Gutiérrez Martín, C. B., Acebes Fernández, V., Aguarón Turrientes, Á, González Fernández, A., Miguélez Pérez, R., et al. (2024). Streptococcus suis research update: serotype prevalence and antimicrobial resistance distribution in swine isolates recovered in Spain from 2020 to 2022. Vet. Sci. 11 (1), 40. doi: 10.3390/vetsci11010040
PubMed Abstract | Crossref Full Text | Google Scholar
Scherrer, S., Biggel, M., Schneeberger, M., Cernela, N., Rademacher, F., Schmitt, S., et al. (2024). Genetic diversity and antimicrobial susceptibility of Streptococcus suis from diseased Swiss pigs collected between 2019 - 2022. Vet. Microbiol. 293, 110084. doi: 10.1016/j.vetmic.2024.110084
PubMed Abstract | Crossref Full Text | Google Scholar
Silva, L. M., Baums, C. G., Rehm, T., Wisselink, H. J., Goethe, R., Valentin-Weigand, P. (2006). Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 115, 117–127. doi: 10.1016/j.vetmic.2005.12.013
PubMed Abstract | Crossref Full Text | Google Scholar
Smith, H. E., Veenbergen, V., van der Velde, J., Damman, M., Wisselink, H. J., Smits, M. A. (1999). The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37, 3146–3152. doi: 10.1128/JCM.37.10.3146-3152.1999
PubMed Abstract | Crossref Full Text | Google Scholar
Thu, I. S. L., Tragoolpua, K., Intorasoot, S., Anukool, U., Khamnoi, P., Kerdsin, A., et al. (2021). Direct Detection of Streptococcus suis from Cerebrospinal Fluid, Positive Hemoculture, and Simultaneous Differentiation of Serotypes 1, 1/2, 2, and 14 within Single Reaction. Pathogens. 10. doi: 10.3390/pathogens10080996
PubMed Abstract | Crossref Full Text | Google Scholar
Tungwongjulaniam, C., Klinman, K., Theerawat, R., Wiratsudakul, A. (2024). A network analysis of the local pig supply chain in a repeated outbreak area of human streptococcosis in Thailand. Zoonoses Public Health. 71, 673–682. doi: 10.1111/zph.13132
PubMed Abstract | Crossref Full Text | Google Scholar
Uruén, C., Fernandez, A., Arnal, J. L., Del Pozo, M., Amoribieta, M. C., de Blas, I., et al. (2024). Genomic and phenotypic analysis of invasive Streptococcus suis isolated in Spain reveals genetic diversification and associated virulence traits. Vet. Res. 55, 11. doi: 10.1186/s13567-024-01267-0
PubMed Abstract | Crossref Full Text | Google Scholar
Uruén, C., Gimeno, J., Sanz, M., Fraile, L., Marín, C. M., Arenas, J. (2023). Invasive Streptococcus suis isolated in Spain contain a highly promiscuous and dynamic resistome. Front. Cell Infect. Microbiol. 13, 1329632. doi: 10.3389/fcimb.2023.1329632
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, M., Du, P., Wang, J., Lan, R., Huang, J., Luo, M., et al. (2019). Genomic epidemiology of streptococcus suis sequence type 7 sporadic infections in the guangxi zhuang autonomous region of China. Pathogens. 8. doi: 10.3390/pathogens8040187
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, L., Sun, J., Zhao, J., Bai, J., Zhang, Y., Zhu, Y., et al. (2024). A CRISPR-Cas12a-based platform facilitates the detection and serotyping of Streptococcus suis serotype 2. Talanta. 267, 125202. doi: 10.1016/j.talanta.2023.125202
留言 (0)