Typhoid fever (typhoid), a life-threatening systemic infection caused predominantly by Salmonella enterica subspecies enterica serotype Typhi (S. Typhi), remains a common infection and a public health concern in resource poor-settings in parts of sub-Saharan Africa and Asia (Kariuki et al., 2021; Kim et al., 2022; Stanaway et al., 2019). An estimated 9 million new typhoid fever cases occur each year, of which 2%–3% results in death even with adequate antibiotics therapy (Pieters et al., 2018; WHO, 2023). Typical symptoms manifest between 1 and 3 weeks postinfection and encompass elevated prolonged fever, headache, malaise, abdominal pain, diarrhea, constipation, hypersplenism, and rose-colored spots on the chest (World Health Organization, 2019). S. Typhi is transmitted via the fecal–oral route in settings with poor standards of sanitation, low levels of hygiene, and inadequate water supply (Crump, 2019; Parry et al., 2010). In addition to inadequate resources, typhoid endemic settings lack a quality public health infrastructure (Dougan and Baker, 2014).
Upon ingestion of contaminated food or water, S. Typhi bacteria that survive the hostile gastric acid-rich environment in the stomach are able to replicate in the new host (Ahirwar et al., 2014; Dougan and Baker, 2014). The typhoid bacilli can invade the intestinal mucosa, typically through microfold (M) cells, and establish an initially clinically undetectable infection involving significant systemic dissemination and a transient primary bacteremia (Dougan and Baker, 2014). S. Typhi also reach the gallbladder hematogenously during primary bacteremia or shortly thereafter through infected hepatic bile entering the gallbladder (Gaines et al., 1968; Hoffman et al., 2023). S. Typhi bacteria can survive, replicate, and evade immune surveillance intracellularly within a modified phagosome known as Salmonella-containing vacuole (SCV) (Dougan et al., 2011; Steele-Mortimer, 2008).
Although the majority of patients recover from typhoid fever after an appropriate treatment, some individuals become asymptomatic carriers and shed the infectious typhoid bacilli intermittently in their feces for an ill-defined period of time after apparent clinical resolution. Since the early twentieth century, asymptomatic carriage has been demonstrated to be a source of transmission of typhoid fever, including in the famous case of Mary Mallon (Marineli et al., 2013). Generally, ~2%–5% of acute typhoid cases fail to clear the infection fully within 1 year and develop asymptomatic chronic carriage (Dongol et al., 2012; Gunn et al., 2014; Parry et al., 2002). Approximately 90% of typhoid chronic carriers have gallstones in their gallbladder (Crawford et al., 2010; Lovane et al., 2016).
Persistent colonization of the gallbladder by S. Typhi is facilitated by formation of biofilms on the surface of cholesterol gallstones (Crawford et al., 2010; Gonzalez-Escobedo et al., 2011). Biofilms are organized three-dimensional multicellular communities encased in self-produced extracellular polymeric substances (EPS) composed of polysaccharides, extracellular DNA [eDNA], proteins, and lipids (Flemming and Wingender, 2010). For S. Typhi to form biofilms on human cholesterol-coated gallstones, a bile-induced EPS and cell-to-cell interaction is required (Crawford et al., 2008). Salmonella-species biofilms have several identified EPS components including cellulose, colanic acid, the Vi antigen, curli fimbriae, the O antigen capsule, and some biofilm-associated proteins (Gonzalez-Escobedo et al., 2011; Gibson et al., 2006; Jonas et al., 2007). Formation of biofilms provides several advantages to the bacteria, including enhanced resistance to antibiotics and the host’s immune response, as well as a stable environment that supports long-term colonization (Steenackers et al., 2012). Biofilms account for 80% of chronic infections in humans, leading to increased rates of hospitalization, high healthcare costs, and increased mortality and morbidity rates (Römling and Balsalobre, 2012). Chronic S. Typhi colonization usually cannot be resolved with antibiotics; gallbladder resection is the only option, although not always effective (Gonzalez-Escobedo et al., 2011). Biofilm formation leads to continuous shedding and reattachment of planktonic cells, followed by bacteria diffusion in urine and feces (Crawford et al., 2010; Gonzalez-Escobedo and Gunn, 2013). Since S. Typhi is a human-restricted pathogen, gallbladder colonization and fecal shedding form a central dogma for further transmission and persistence of typhoid fever.
In the gallbladder, S. Typhi is exposed to bile, a complex digestive secretion composed of bile acids, bilirubin, phospholipids, and cholesterol that exhibit strong antimicrobial properties (An et al., 2022; Staley et al., 2017). The molecular mechanisms involved in establishing the carrier state are poorly understood; however, S. Typhi is thought to undergo genetic changes within the gallbladder as an adaptive mechanism (Gonzalez-Escobedo et al., 2011; Neiger et al., 2019; Thanh Duy et al., 2020).
Although it is widely accepted that S. Typhi carriers contribute to typhoid transmission in endemic settings, little progress has been made in understanding the typhoid carrier state. The current study aimed at identifying the genetic differences in longitudinal clinical S. Typhi isolates from carriers, in a typhoid endemic setting in Nairobi, Kenya.
2 Methods2.1 Source of bacterial strainsS. Typhi strains were isolated from the blood and stool samples of six patients residing in four different households in Mukuru, an informal settlement in Nairobi, Kenya, known for being typhoid-endemic. A typhoid fever patient, along with at least one household contact, provided stool samples for the detection of S. Typhi shedding (Supplementary Table S1). The laboratory methods for isolation and identification of the isolates are described in our previous publication (Muturi et al., 2024). Briefly, blood samples were inoculated into BACTEC™ culture vials and incubated in a BACTEC blood culture system (Becton, Dickinson and Company, New Jersey, USA) before subculturing on XLD agar. Stool samples were enriched in selenite fecal (SF) broth prior to culturing on XLD agar. S. Typhi culture-positive samples were identified using biochemical tests with the Analytical Profile Index 20E (API 20E) system and confirmed by polymerase chain reaction (PCR) amplification of a 1,278-bp fragment of the VI region of the flagellin gene, using primers tviB-F (5′-TCAGCGACTTCTGTTCTATTCAAGTAAGAAAGGGGTACGG-3′) and tviB-R (5′-GCTCCTCACTGACGGACGTGCGAACGTCGTCTAGATTATG-3′). A total of 22 isolates were collected, comprising 2 from blood samples and 20 from stool samples. Nineteen of the isolates (19/22) came from four patients diagnosed with cholelithiasis. Of these patients, three exhibited typhoid symptoms and continued shedding S. Typhi post-treatment. Gallstones were confirmed via ultrasound, a standard imaging procedure for assessing gallbladder diseases. One patient with gallstones was asymptomatic but continued to shed S. Typhi and lived in the same household (household B) as an acute typhoid fever case who did not have gallstones. In household A, one contact without cholelithiasis shed S. Typhi once, whereas the index case, who had gallstones, continued shedding S. Typhi after antibiotic treatment. The index case refers to the first identified individual infected with S. Typhi and presenting typhoid fever symptoms in each household.
2.2 Whole-genome sequencingDNA extracted from S. Typhi strains using GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, Missouri, United States) was prepared for whole-genome sequencing by SeqCoast Genomics (Portsmouth, New Hampshire, United States) using an Illumina DNA Prep Tagmentation kit and unique dual indexes. Sequencing was performed on the Illumina NextSeq 2000 platform using a 300-cycle flow cell kit to produce 2 × 150-bp paired reads as previously described (Grant et al., 2023). PhiX control, 1%–2%, was spiked into the run to support optimal base calling. Read demultiplexing, read trimming, and run analytics were performed using DRAGEN v3.10.12, an on-board analysis software on the NextSeq 2000.
2.3 Genome assembly and annotationQuality-trimming of the reads was done using Trimmomatic (version 0.39) (Bolger et al., 2014). Read error was corrected using SPAdes (v. v3.13.1) (Bankevich et al., 2012). The reads were assembled into contigs using SPAdes (wrapped in Unicycler). Read mapping was done using Bowtie2 and SAMtools (wrapped in Unicycler, version 0.4.4) (Langmead and Salzberg, 2012; Li et al., 2009; Wick et al., 2017). Pilon (v1.24) (wrapped in Unicycler) was used in polishing of each assembly (Walker et al., 2014). Gene prediction and functional annotation were performed using BAKTA (version 1.5.1) (Schwengers et al., 2021). The annotation pipeline was as follows: prediction of protein-coding genes using Prodigal, tRNA identification using tRNAscan-SE (Chan and Lowe, 2019), tRNA and tmRNA identification using Aragorn (Laslett and Canback, 2004), prediction of rRNA sequences using Infernal and the Rfam database (Nawrocki and Eddy, 2013), CRISPR prediction using PILER-CR (Edgar, 2007), antimicrobial resistance gene identification using AMRFinderPlus (Feldgarden et al., 2021), prediction of signal peptides using DeepSig (Savojardo et al., 2018), prediction of transposases using ISFinder (Siguier et al., 2006), and computation of codon usage biases for each amino acid using the codonUsage.py script (https://github.com/Arkadiy-Garber/BagOfTricks) (Garber, 2024).
2.4 Genotype identification and bacteria clusteringIdentification and clustering of S. Typhi genotypes was performed using Pathogenwatch (https://pathogen.watch/), a web-based platform with several different components (Argimón et al., 2021; Wong et al., 2016). The platform provides compatibility with S. Typhi typing information for MLST (Sharma et al., 2016), in silico serotyping (SISTR) (Yoshida et al., 2016), and an SNP genotyping scheme (GenoTyphi) (Wong et al., 2016). S. Typhi assemblies (in fasta format) were uploaded to the platform (https://pathogen.watch/upload/fasta) for the analysis. A collection of uploaded sequences was created in Pathogenwatch to generate a phylogenetic tree. S. Typhi genotype 2.1 (GenBank Accession: SAMEA2158302) was used as the outgroup. The phylogenetic tree generated by Pathogenwatch (Newick format) was downloaded and exported to Microreact (https://microreact.org) for visualization. Associated metadata in CSV format was also exported to Microreact, and a phylogenetic tree in Newick format, reflecting branch lengths corresponding to SNP/genetic distances, was obtained.
2.5 Antimicrobial susceptibility testingAntimicrobial susceptibility testing was performed using the disk diffusion technique (Reller et al., 2009) for all antimicrobials commonly used in Kenya for typhoid fever treatment including ampicillin (10 µg), tetracycline (30 µg), co-trimoxazole (25 µg), chloramphenicol (30 µg), amoxicillin–clavulanate (20/10 μg), cefpodoxime (30 µg), ceftazidime (30 µg), ceftriaxone (30 µg), cefotaxime (30 µg), azithromycin (15 μg), ciprofloxacin (5 µg), nalidixic acid (10 µg), kanamycin (30 μg), and gentamicin (10 μg). The diameter of the zone of inhibition was measured after 18 h–24 h, and results were interpreted according the Clinical and Laboratory Standards Institute (CLSI), guidelines for Salmonella (Clinical and Laboratory StandardsInstitute, 2023).
2.6 Screening of antimicrobial resistance genesNCBI Antimicrobial Resistance Gene Finder Plus (AMRFinderPlus, v3.12.8) (https://github.com/ncbi/amr/wiki) was used to identify acquired antimicrobial resistance genes and known resistance-associated point mutations in S. Typhi-assembled nucleotide sequences (Feldgarden et al., 2021).
2.7 Variant callingTo identify genetic variations in the longitudinal clinical isolates relative to acute isolates (S. Typhi isolated during typhoid fever diagnoses), S. Typhi strains isolated before each typhoid index case was treated with antibiotics were used as the reference genome and were compared with those isolated after treatment (follow-up isolates), and/or those isolated from household contacts using breseq v0.38.2 (https://github.com/barricklab/breseq), a computational pipeline for finding mutations relative to a reference sequence in short-read DNA (Deatherage and Barrick, 2014). The variant calling pipeline was as follows: quality-filtering of raw reads using Trimmomatic v0.39 (Bolger et al., 2014), mapping of reads against a reference genome (first strain isolated from the index case), analysis of possible mutations based on mapping data, identification of mutations, and tabular summaries of mutation profile across samples (Langmead and Salzberg, 2012).
2.8 Plasmid identificationTo identify plasmids carrying acquired AMR genes in the isolated S. Typhi strains, a plasmid detection tool PLASMe (v1.1) was used (https://github.com/HubertTang/PLASMe). The tool uses the alignment component in PLASMe to identify closely related plasmids whereas diverged plasmids are predicted using order-specific Transformer models (Tang et al., 2023). Assembled S. Typhi genomes were used for this analysis.
2.9 In vitro biofilm formation assaysBecause of the importance of biofilm formation on gallstones in chronic carriage (Crawford et al., 2010; Gunn et al., 2014), the biofilm-forming ability of all the 22 human S. Typhi isolates was tested under gallbladder simulating conditions as previously described (González et al., 2018). Briefly, S. Typhi biofilms were grown on non-treated polystyrene 96-well plates (Corning, Kennebunkport, ME). To simulate growth conditions on gallstones, wells in two plates were precoated with cholesterol by adding a solution of 5 mg/mL in 1:1 isopropanol:ethanol and air-dried overnight (Crawford et al., 2008). A pure colony of S. Typhi on an XLD agar plate was cultured in Tryptone Soy Broth (TSB). Overnight (O/N) cultures in broth were normalized to OD600 = 0.8 and diluted 1:2,500 in TSB or TSB containing 2.5% human bile, and 100 µL/well was dispensed into the plates. The plates were incubated at 25°C in a Fisherbrand™ nutating mixer (Thermo Fisher Scientific; Hampton, NH) at 24 rpm for 96 h. Media (TSB or TSB containing bile) were changed after every 24 h for consistent S. Typhi biofilm growth. Plates were emptied on the fourth day and washed twice before heat fixing at 60°C for 1 h. The biofilms were stained using a crystal violet solution and acetic acid (33%) used to elute crystal violet before reading the OD570. GraphPad Prism 9.5 was used to analyze the biofilm formation results. One-way analysis of variance (ANOVA) was used to test the level of significance in biofilm formation between the different S. Typhi sub-lineages and in different conditions, i.e., biofilms in absence of cholesterol and bile, in cholesterol-coated plates in absence of bile, and in presence of cholesterol and bile. Student’s t-test was used to test the level of significance in biofilm formation in strains isolated before treatment vs. last strains shed by the patient, P-values less than 0.05 (P<0.05) were considered significant.
2.10 Ethical statement2.10.1 Ethical approvalThis study was conducted in accordance with the ethical standards and guidelines of Scientific and Research Unit (Approval No. SERU4227) of Kenya Medical Research Institute (KEMRI).
2.10.2 Informed consentInformed consent was obtained from all participants involved in the study.
2.10.3 Laboratory protocolsThe biofilm samples were handled following strict laboratory protocols to ensure participant safety and environmental compliance.
3 Results3.1 Genotype identification and clustering treeGiven the highly structured nature of the S. Typhi population, with numerous subclades associated with specific geographical regions and antimicrobial resistance patterns, the genotypes responsible for asymptomatic carriage in the current study were identified using a combination of three methods employed by Pathogenwatch for S. Typhi typing: multi-locus sequence typing (MLST), in silico serotyping (SISTR), and the SNP genotyping scheme (GenoTyphi). Among the 22 bacterial isolates, which include two from blood samples and 20 from stool samples collected at various time points from typhoid fever patients and asymptomatic carriers of S. Typhi (Table 1), all were identified as genotype 4.3.1 (S. Typhi Haplotype 58 [H58]). Of these, 11 isolates from households A and B were classified under lineage 4.3.1.1, whereas the remaining 11 isolates from households C and D fell under lineage 4.3.1.2. (Figure 1). Lineage 4.3.1.2 was further divided into two sub-lineages: 4.3.1.2.EA2 (four isolates from the index case in household C) and 4.3.1.2.EA3 (seven isolates from the index case in household D). Each patient shed S. Typhi belonging to only one lineage or sub-lineage. From household A, isolates (i)–(v) were from the index case whereas isolate (vi) was from a household contact. Isolates (i) and (ii), household B, were from the index case whereas (iii), (iv), and (v) were from an asymptomatic household contact living with the index case. All household C isolates belonged to sub-lineage 4.3.1.2.EA2 whereas all household D isolates belonged to sub-lineage 4.3.1.2.EA3. Longitudinal S. Typhi isolates were collected at different time points as shown in Table 1.
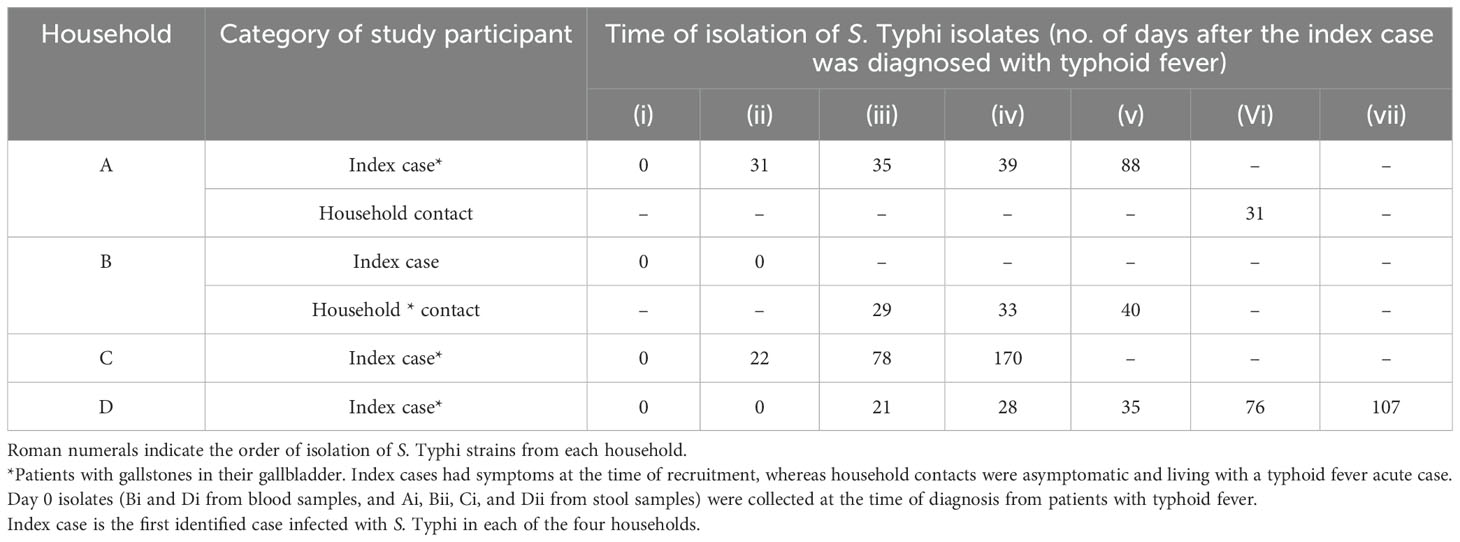
Table 1. Time of isolation/shedding of S. Typhi.
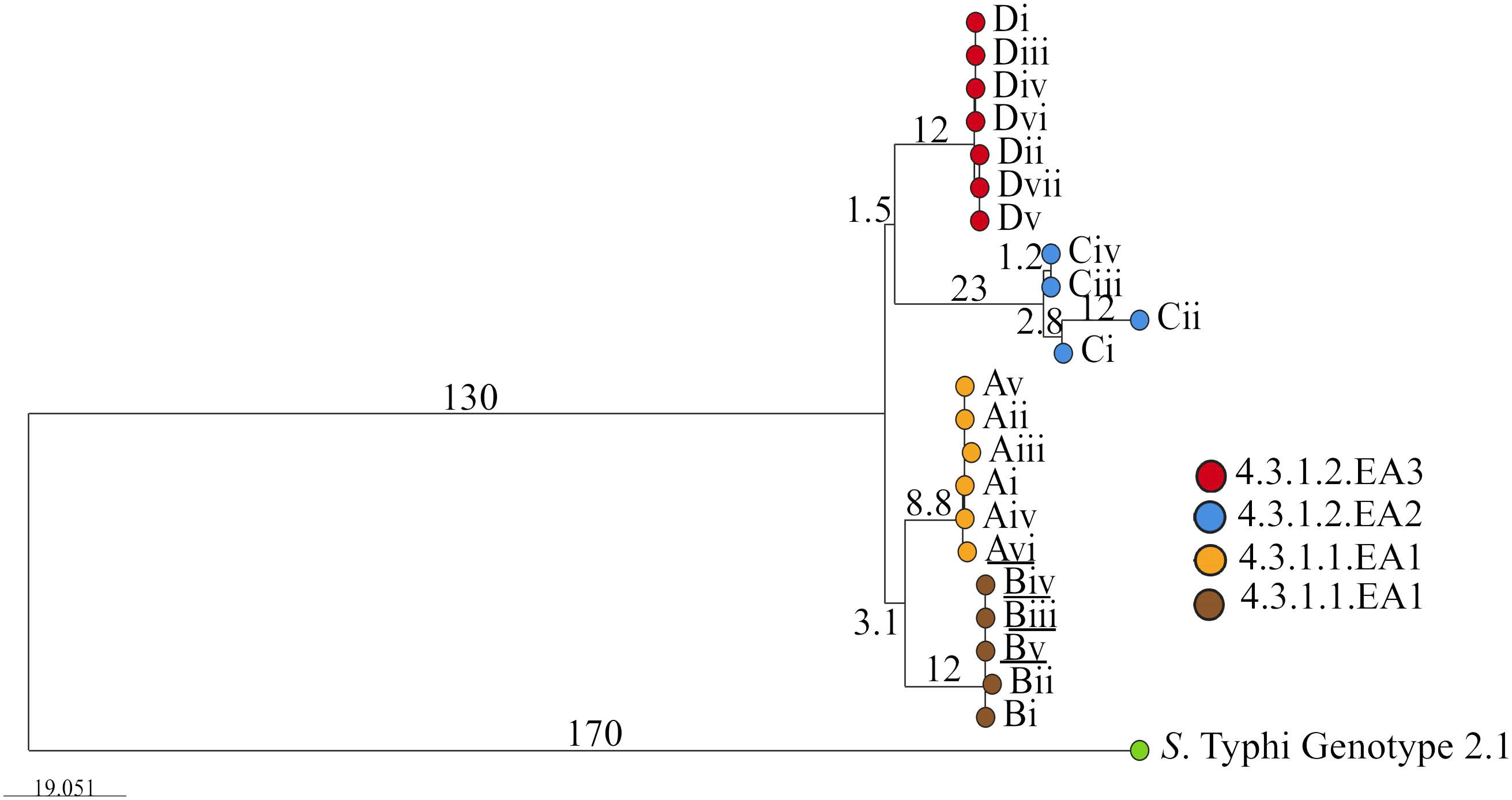
Figure 1. Phylogenetic tree showing the evolutionary relationship between S. Typhi isolated from the study participants living in the different households. Roman numbers indicate specific isolates from the different households (A, B, C and D). Branch tips are color-coded according to the household of isolation. Underlined are isolates from household contacts (Avi, Biii, Biv, and Bv). The bar represents branch length scale bar, indicating evolutionary distance. Branch length corresponds to SNP/genetic distances. The tree can be visualized here https://microreact.org/project/qkEmefYaRCmJRpBKgJnzDS-s-typhi-isolated-from-typhoid-acute-cases-and-asymptomatic-carriers-in-kenya.
3.2 Antimicrobial resistance genesDifferent antimicrobial resistance patterns were observed in the isolated S. Typhi strains. The seven isolates belonging to sub-lineage 4.3.1.2.EA3 (from household D) were multidrug resistant, all expressing the following acquired antimicrobial resistance genes: sul1, dfrA7, catA1, aph(6)-Id, aph(3”)-Ib, sul2, and blaTEM-1, and a point mutation in the Quinolone Resistance Determining Region (QRDR) of gyrA (gyrA S83Y). Phenotypic susceptibility data showed that these seven isolates were resistant to ampicillin, chloramphenicol, trimethoprim–sulfamethoxazole, and nalidixic acid but non-susceptible to ciprofloxacin. The four sub-lineage 4.3.1.2.EA2 isolates (S. Typhi strains from Household C) had a gyrB S464F mutation in the QRDR, and all were non-susceptible to nalidixic acid and ciprofloxacin according to phenotypic susceptibility results. The third group, lineage 4.3.1.1, had six strains (household A isolates) with a gyrB S464F mutation, also demonstrating non-susceptibility to nalidixic acid and ciprofloxacin. The other five lineage 4.3.1.1 strains (household B isolates) had gyrA S83F mutations in the QRDR and showed resistance to nalidixic acid and non-susceptibility to ciprofloxacin (Table 2). S. Typhi isolates carrying genes that confer resistance to ampicillin, sulfamethoxazole–trimethoprim, and chloramphenicol were classified as multidrug resistant (MDR), whereas those lacking these genes were classified as non-MDR.
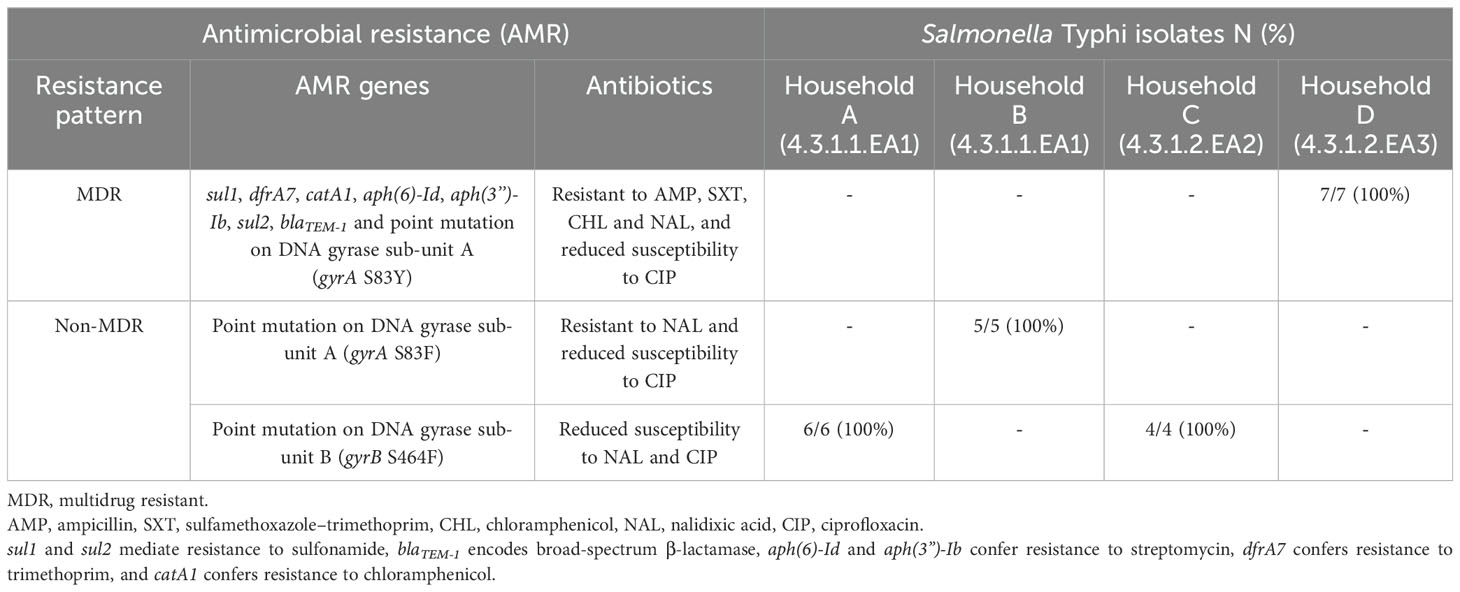
Table 2. Antibiotic resistance profiles in isolated S. Typhi.
3.3 Variant callingComparative genome sequencing of the S. Typhi isolates revealed several key mutations across different sub-lineages. In the sub-lineage 4.3.1.2.EA3 isolates from household D, a missense mutation in the treB gene (treB A383T) was identified in three out of seven isolates. Additionally, a second missense mutation in the tviE gene (tviE P263S), which codes for the Vi polysaccharide biosynthesis protein TviE, was observed in four out of seven isolates. Notably, two of these isolates carried both mutations. A nonsense mutation (E237*) was also found in 1/7 of the isolates, affecting a locus coding for an integrase/transposase family protein similar to DDE-type integrase/transposase/recombinase in S. Typhi CT18 (GenBank: MEM6073260.1). The first follow-up isolate from household C (sub-lineage 4.3.1.2.EA2, isolate Cii) exhibited a total of 16 mutations. This included seven silent mutations, eight missense mutations, and a deletion in the mutL gene, which encodes the DNA mismatch repair endonuclease MutL (Table 3 and Supplementary Table S2 and Supplementary File 3). The second and third follow-up isolates from the same patient (Ciii and Civ) showed two silent mutations, one in the yccC gene, coding for a putative membrane protein, and another in the tehA gene, coding for a dicarboxylate transporter/tellurite-resistance protein (tehA A124A and yccC R199R mutations respectively). These isolates also had missense mutations in the amiA gene, which encodes N-acetylmuramoyl-L-alanine amidase (amiA V145A), and an additional mutation in the hpaX gene (hpaX K124E), encoding 4-hydroxyphenylacetate permease. Notably, the second follow-up isolate (Ciii) had an additional missense mutation (D78G) in a locus coding for phage baseplate assembly protein V (GenBank: MEM6147547.1). In the follow-up isolates from households A and B, which belong to sub-lineage 4.3.1.1, several mutations were detected. The first follow-up S. Typhi isolate from the index case in household A showed no mutations. However, the second follow-up sample (Aiii) had a single nucleotide polymorphism in the crl gene (crl L38P), which encodes a sigma factor-binding protein. The third and fourth follow-up isolates from the same patient (Aiv and Av) each had a silent mutation in the tnpA gene (tnpA Y41Y), which codes for IS200/IS605 family transposase. Additionally, an isolate from a household contact in the same household (Avi) had a missense mutation in the treB gene (treB A383T). From household B, S. Typhi isolated from the stool of the index case before treatment (isolate Bii) had a missense mutation in the waaK gene (waaK P167L), which encodes lipopolysaccharide N-acetylglucosaminyltransferase. This mutation was not present in the strain isolated from the patient’s blood sample collected on the same day.
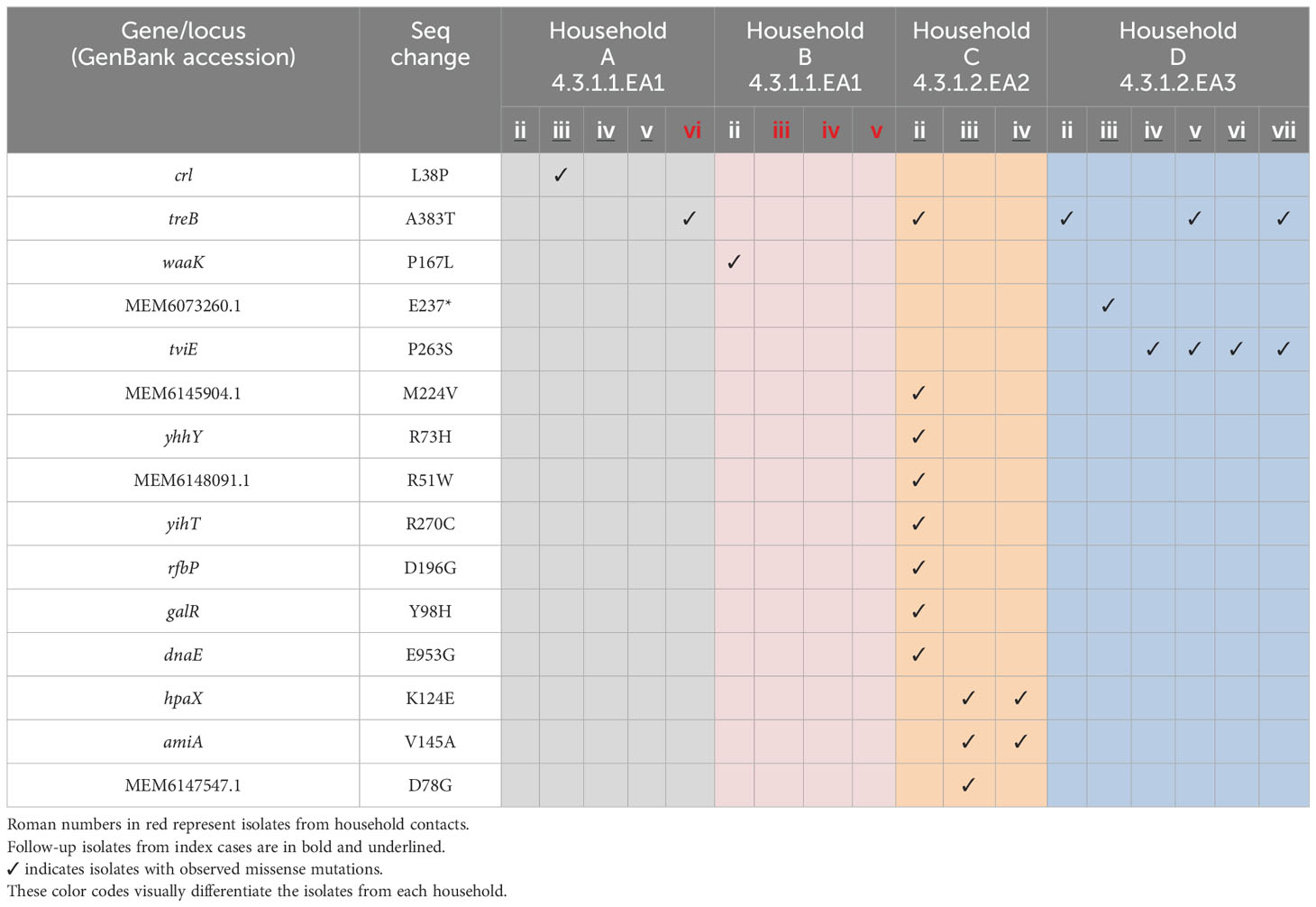
Table 3. Missense mutations in S. Typhi strains isolated from index cases after apparent clinical resolution and from asymptomatic household contacts.
3.4 Plasmid identificationS. Typhi genotype 4.3.1 lineages/sub-lineages identified in this study were found to contain plasmids. Genes conferring multidrug resistance in S. Typhi were detected in two different plasmids in sub-group 4.3.1.2.EA3 (household D) strains. The antimicrobial resistance genes sul1, dfrA, catA1, aph(6)-Id, aph(3”)-Ib, and sul2 were detected in a plasmid with sequences corresponding to S. Typhi strain 311189_252186 plasmid pHCM1, whereas blaTEM-1 was detected in a plasmid with sequences corresponding to E. coli NES6 plasmid pN-ES-6-1 (Table 4). The two plasmids were not detected in non-MDR S. Typhi Isolates.
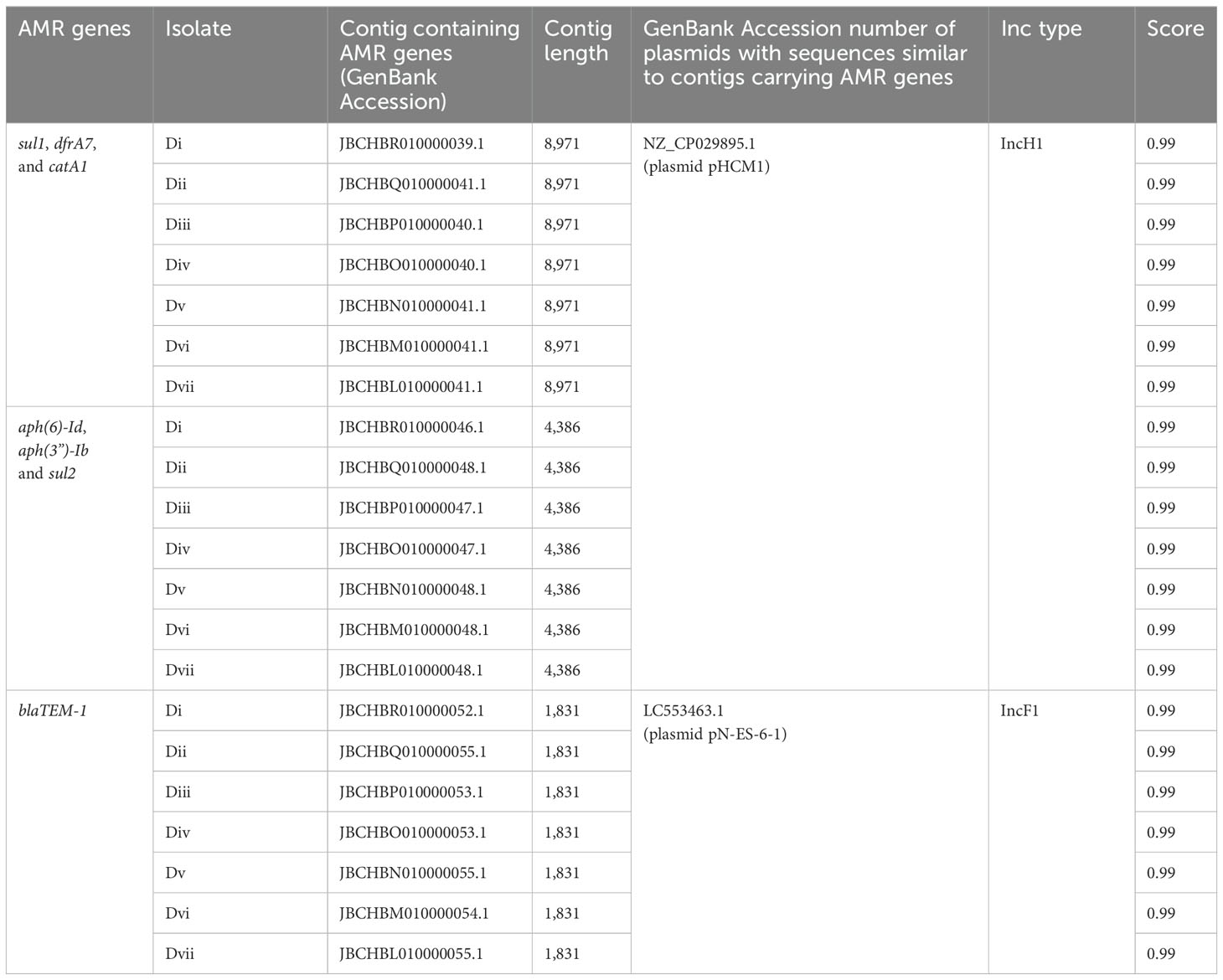
Table 4. Contigs carrying AMR genes in household D S. Typhi isolates and plasmids with similar sequences from the NCBI public database.
3.5 Biofilm formationWe wanted to determine if biofilm-forming ability is correlated with the stage of typhoid fever/carriage, antimicrobial resistance, or the presence of plasmids. Biofilms were examined in gallbladder-simulating conditions. There was varying ability to form biofilms under in vitro conditions in the S. Typhi strains tested. All isolates formed weak biofilms in absence of both cholesterol and bile (OD570 below 0.3) and significantly strong biofilms in the presence of cholesterol and 2.5% human bile (Figure 2A). Differences in biofilm forming ability were observed across the identified genotype 4.3.1 lineages/sub-lineages. The sub-lineage 4.3.1.2.EA2 formed the strongest biofilms (OD570 slightly above 2.0), whereas sub-lineage 4.3.1.2.EA3 formed relatively weak biofilms (OD570 below 0.5) even in the presence of cholesterol and human bile (Figure 2A). However, there was no statistical significance in biofilm forming ability in strains isolated during the symptomatic vs. asymptomatic stage in each of the three sub-groups of S. Typhi (Figure 2C). The weak biofilm-forming isolates from sub-lineage 4.3.1.2.EA3 had acquired antimicrobial resistance (AMR) genes encoded on plasmids. Sub-lineages 4.3.1.2.EA2 and 4.3.1.1.EA1 did not have identifiable acquired AMR genes, but isolates in these subgroups formed significantly stronger biofilms in the presence of both cholesterol and human bile (Figure 2B). Strains from symptomatic patients and asymptomatic carriers exhibited no statistically significant difference in biofilm-forming ability across the three S. Typhi subgroups (Figure 2C).
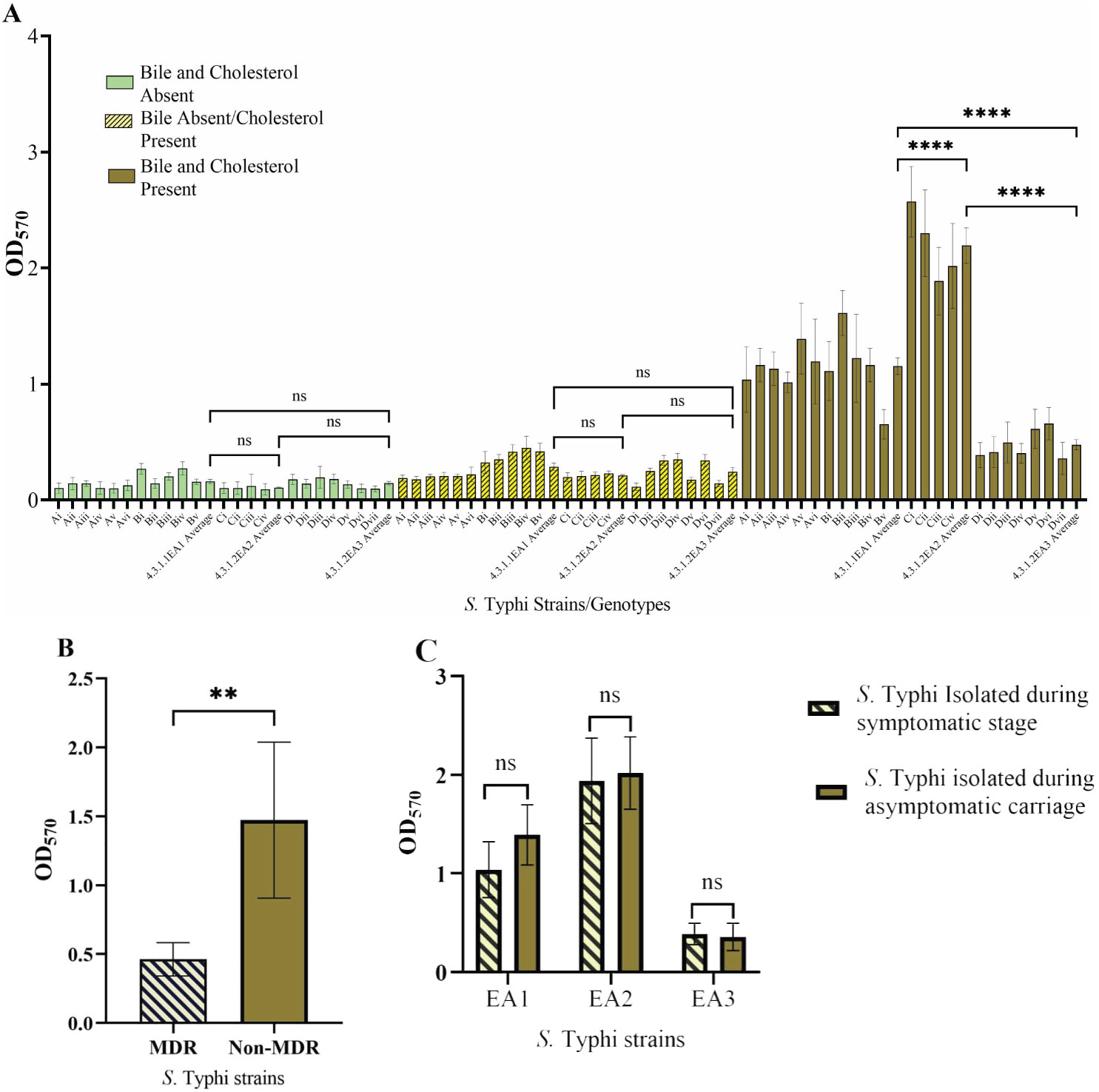
Figure 2. Salmonella Typhi biofilms. (A) Quantity of biofilms after growth in presence/absence of cholesterol and/or bile. (B) Biofilm formation by MDR S. Typhi strains vs. non-MDR strains. (C) Comparison of biofilms formed by S. Typhi strains isolated before treatment vs. after treatment. Error bars represent SEM, ****P<0.001; **P<0.05; nsP≥0.5.
4 DiscussionAlthough typhoid fever has largely been eliminated in high-income countries, it remains a major global public health concern especially among low- and middle-income countries (Kim et al., 2022). Haplotype 58 (H58), which is associated with antimicrobial resistance, has also been reported from other parts of sub-Saharan Africa and Southeast Asia (Maes et al., 2020; Pragasam et al., 2020). In this study, H58 (genotype 4.3.1) was identified as the single genotype shed by four cholelithiasis patients living in a typhoid endemic setting in Nairobi, Kenya. S. Typhi H58 is the most dominant genotype in many parts of Southeast and South Asia as well as in East Africa and has spread globally (Park et al., 2018; Wong et al., 2015). Three H58 east African subgroups (4.3.1.1.EA1, 4.3.1.2.EA2, 4.3.1.2.EA3) previously reported circulating in the current study setting by our group (Kariuki et al., 2021) were the main lineages/sub-lineages shed by the cholelithiasis patients. The most abundant subgroup was 4.3.1.1.EA1 with 11/22 (50%) isolates, originating from individuals living in two different households. In one of these households, an acute case shed an S. Typhi belonging to the same sub-group as an asymptomatic household contact who was also diagnosed with gallstones. This suggests possible transmission of the pathogen by the carrier to the household contact (household B). From a different household, a typhoid patient also diagnosed with gallstones continued to shed sub-lineage 4.3.1.2.EA2, whereas in the fourth household, S. Typhi sub-lineage 4.3.1.2.EA3 strains were isolated from stool samples collected from an acute case after treatment. Unlike the other two sub-groups, the sub-lineage 4.3.1.2.EA3 strains had MDR genes, showing resistance to ampicillin, sulfamethoxazole–trimethoprim, and chloramphenicol. All 22 S. Typhi isolates had point mutations in the QRDR, conferring reduced susceptibility to ciprofloxacin, a drug of choice for treating typhoid fever. There was no variation noted in antimicrobial resistance patterns among strains isolated from the patients in the same household.
Multidrug resistance genes were not detected in 4.3.1.1.EA1 and 4.3.1.2.EA2 S. Typhi genomes, but the strains belonging to these subgroups formed significantly stronger biofilms as compared with the MDR sub-lineage 4.3.1.2.EA3 strains. Biofilms act as a physical barrier protecting bacteria from killing by antimicrobials including antibiotics. A previous study demonstrated the role of biofilms in protecting Salmonella from ciprofloxacin (González et al., 2018). We hypothesize that the 4.3.1.1.EA1 and 4.3.1.2.EA2 sub-lineages form better biofilms to counteract the absence of antimicrobial resistance factors or, conversely, that lineage 4.3.1.2.EA3 has lost biofilm-related genes because it possesses acquired genes (in plasmids) encoding strong antimicrobial resistance. To the best of our knowledge, this is the first study comparing biofilm forming ability in different S. Typhi lineages. The mechanism leading to differences in biofilm formation in isolates from the same genotype will need to be further investigated.
Genetic variations were observed in S. Typhi from asymptomatic carriers, with a treB A383T point mutation being observed in at least one isolate from each of the four households. The treB gene codes for PTS trehalose transporter subunit IIBC. As seen in Table 3, household B, some of the mutations observed in the first follow-up isolate were not detected in S. Typhi strains isolated during the consecutive follow-ups. However, some mutations were observed in more than one strain isolated from the same patient. From the patient shedding sub-lineage 4.3.1.2.EA3 strains, the tviE P263S mutation was observed in the fourth isolate and all strains isolated thereafter. This suggests that some mutations are retained in the population during asymptomatic carriage, whereas others are not. Mutations that confer a selective advantage to S. Typhi during carriage are likely maintained, whereas those that negatively impact the pathogen are eliminated. Additionally, changes in environmental conditions during shedding from the gallbladder may alter the selective pressure on mutations, resulting in the retention or loss of specific mutations. Although no strain was isolated directly from gallbladder in our study, mutations in the tviE gene were also observed in S. Typhi gallbladder genome sequences in a previous study (Thanh Duy et al., 2020). The tviE gene facilitates the polymerization and translocation of the Vi capsule (Virlogeux et al., 1995). Vi capsular polysaccharide, an antiphagocytic capsule, covers the surface of S. Typhi allowing it to selectively evade phagocytosis by human neutrophils whereas promoting human macrophage phagocytosis (Zhang et al., 2022). This crucial virulence factor in S. Typhi (Vi) also plays a key role in the development of vaccines against typhoid fever (Hu et al., 2017). However, additional research will be required to understand if this mutation alters the expression of Vi antigen to benefit S. Typhi pathogenesis or chronic carriage. Both nonsense and non-synonymous mutations have been previously reported in gallbladder S. Typhi isolates, particularly in genes encoding hypothetical proteins, membrane lipoproteins, transport/binding proteins, surface antigens, and carbohydrate degradation enzymes (Thanh Duy et al., 2020).
The primary limitation of this study is the unavailability of gallbladder isolates from patients shedding S. Typhi for comparison with those obtained from stool and blood samples. Additionally, there were no S. Typhi isolates from other genotypes available for comparative analysis.
5 ConclusionsThe resistance patterns in S. Typhi did not change during the duration of asymptomatic carriage in study participants, but these individuals continued to shed and transmit drug-resistant strains of this pathogen. This included strains isolated from the patients in the same household, suggesting that asymptomatic typhoid carriers are responsible for the transmission and persistence of drug-resistant S. Typhi in the study setting. No specific set of AMR genes was linked to asymptomatic carriage. Mutations in S. Typhi were observed to occur during carriage including those in the Vi antigen locus. Sub-lineages analyzed in this study that were not multidrug resistant showed the ability to form stronger biofilms than the multidrug resistant strains. This study provides some insights into mutations, drug resistance, and biofilm formation during typhoid carriage, and this information may be used to influence public health approaches aimed at reducing carriage, persistence, and transmission of S. Typhi.
Data availability statementThe data presented in the study are deposited in the NCBI GenBank repository, accession number PRJNA1101423.
Ethics statementThe studies involving humans were approved by The Scientific and Ethics Review Unit (SERU) of Kenya Medical Research Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributionsPM: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. PW: Supervision, Writing – review & editing. MW: Supervision, Writing – review & editing. CM: Project administration, Writing – review & editing. SMK: Methodology, Writing – review & editing. MMM: Formal analysis, Writing – review & editing. MM: Project administration, Writing – review & editing. JFG: Formal analysis, Investigation, Writing – review & editing. SK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. JSG: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the National Institute of Health (NIH) National Institute of Allergy and Infectious Diseases (Grants: R01 AI099525 and R01 AI116917).
AcknowledgmentsWe thank Darius Ideke (Kenya Medical Research Institute) and Victoria Sadowski (Abigail Wexner Research Institute at Nationwide Children’s Hospital) and Christine Sun (Ohio State University) for their technical support.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1468866/full#supplementary-material
ReferencesAhirwar, S. K., Pratap, C. B., Patel, S. K., Shukla, V. K., Singh, I. G., Mishra, O. P., et al. (2014). Acid exposure induces multiplication of salmonella enterica serovar typhi. J. Clin. Microbiol. 52, 4330–4333. doi: 10.1128/jcm.02275-14
PubMed Abstract | Crossref Full Text | Google Scholar
An, C., Chon, H., Ku, W., Eom, S., Seok, M., Kim, S., et al. (2022). Bile acids: major regulator of the gut microbiome. Microorganisms 10, 1792. doi: 10.3390/microorganisms10091792
PubMed Abstract | Crossref Full Text | Google Scholar
Argimón, S., Yeats, C. A., Goater, R. J., Abudahab, K., Taylor, B., Underwood, A., et al. (2021). A global resource for genomic predictions of antimicrobial resistance and surveillance of salmonella typhi at pathogenwatch. Nat. Commun. 12, 2879. doi: 10.1038/s41467-021-23091-2
PubMed Abstract | Crossref Full Text | Google Scholar
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
PubMed Abstract | Crossref Full Text | Google Scholar
Chan, P. P., Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Gene Prediction, Kollmar, M. (eds). Methods in Molecular Biology, vol 1962. (New York, NY: Humana). doi: 10.1007/978-1-4939-9173-0_1
PubMed Abstract | Crossref Full Text | Google Scholar
Clinical and Laboratory Standards Institute. (2023). “CLSI supplement M100,” in Performance Standards for Antimicrobial Susceptibility Testing, 33rd (Clinical and Laboratory Standards Institute, USA).
Crawford, R. W., Gibson, D. L., Kay, W. W., Gunn, J. S. (2008). Identification of a bile-induced exopolysaccharide required for salmonella biofilm formation on gallstone surfaces. Infection Immun. 76, 5341–5349. doi: 10.1128/iai.00786-08
PubMed Abstract | Crossref Full Text | Google Scholar
Crawford, R. W., Rosales-Reyes, R., Ramirez-Aguilar, M. D. L. L., Chapa-Azuela, O., Alpuche-Aranda, C., Gunn, J. S. (2010). Gallstones play a significant role in salmonella spp. Gallbladder colonization and carriage. Proc. Natl. Acad. Sci. 107, 4353–4358. doi: 10.1073/pnas.1000862107
PubMed Abstract | Crossref Full Text | Google Scholar
Deatherage, D. E., Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Eng. Analyzing Multicellular Systems: Methods Protoc. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
PubMed Abstract | Crossref Full Text | Google Scholar
Dongol, S., Thompson, C. N., Clare, S., Nga, T. V. T., Duy, P. T., Karkey, A., et al. (2012). The microbiological and clinical characteristics of invasive salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PloS One 7, e47342. doi: 10.1371/journal.pone.0047342
PubMed Abstract | Crossref Full Text | Google Scholar
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11, 12728. doi: 10.1038/s41598-021-91456-0
PubMed Abstract | Crossref Full Text | Google Scholar
Gaines, S., Sprinz, H., Tully, J. G., Tigertt, W. D. (1968). Studies on infection and immunity in experimental typhoid fever: VII. The distribution of salmonella typhi in chimpanzee tissue following oral challenge, and the relationship between the numbers of bacilli and morphologic lesions. J. Infect. Dis. 118, 293–306. doi: 10.1093/infdis/118.3.293
PubMed Abstract | Crossref Full Text | Google Scholar
Gibson, D. L., White, A. P., Snyder, S. D., Martin, S., Heiss, C., Azadi, P., et al. (2006). Salmonella produces an O-antigen capsule regulated by agfD and important for environmental persistence. J. Bacteriol. 188, 7722–7730. doi: 10.1128/jb.00809-06
PubMed Abstract | Crossref Full Text | Google Scholar
González, J. F., Alberts, H., Lee, J., Doolittle, L., Gunn, J. S. (2018). Biofilm formation protects salmonella from the antibiotic ciprofloxacin in vitro and in vivo in the mouse model of chronic carriage. Sci. Rep. 8, 2225. doi: 10.1038/s41598-017-18516-2
PubMed Abstract | Crossref Full Text | Google Scholar
Gonzalez-Escobedo, G., Gunn, J. S. (2013). Identification of Salmonella enterica Serovar Typhimurium Genes Regulated during Biofilm Formation on Cholesterol Gallstone Surfaces. Infection Immun. 81, 3770–3780. doi: 10.1128/IAI.00647-13
PubMed Abstract | Crossref Full Text | Google Scholar
Gonzalez-Escobedo, G., Marshall, J. M., Gunn, J. S. (2011). Chronic and acute infection of the gall bladder by salmonella typhi: understanding the carrier state. Nat. Rev. Microbiol. 9, 9–14. doi: 10.1128/iai.00647-13
PubMed Abstract | Crossref Full Text | Google Scholar
Grant, N. A., Donkor, G. Y., Sontz, J. T., Soto, W., Waters, C. M. (2023).Deployment of a vibrio cholerae ordered transposon mutant library in a quorum-competent genetic background. Available online at: https://www.biorxiv.org/content/10.1101/2023.10.31.564941v2.full (Accessed July 10, 2024).
留言 (0)