Wheat is a crop that has played a key role in global food security for many centuries (Erenstein et al., 2022). In Serbia, wheat is the second most widely grown cereal after maize, and the majority of production is grown in the province of Vojvodina (southern parts of the Pannonian Plain), where the climate is particularly favorable for growing winter wheat. More than 50 different viruses are known to infect wheat worldwide, causing severe symptoms with potentially grave impact on yield and quality (Ordon et al., 2009; Liu et al., 2020).
Over the past several decades, the wheat dwarf disease caused by wheat dwarf virus (WDV, Mastrevirus hordei, genus Mastrevirus, family Geminiviridae) has spread more extensively in many cereal-growing areas across Europe, Africa and Asia, causing huge economic losses when its presence reaches epidemic proportions (Ekzayez et al., 2011; Liu et al., 2012; Schubert et al., 2014; Wang et al., 2014; Parizipour et al., 2017; Liu et al., 2020).
Typical symptoms induced by WDV include dwarfing and leaf yellowing, along with reduced headings. The virus may cause yield losses of up to 100% (Jones, 2021), primarily in wheat and barley crops, but it has also been isolated from rye, triticale, and several wild grasses (Lindblad et al., 1999; Köklü et al., 2007; Schubert et al., 2007; Ramsell et al., 2009). It is transmitted by leafhoppers of the genus Psammotettix (family Cicadellidae) in a circulative, non-propagative manner, with P. alienus being considered the most efficient vector in many regions (Lemmetty and Huusela-Veistola, 2005; Wang et al., 2014; Ekzayez et al., 2011). The virus can neither be transmitted by contact between plants, nor by soil, pollen or seeds (Vacke et al., 2004).
Wheat dwarf virus has particles with a characteristic twinned morphology and its genome consists of a monopartite, circular, single-stranded DNA of about 2.6–2.8 kb in length. The WDV genome encodes four proteins, in both the virion- and complementary-sense strands. The virion-sense ORFs encode the capsid protein (CP, gene V1) and the movement protein (MP, gene V2) involved in cell-to-cell movement. The complementary-sense ORFs encode replication-associated proteins (Rep and RepA), expressed from genes C1 and C2. The genome also has long and short intergenic regions (LIR and SIR, respectively), which contain regulatory elements for viral replication and transcription (Dickinson et al., 1996; Kvarnheden et al., 2002; Liu et al., 2014). WDV was initially divided into two main groups: wheat- (WDV-W group) and barley-adapted forms (WDV-B group). Later, phylogenetic analysis identified six strains (A–F) based on sequence similarity between isolates and the phylogenetic relationship; strains A and F originated mainly from barley and were assigned to WDV-B, while strains B–E were assigned to WDV-W specific group (Lindsten and Vacke, 1991; Muhire et al., 2013; Schubert et al., 2014; Wu et al., 2015). Wheat strains show a high level of mutual identity (>98%), while WDV-B isolates are more variable and nucleotide identity among them exceeds 94% (Tóbiás et al., 2011). These two groups of isolates also show differences in their host range: isolates belonging to the wheat-adapted group infect wheat, barley, rye, triticale, and many wild grasses, whereas isolates from the barley-adapted group infect almost exclusively barley and rarely wheat (Köklü et al., 2007; Schubert et al., 2007; Kundu et al., 2009; Wu et al., 2015).
Regardless of the importance of wheat as a crop in Serbia, few studies have been conducted so far to determine the presence of viruses in major cereal-producting areas in our country. Apart from wheat streak mosaic virus (WSMV, Tritimovirus tritici), which has been the most frequently isolated virus of wheat in Serbia (Šutić and Tošić, 1964; Vučurović et al., 2020), brome mosaic virus (BMV, Bromovirus BMV), as well as barley yellow dwarf virus-PAV and -MAV (BYDV-PAV, Luteovirus pavhordei and BYDV-MAV, Luteovirus mavhordei) and cereal yellow dwarf virus-RPV (CYDV-RPV, Polerovirus CYDVRPV) (Tošić, 1971; Balázs et al., 1990) have also been detected. Interestingly, an investigation conducted by Vučurović et al. (2020) failed to detect the presence of any of these viruses in a substantial percentage of symptomatic wheat samples. In Serbia, WDV was detected for the first time in 2017 (Ristić et al., 2019), but there was later no further information on its occurrence and distribution in our country. Therefore, the aim of this study was to evaluate its distribution in Serbia. Since no sequence data had been available for Serbian WDV isolates, this study also aimed to determine the complete genomic sequences of the isolates collected from different regions in Serbia. The study expands the general knowledge of the molecular diversity and population structure of WDV isolates originating from Serbia and reveals their genetic relationship to those from other parts of the world.
Materials and methods Plant collections and virus detectionPlants with typical dwarfing symptoms were observed in many winter wheat crops across Serbia from March to May of 2019. A total of 161 samples were collected after visual inspection of 21 locations in nine administrative districts of Serbia. Before sampling, disease incidence was estimated in each field by counting plants that exhibits virus-like symptoms in a random batch of 100 plants in four replicates. The sampling method was adjusted to field size. Up to 10 obviously symptomatic plants were selected randomly in small plots (up to 0.5 ha). In contrast, more than 10 symptomatic plants were also collected randomly in large plots (2–3 ha) by moving across the field according to “X model” (Table 1). The samples obtained from leaves of a symptomatic plant were initially tested for WDV and other most common wheat viruses using a double-antibody sandwich (DAS)-ELISA test with commercial polyclonal antisera (Loewe Biochemica, Germany): WSMV, BYDV-PAV and -MAV, BMV, and wheat spindle streak mosaic virus (WSSMV), according to label instructions. Absorbance at 405 nm was measured with an ELISA microplate reader (DASsrl, Italy) and samples showing average optical densities (OD) twice as high as the average of the negative control were considered positive. Commercial positive and negative controls for the viruses mentioned, which are available in commercial kits (Loewe Biochemica, Germany), were included in each ELISA.
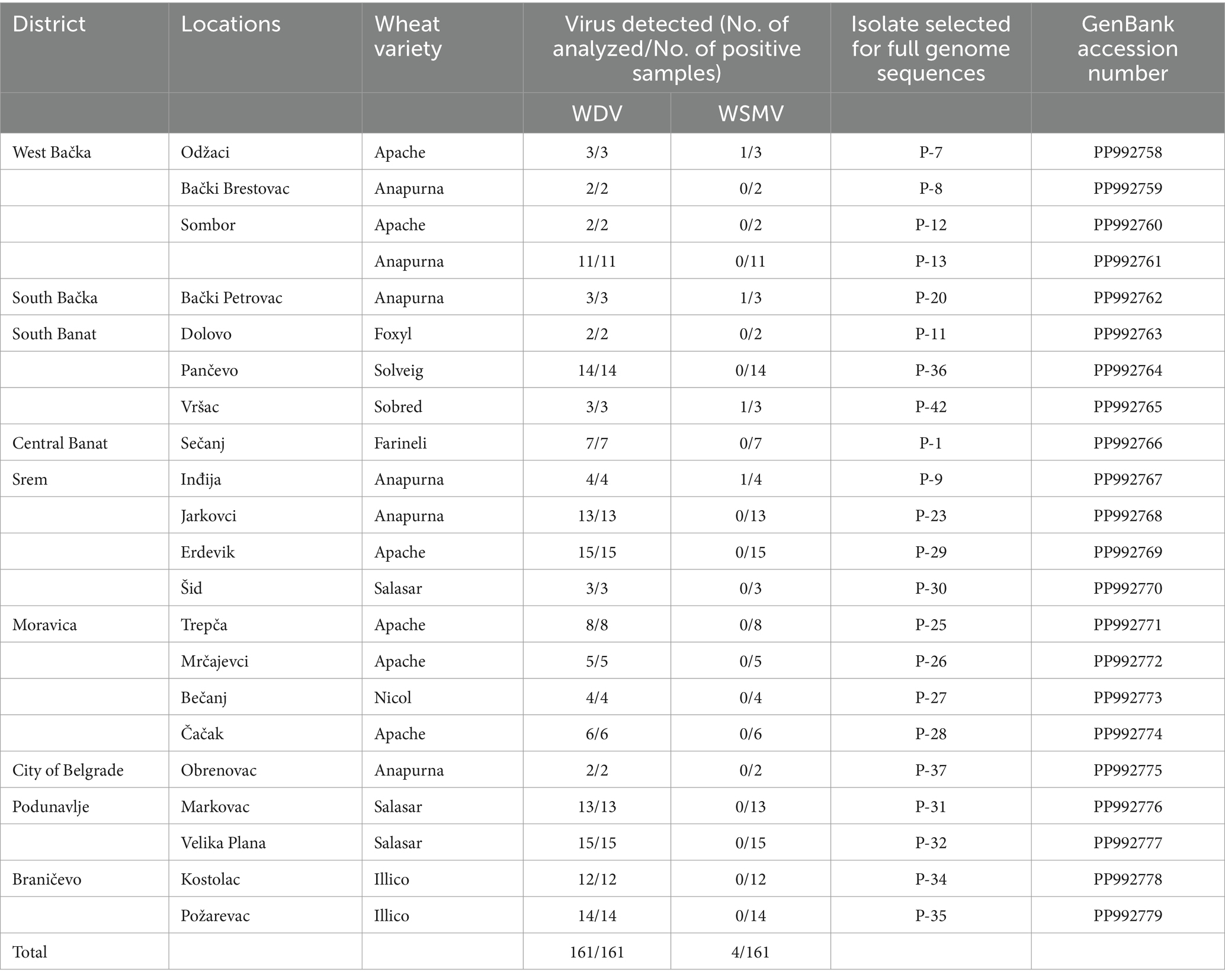
Table 1. Detection of WDV infection by ELISA test in winter wheat crops of various Serbian districts in 2019 and isolates analyzed in this study.
A total of 22 ELISA-positive samples with a single WDV infection originating from different locations or wheat varieties were randomly selected for molecular characterization based on the complete genome sequence. A previously identified WDV isolate, designated as P1-17 (Ristić et al., 2019), was also included in the study.
DNA extraction, rolling circle amplification, and PCRTo obtain the complete genome of selected isolates, several overlapping segments of the WDV genome were amplified using a combination of different primers listed in Table 2. Total nucleic acids were isolated from 100 mg of symptomatic leaves using a cetyltrimethylammonium bromide (CTAB) method (Li et al., 2008) and used for PCR amplification, as well as in rolling circle amplification (RCA) to enrich the viral DNA present in the sample. The RCA reaction was performed using the REPLI-g Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. The RCA product was diluted 1:20 and used for amplification of the target DNA fragments. The PCR amplification was performed in a 25 μL reaction using PCR Master Mix (ThermoFisher Scientific Inc., United States) containing 12.5 μL of Master mix (2x), 2.5 μL of each primer (10 μM), 1 μL of template DNA, and 6.5 μL of molecular-grade water. PCR reactions were performed in an Eppendorf Mastercycler X50l (Eppendorf, Germany) as follows: initial denaturation at 94°C for 2 min, and the final extension at 72°C for 10 min. The annealing temperature and number of cycles are given for each primer pair in Table 2. Negative control was included by replacing template DNA with molecular-grade water, while the Serbian P1-17 WDV isolate was used as a positive control. PCR products were separated by electrophoresis in 1% agarose gel run in 1xTBE buffer and stained with ethidium bromide.
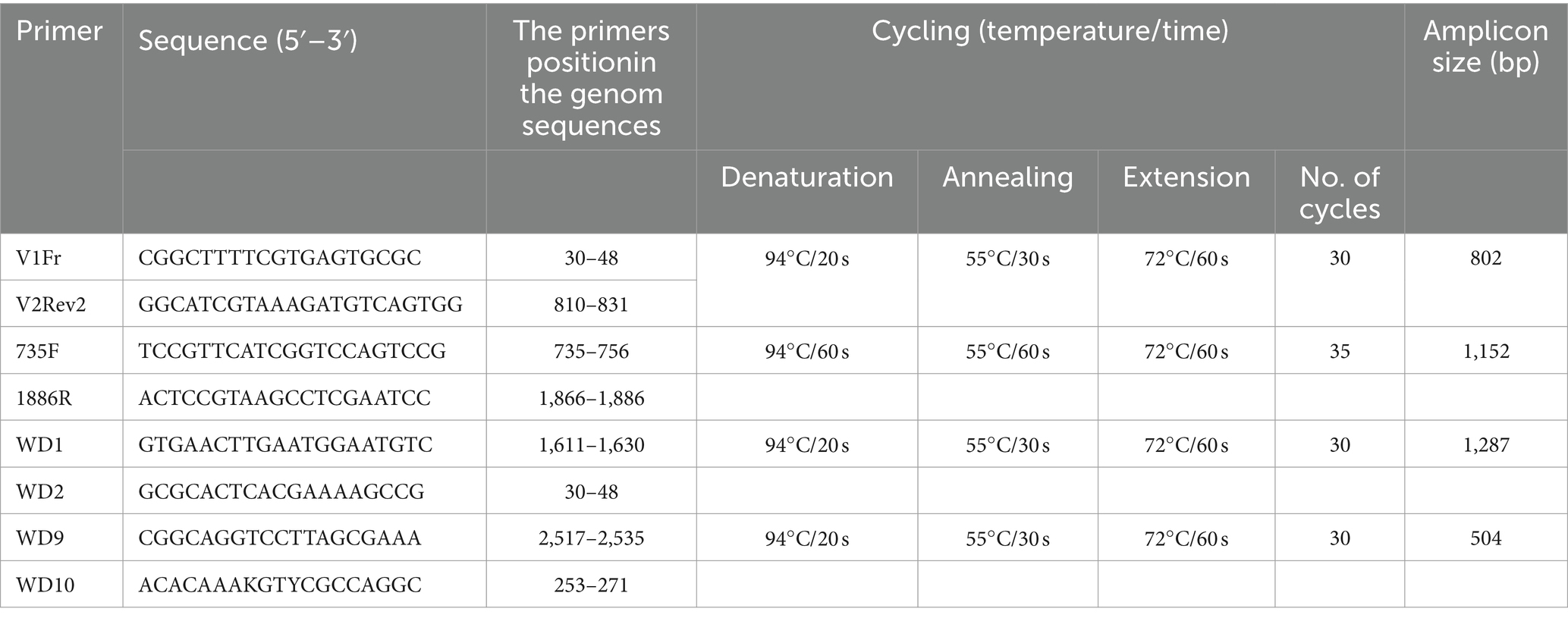
Table 2. Primers used for amplification and sequencing.
Sequence analysis and construction of phylogenetic treesProducts of an expected size, obtained in PCR assays were purified with a QIAquick PCR Purification Kit (Qiagen) and at least three samples of each amplicon were sent for sequencing in both directions on an automated sequencer (Macrogen-Europe BV) using the same primers as in amplifications. For each isolate, sequence data were analyzed and assembled using the ClustalW program (Thompson et al., 1994) implemented in MEGA X software (Kumar et al., 2018), and complete sequences of the WDV genome of isolates were obtained. All sequences generated in this study were deposited in the GenBank database and assigned accession numbers (Table 1). The obtained sequences were compared with each other by calculating nucleotide identities (nt) using the p-distance model implemented in MEGA X software (Kumar et al., 2018). All Serbian sequences were also compared with previously reported isolates available in the GenBank using BLAST algorithm.
A phylogenetic tree was inferred using the maximum parsimony method implemented in MEGA X. The bootstrap method (1,000 replicates) was used to assess the reliability of individual nodes in the phylogenetic tree, and bootstrap values <70% were omitted. The sequence AM296025 of a German isolate of oat dwarf virus (ODV) was used as outgroup sequence. The phylogenetic trees were visualized using iTOL v6 (Letunic and Bork, 2024). Intra- and intergroup diversity was calculated using Diversity Estimation analysis and K2 + G evolutionary model, both implemented in MEGA X.
Recombination analysisRecombination analysis was carried out using the Recombination Detection Program v.4.16 (RDP4). Recombination events and recombination break points were analyzed using the RDP, GENECONV, BootScan, MaxChi, Chimera, SiScan, and 3Seq methods implemented in the RDP4 program (Martin et al., 2015). Default parameter values were used, and a sequence was considered recombinant if the recombination signal was supported by at least five methods with p values ≤0.05.
Results Symptoms and virus detection using DAS-ELISADuring visual inspection of winter wheat fields of different varieties over the period from March to May 2019, symptoms resembling those caused by WDV were observed at various locations throughout Serbia. The first symptoms included various types of leaf yellowing (Figure 1A) and necrotic leaf spots, as well as different degrees of dwarfing (Figure 1B), and reduced heading (Figure 1C). Later, these plants decayed due to necrosis and there were large yield losses in many locations regardless of wheat variety. The average estimated disease incidence mostly ranged between 40 and 60%, but it was extremely high (up to 100%) in some fields.

Figure 1. Symptoms of wheat dwarf (WDV) infection on wheat: (A) yellowing of leaf; (B) severe yellowing and plant dwarfism; (C) yellowing, necrosis, and reduced heading formation.
The results of DAS-ELISA test revealed that WDV was present in all analyzed samples collected at 21 locations in all of the nine inspected districts (Table 1; Figure 2), indicating that WDV was common and widely distributed throughout Serbia. In a single infection, WDV was detected in 97.52% of the samples tested. The presence of WSMV was proved in 2.48% of the samples tested at four locations and the virus was in mixed infection with WDV. No other viruses were detected.
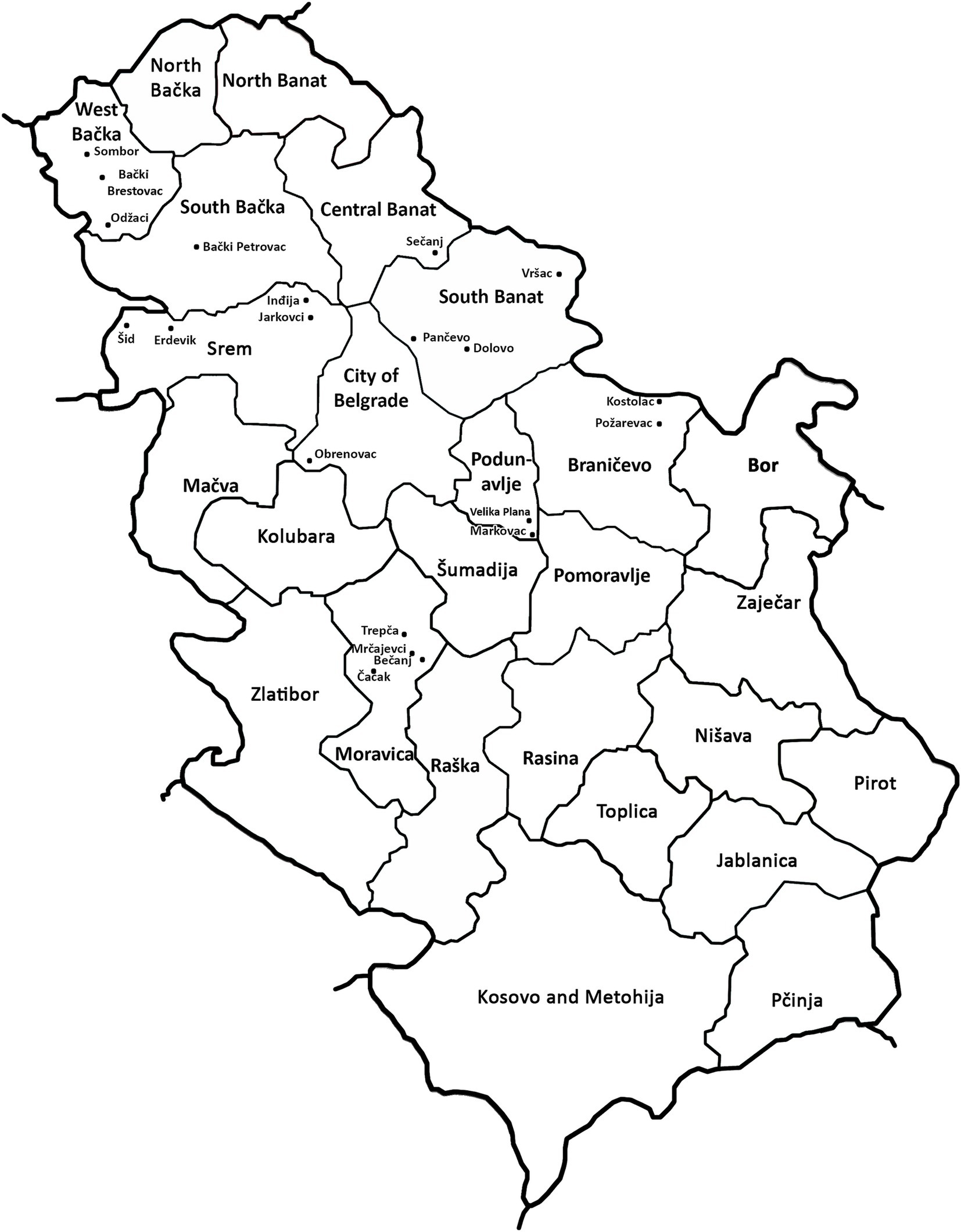
Figure 2. Locations with wheat dwarf virus (WDV) infection in Serbia in the spring of 2019.
Genetic diversity of Serbian WDV isolatesAll 22 selected isolates collected during this study, as well as the previously identified isolate P1-17 (PP992780), were fully sequenced and submitted to GenBank. Details of the origin of selected samples and accession numbers in GenBank are given in Table 1. Analyses of the WDV complete genome sequence showed high levels of identity among the Serbian WDV isolates originating from wheat, which exhibited nt identities ranging from 98.3 to 99.9%. Isolates P-26 (Mrčajevci) and P-31 (Markovac) showed the highest percentage of nt identity with isolates P-32 (Velika Plana) (99.9%), while P-7 (Odžaci) was the most distant from isolates P-12 (Sombor) (98.3%). Also, isolate P-7 was the most distant from all other Serbian WDV isolates and shared 98.3–99.4% nt identity with theme. BLAST results revealed that Serbian WDV isolates shared the highest nt identity of 98.95–99.78% with those available in GenBank. Most Serbian isolates showed the highest nt identity of 98.95–99.78% with the Ukraine isolate FN806784. Isolates P-7 shared the highest nt identity of 99.75% with the Czech isolate FJ546188, while the sequence of isolate P-12 had the highest nucleotide identity of 99.42% with one isolate each from the Czech Republic and Germany (FJ546191 and AM296023, respectively).
It is noteworthy that none of the sampled plants showed evidence of mixed infections with different WDV isolates characterized by multiple peaks in the sequencing ladder, and none of the seven recombination detection programs showed possible recombination events among the examined WDV isolates.
WDV phylogenetic analysisA maximum parsimony tree (Figure 3) was constructed using the whole genome sequences of the WDV isolates identified in this study and selected sequences of 40 previously characterized isolates retrieved from GenBank (Supplementary Table S1). The isolates were divided into two groups, wheat- and barley-adapted, with a genetic diversity between them of 0.197 ± 0.010. The barley-adapted group (0.039 ± 0.003) comprises clades A and F, while clades B, C, D, and E belong to the wheat-adapted group (0.032 ± 0.002). Genetic diversity among the six clades of WDV isolates ranged from 0.049 ± 0.004 to 0.209 ± 0.011, while the diversity within each clade was: 0.036 ± 0.003 (A), 0.009 ± 0.001 (C), 0.036 ± 0.003 (E), and 0.007 ± 0.001 (F). European isolates originating from barley were grouped into clades A and F. Clade C comprised wheat isolates from Hungary, clades D and B contained a barley isolate each from Iran, whereas clade E consisted of European and Chinese isolates from different hosts, including wheat. The 23 Serbian WDV isolates were grouped into clade E, together with other European WDV isolates manly from wheat. However, the sequence of isolate P-7 showed some diversity and belonged to a different subgroup within clade E, showing a closer phylogenetic relationship with the Czech isolate CZ1561 than with other Serbian WDV isolates.
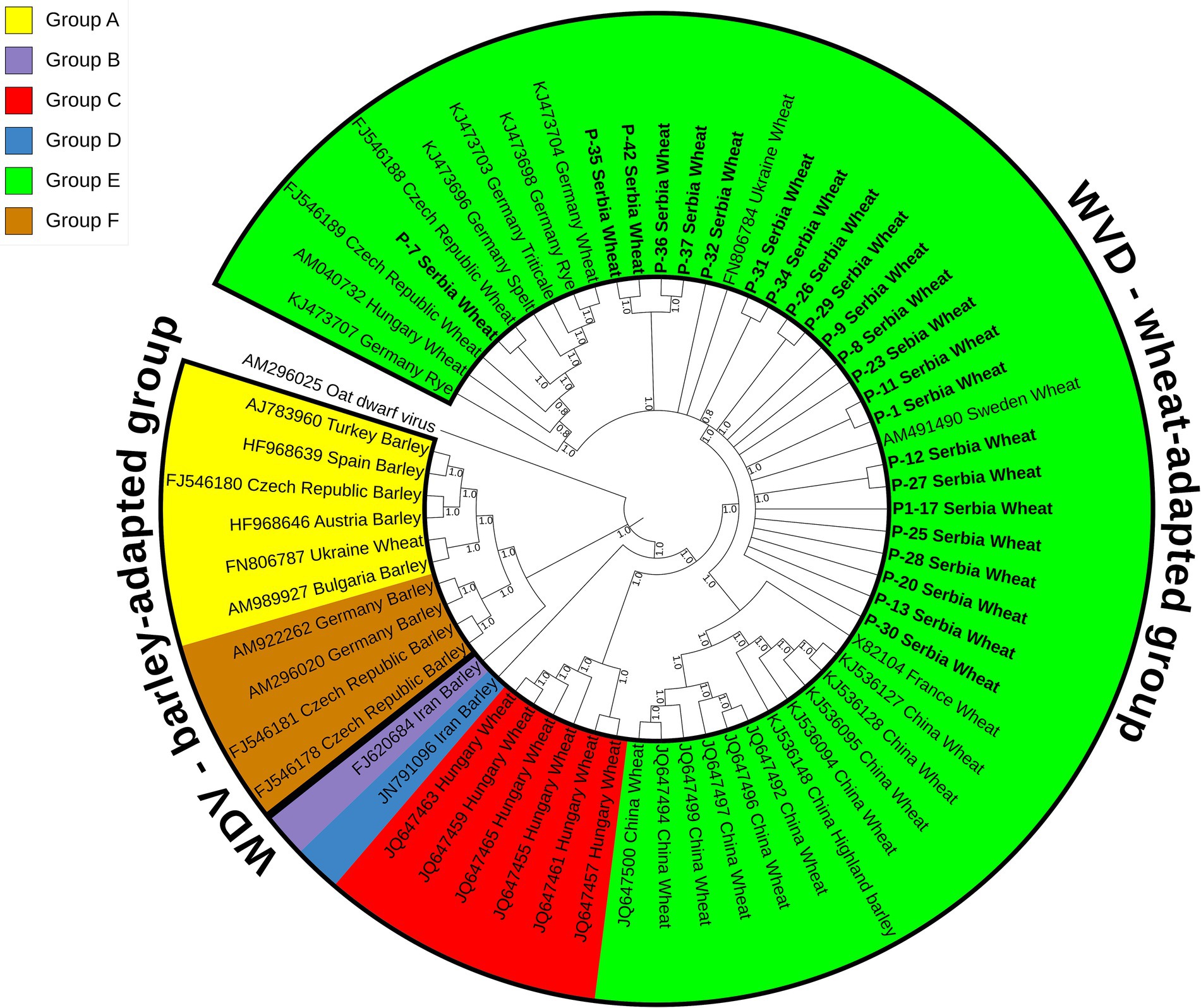
Figure 3. Maximum parsimony tree based on 63 complete sequences of wheat dwarf virus (WDV) isolates constructed using MEGA X and bootstrap analysis with 1,000 replicates. Bootstrap values of 70% and above are shown at nodes. Clade A–F were labeled in different colors in the tree. The isolates from this study are bolded and underlined.
DiscussionMore than 50 different viruses are known to occur in wheat (Ordon et al., 2009), including WDV as the causal agent of wheat dwarf disease. Many studies have shown that WDV is the most commonly detected and widely distributed virus infecting cereals in many European countries (Áy et al., 2008; Kundu et al., 2009; Tóbiás et al., 2011; Schubert et al., 2014; Trzmiel, 2018). The increased WDV distribution and occurrence in areas where the virus had not been previously reported are attributed to changes in agricultural practices, global climate change and the use of cereal varieties with high susceptibility to WDV (Pfrieme et al., 2023). Accordingly, WDV had been expected to appear in Serbia and, indeed, the virus was detected for the first time in 2017 (Ristić et al., 2019).
After this single record, the virus presence reached epidemic proportions 2 years later, in 2019, when symptoms of yellowing and severe dwarfism (Figure 1) were observed in many winter wheat crops across Serbia. To confirm the presence of WDV and determine the possibility of mixed infection, all samples were serologically analyzed for the presence of five economically important wheat viruses. Serological analyses of wheat samples revealed the presence of WDV in 100% of tested samples collected at 21 locations in nine districts, indicating that WDV is a widespread and prevalent wheat virus in our country. In addition to this virus, WSMV was also detected at a much lower frequency and always in mixed infections with WDV (Table 1). Many other studies (Kundu et al., 2009; Tóbiás et al., 2011; Schubert et al., 2014) have also shown that WDV is one of the most dangerous and frequently identified cereal viruses in different parts of Europe. WDV is considered one of the most widespread wheat viruses, causing major economic damage in Hungary (Áy et al., 2008). In Poland, WDV is a serious problem in the production of various cereals, including triticale, wheat, barley, and rye (Trzmiel, 2018). In Iran, WDV epidemics also occur regularly in various cereal crops as well as in several grassy weeds (Parizipour et al., 2017).
Analyses of the complete genome sequences of 23 viral isolates originating from different locations or wheat varieties (Table 1; Figure 2) allowed to present the first molecular characterization of WDV from Serbia. The sequence diversity established in this study showed that the Serbian WDV isolates were highly homologous to each other regardless geographic origin and to the corresponding sequences of wheat-infecting WDV isolates from other parts of the world, indicating the presence of wheat-adapted forms of WDV in Serbia. Similar low genetic variation was also observed in WDV-wheat strains from Germany (Schubert et al., 2007), the Czech Republic (Kundu et al., 2009), Sweden (Ramsell et al., 2008), and Ukraine (Tóbiás et al., 2011). This is not surprising given that it was previously shown that wheat isolates had at least 98.3 to 98.8% sequence similarity with the respective strains, whereas barley WDV isolates had at least 94% similarity (Pfrieme et al., 2023). However, isolate P-7 showed a closer relationship to the Czech isolate CZ1561 than to other Serbian WDV isolates, especially when compared to isolate P-8 collected at the same location, indicating that the virus might have been introduced more than once into Serbia or might have been altered in some cases by the arrival of new WDV isolates.
Phylogenetic analyses (Figure 3) based on the whole genome sequences revealed two distinct groups, the wheat- and the barley-adapted, consistent with previous phylogenetic studies (Tóbiás et al., 2011; Muhire et al., 2013; Wu et al., 2015; Mishchenko et al., 2022). The Serbian WDV sequences clustered in clade E within the wheat-adapted group, suggesting a possible common origin, but some separate groupings of isolate P-7 showed a degree of genomic variation. This finding is interesting considering that the phylogenetic relationships of WDV isolates are usually highly dependent on the geographic origin of the virus (Köklü et al., 2007). However, as noted by Tóbiás et al. (2011), the occurrence and behavior of vectors, plant varieties grown in any given area, agricultural practices and, most importantly, the original source of the virus in a country should also be considered when assessing the spreading, as well as evolutionary divergence of WDV.
Even though natural infection with both strains, WDV-W and WDV-B, has been reported in wheat (Tóbiás et al., 2011), we did not detect the WDV-B strain in Serbia. Moreover, while symptoms on wheat are severe and intense, no symptoms have been observed on barley. In addition, none of the seven recombination detection programs showed any possible recombination events among the WDV isolates examined, although several new types of recombinants have been identified between wheat and barley strains and within autologous groups (Wu et al., 2015; Parizipour et al., 2017).
Symptoms typical of WDV, high WDV frequency, significantly low frequency of mixed infection with WSMV, and absence of other viruses, including those that also cause dwarfism, converged toward a conclusion that the causal agent of the disease that appeared in extremely high infection rates in wheat crops in Serbia in 2019 was WDV. After the 2019 epidemic, no symptoms of yellowing and dwarfing of wheat were seen and WDV was not detected in field surveys of winter wheat in Serbia for 4 years (unpublished data). A similar situation was reported in many other countries, including Sweden (Lindblad and Waern, 2002), Austria (Schubert et al., 2014) and Hungary (Tóbiás et al., 2011). WDV incidence in wheat and other cereals has been reported to differ greatly from year to year and even field to field (Manurung et al., 2004; Širlova et al., 2005).
Although the determination of the vectors present was not performed in this study, it is already known that the vector, P. alienus, is present and widespread in our country (Cvrković et al., 2010; Jović et al., 2010; Jakovljević et al., 2013). It is assumed that epidemics are triggered by the use of cereal varieties with high susceptibility to WDV in combination with favorable conditions that lead to high vector incidence. However, further studies on the population density and activity of leafhoppers under local agroecological conditions are needed to develop appropriate measures and improve the ability to control the virus and its vectors to minimize the risks of WDV epidemics and crop losses, as resistant wheat and barley varieties are not yet available.
Data availability statementThe datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributionsIS: Conceptualization, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. KZ: Formal analysis, Investigation, Writing – review & editing. DR: Formal analysis, Investigation, Writing – review & editing. IV: Formal analysis, Investigation, Writing – review & editing. BK: Conceptualization, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by grants 451-03-65/2024-03/200116 and 451-03-66/2024-03/200010 of the Ministry of Science, Technological Development and Innovation, Republic of Serbia.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1469453/full#supplementary-material
Footnotes ReferencesÁy, Z., Kerényi, Z., Takács, A., Papp, M., Petróczi, I., Gáborjányi, R., et al. (2008). Detection of cereal viruses in wheat (Triticum aestivum L.) by serological and molecular methods. Cereal Res. Commun. 36, 215–224. doi: 10.1556/CRC.36.2008.2.2
Crossref Full Text | Google Scholar
Balázs, F., Mesterházy, Á., and Nyitrai, Á. (1990). Virus diseases of barley and wheat in Vojvodina, Yugoslavia. Cereal Res. Commun. 18, 199–202.
Cvrković, T., Mitrović, M., Jović, J., Krnjajić, S., Krstić, O., and Toševski, I. (2010). Diversity of cicads (Hemiptera: Auchenorrhyncha) in Serbian vineyards. Plant Prot. 61, 217–232.
Dickinson, V. J., Halder, J., and Woolston, C. J. (1996). The product of maize streak virus ORF V1 is associated with secondary Plasmodesmata and is first detected with the onset of viral lesions. Virology 220, 51–59. doi: 10.1006/viro.1996.0285
PubMed Abstract | Crossref Full Text | Google Scholar
Ekzayez, A. M., Kumari, S. G., and Ismail, I. (2011). First report of wheat dwarf virus and its vector (Psammotettix provincialis) affecting wheat and barley crops in Syria. Plant Dis. 95:76. doi: 10.1094/PDIS-09-10-0628
PubMed Abstract | Crossref Full Text | Google Scholar
Erenstein, O., Jaleta, M., Mottaleb, K. A., Sonder, K., Donovan, J., and Braun, H. J. (2022). “Global trends in wheat production, consumption and trade” in Wheat Improvement. eds. M. P. Reynolds and H. J. Braun (Cham, Switzerland: Springer), 47–66.
Jakovljević, M., Kosovac, A., Krstić, O., Mitrović, M., Jović, J., Toševski, I., et al. (2013). Diversity of Auchenorrhyncha species of subfamily Deltocephalinae in Serbian agroecosystems and potential phytoplasma vectors. Plant Prot. 64, 134–143.
Jović, J., Cvrković, T., Mitrović, M., Krstić, O., Krnjajić, S., and Toševski, I. (2010). Composition and structure of planthoppers and leafhoppers assemblages in South Banat maize fields. Plant Prot. 61, 233–247.
Köklü, G., Ramsell, J. N. E., and Kvarnheden, A. (2007). The complete genome sequence for a Turkish isolate of wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes 34, 359–366. doi: 10.1007/s11262-006-0029-0
PubMed Abstract | Crossref Full Text | Google Scholar
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
PubMed Abstract | Crossref Full Text | Google Scholar
Kundu, J. K., Gadiou, S., and Cervena, G. (2009). Discrimination and genetic diversity of wheat dwarf virus in the Czech Republic. Virus Genes 38, 468–474. doi: 10.1007/s11262-009-0352-3
PubMed Abstract | Crossref Full Text | Google Scholar
Letunic, I., and Bork, P. (2024). Interactive tree of life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
PubMed Abstract | Crossref Full Text | Google Scholar
Li, R., Mock, R., Huang, Q., Abad, J., Hartung, J., and Kinard, G. (2008). A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods 154, 48–55. doi: 10.1016/j.jviromet.2008.09.008
PubMed Abstract | Crossref Full Text | Google Scholar
Lindblad, M., Sandgren, M., and Sigvald, R. (1999). “Epidemiology and control of wheat dwarf” in VIIth International Plant Virus Epidemiology Symposium Proceedings.
Lindblad, M., and Waern, P. (2002). Correlation of wheat dwarf incidence to winter wheat cultivation practices. Agric. Ecosyst. Environ. 92, 115–122. doi: 10.1016/S0167-8809(01)00302-4
Crossref Full Text | Google Scholar
Lindsten, K., and Vacke, J. (1991). A possible barley adapted strain of wheat dwarf virus (WDV). Acta Phytopathol. Entomol. Hung. 26, 175–180.
Liu, Y., Jin, W., Wang, L., and Wang, X. (2014). Replication-associated proteins encoded by wheat dwarf virus act as RNA silencing suppressors. Virus Res. 190, 34–39. doi: 10.1016/j.virusres.2014.06.014
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, Y., Khine, M. O., Zhang, P., Fu, Y., and Wang, X. (2020). Incidence and distribution of insect-transmitted cereal viruses in wheat in China from 2007 to 2019. Plant Dis. 104, 1407–1414. doi: 10.1094/PDIS-11-19-2323-RE
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, Y., Wang, B., and Vida, G., Karolyi–Cseplo, M., Wu, B., Wu, Y., and Wang, X. (2012). Genomic analysis of the natural population of wheat dwarf virus in wheat from China and Hungary. J. Integr. Agric. 11, 2020–2027. doi: 10.1016/S2095-3119(12)60459-6
Crossref Full Text | Google Scholar
Manurung, B., Witsack, W., Mehner, S., Grüntzig, M., and Fuchs, E. (2004). The epidemiology of wheat dwarf virus in relation to occurrence of the leafhopper Psammotettix alienus in middle-Germany. Virus Res. 100, 109–113. doi: 10.1016/j.virusres.2003.12.019
PubMed Abstract | Crossref Full Text | Google Scholar
Martin, D. P., Murrell, B., Golden, M., Khoosal, A., and Muhire, B. (2015). RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:vev003. doi: 10.1093/ve/vev003
PubMed Abstract | Crossref Full Text | Google Scholar
Mishchenko, L. T., Dunich, A. A., Mishchenko, I. A., Dashchenko, A. V., Kozub, N. O., Kyslykh, T. M., et al. (2022). Wheat dwarf virus in Ukraine: occurrence, molecular characterization and impact on the yield. J. Plant Dis. Prot. 129, 107–116. doi: 10.1007/s41348-021-00552-w
Crossref Full Text | Google Scholar
Muhire, B., Martin, D. P., Brown, J. K., Navas-Castillo, J., Moriones, E., Zerbini, F. M., et al. (2013). A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch. Virol. 158, 1411–1424. doi: 10.1007/s00705-012-1601-7
PubMed Abstract | Crossref Full Text | Google Scholar
Ordon, F., Habekuss, A., Kastirr, U., Rabenstein, F., and Kühne, T. (2009). Virus resistance in cereals: sources of resistance, genetics and breeding. J. Phytopathol. 157, 535–545. doi: 10.1111/j.1439-0434.2009.01540.x
Crossref Full Text | Google Scholar
Parizipour, M. H. G., Schubert, J., Behjatnia, S. A. A., Afsharifar, A., Habekuß, A., and Wu, B. (2017). Phylogenetic analysis of wheat dwarf virus isolates from Iran. Virus Genes 53, 266–274. doi: 10.1007/s11262-016-1412-0
PubMed Abstract | Crossref Full Text | Google Scholar
Pfrieme, A. -K., Will, T., Pillen, K., and Stahl, A. (2023). The past, present, and future of wheat dwarf virus management—a review. Plan. Theory 12:3633. doi: 10.3390/plants12203633
PubMed Abstract | Crossref Full Text | Google Scholar
Ramsell, J. N. E., Boulton, M. I., Martin, D. P., Valkonen, J. P. T., and Kvarheden, A. (2009). Studies on the host range of the barley strain of wheat dwarf virus using an agroinfectious viral clone. Plant Pathol. 58, 1161–1169. doi: 10.1111/j.1365-3059.2009.02146.x
Crossref Full Text | Google Scholar
Ramsell, J. N. E., Lemmetty, A., Jonasson, J., Andersson, A., Sigvald, R., and Kvarnheden, A. (2008). Sequence analyses of wheat dwarf virus isolates from different hosts reveal low genetic diversity within the wheat strain. Plant Pathol. 57, 834–841. doi: 10.1111/j.1365-3059.2008.01862.x
Crossref Full Text | Google Scholar
Ristić, D., Vučurović, I., Starović, M., Stanković, I., Vučurović, A., Petrović, B., et al. (2019). “Wheat dwarf virus—a newly emerging pathogen for wheat crops in Serbia” in VIII Congress on Plant Protection: Integrated Plant Protection for Sustainable Crop Production and Forestry Proceedings.
Schubert, J., Habekuss, A., Kazmaier, K., and Jeske, H. (2007). Surveying cereal-infecting geminiviruses in Germany–diagnostics and direct sequencing using rolling circle amplification. Virus Res. 127, 61–70. doi: 10.1016/j.virusres.2007.03.018
Crossref Full Text | Google Scholar
Schubert, J., Habekuss, A., Wu, B. L., Thieme, T., and Wang, X. F. (2014). Analysis of complete genomes of isolates of the wheat dwarf virus from new geographical locations and descriptions of their defective forms. Virus Genes 48, 133–139. doi: 10.1007/s11262-013-0989-9
PubMed Abstract | Crossref Full Text | Google Scholar
Širlova, L., Vacke, J., and Chaloupkova, M. (2005). Reaction of selected winter wheat varieties to autumnal infection with wheat dwarf virus. Plant Prot. Sci. 41, 1–7. doi: 10.17221/2732-PPS
Crossref Full Text | Google Scholar
Šutić, D., and Tošić, M. (1964). Wheat streak mosaic virus in Yugoslavia. Plant Prot. 79, 307–314.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
PubMed Abstract | Crossref Full Text | Google Scholar
Tóbiás, I., Shevchenko, O., Kiss, B., Bysov, A., Snihur, H., Polischuk, V., et al. (2011). Comparison of the nucleotide sequences of wheat dwarf virus (WDV) isolates from Hungary and Ukraine. Pol. J. Microbiol. 60, 125–131. doi: 10.33073/PJM-2011-017
PubMed Abstract | Crossref Full Text | Google Scholar
Tošić, M. (1971). Virus diseases of wheat in Serbia: I. Isolation and determination of the wheat streak mosaic virus and brome mosaic virus 1. J. Phytopathol. 70, 145–162. doi: 10.1111/j.1439-0434.1971.tb03472.x
留言 (0)