Urinary tract infections (UTIs) are some of the most common bacterial infections that affect human beings (Stamm and Norrby, 2001). These infections are classically defined by the invasion and growth of bacteria in the urinary tract, accompanied by clinical symptoms. Among the most common etiological agents are uropathogenic Escherichia coli (UPEC) and Proteus mirabilis, which can cause both uncomplicated and complicated UTIs (Foxman, 2014). Those bacteria can survive in the urinary tract using several pathogenic mechanisms such as biofilm formation, uroepithelial cell invasion, adhesins, toxins, and siderophores (Hannan et al., 2012; Schaffer and Pearson, 2015). UPEC and P. mirabilis have different strategies for invading and persisting in the bladder. UPEC can invade the uroepithelial cell and form intracellular bacterial communities (IBC), providing the bacteria the ability to survive TLR4-mediated expulsion, cell exfoliation, urination, inflammation, and antimicrobial treatments (Rosen et al., 2007; Schwartz et al., 2011). On the other side, P. mirabilis produces MR/P pili that facilitate biofilm formation and colonization of the bladder and kidneys, being also crucial for catheter-associated biofilm formation (Armbruster and Mobley, 2012). The production of the urease enzyme provides a nitrogen source for P. mirabilis, and is highly important in pathogenesis (Milo et al., 2021). It is also important to consider that urine represents a moderately oxygenated environment, with high-osmolarity, and iron-limitation (Zunino et al., 1994).
Some of the main virulence mechanisms that allow these bacteria to colonize the urinary tract are flagella, fimbriae, and iron-uptake systems (Govindarajan and Kandaswamy, 2022). Flagella are mainly related to motility, contributing to bacterial spread during UTI, with biofilm formation (Zunino et al., 1994; Scavone et al., 2023), while improving bacterial fitness (Lane et al., 2005). Fimbriae play a crucial role in facilitating adhesion among bacteria, and to different abiotic and biotic surfaces, as uroepithelial cells (Mendoza-Barberá et al., 2023; Shanmugasundarasamy et al., 2022). In clinical UPEC strains, FimH adhesin (type I fimbriae) is widely distributed in pediatric patients presenting complicated UTIs, and interacts with uroplakin proteins in the bladder allowing entry into the eukaryotic cell (Luna-Pineda et al., 2016). In P. mirabilis, the adherence of the bacterium to epithelial and surfaces is mediated by 17 different fimbriae, among which MR/P fimbriae is the most prominent (Armbruster et al., 2018). As the urinary tract is iron-restricted, iron scavenging uptake systems are induced during infection (Himpsl et al., 2010). The iron scavenging systems play a crucial role in the bacteria growth and colonization; increasing iron concentrations could improve biofilm formation and enhance the bacterial resistance to antibiotics (Govindarajan et al., 2022). UPEC can acquire iron from the environment using three classes of systems: siderophores, hemophores (or heme binding and uptake systems), and direct ferrous iron (FeII)-uptake systems (Subashchandrabose and Mobley, 2015). P. mirabilis has developed 21 iron acquisition systems including siderophore-based mechanisms, ferrous iron transport, metal-type ABC transporters, among others (Chakkour et al., 2024).
Gram-negative bacteria naturally produce extracellular outer membrane vesicles (OMVs) (Kim et al., 2015), spherical bilayered particles released from the bacterial outer membrane, showing a diameter size ranging from 20 to 250 nm (Kulp and Kuehn, 2010). It has been also reported that OMVs might have different functions depending on their content and/or their target cells (Ellis and Kuehn, 2010). OMVs production has also been related to the bacterial stress response (McBroom and Kuehn, 2007). Bacterial OMVs can mediate antibiotic resistance, nutrient acquisition, and several bacteria-bacteria interactions, such as killing of competing bacteria (Lee et al., 2016). On the other hand, OMVs can participate in bacteria-host interactions, such as promoting inflammatory response, facilitating adhesion to eukaryotic cells, and delivering several virulence factors (Lee et al., 2016; Magaña et al., 2024; Charpentier et al., 2023).
Previously reported E. coli OMVs proteomic composition, showed that OMVs contain mainly outer membrane and periplasmic proteins (Lee et al., 2007). The presence of inner membrane proteins was low, suggesting that a specific sorting mechanism for vesicular proteins exists (Lee et al., 2007; Aguilera et al., 2014).
Despite the large amount of research regarding the potential functions of OMVs, their role in uropathogenic infections and their pathophysiological functions remain unclear. To further understand the role of OMVs in UTI, in this study, we carried out an extensive physicochemical characterization, and analyzed the total proteome of purified OMVs derived from UPEC and P. mirabilis strains obtained from cultures in Luria-Bertani broth and artificial urine. So far, this is the first proteomic characterization of P. mirabilis OMV.
2 Materials and methods 2.1 Bacterial strains and culture conditionsTwo uropathogenic bacterial strains capable of forming biofilm were used in the present study: Escherichia coli U144 and Proteus mirabilis 2921. They were isolated from urine samples from patients with UTI and previously characterized in our laboratory (González et al., 2017; Zunino et al., 2000). Both strains were stored in glycerol at -80°C and recovered in Luria-Bertani agar (LA) at 37°C. The bacterial samples used in the present analyses were obtained previously and were anonymized.
2.2 Outer membrane vesicle isolation and purificationOuter membrane vesicles (OMVs) isolation and purification were performed as previously described (Tashiro et al., 2017). For OMV isolation, the bacterial strains were grown in Luria-Bertani (LB) broth and artificial urine (AU: in g/L; CaCl2, 0.49; MgCl2, 0.65; NaCl, 4.6; Na2SO4, 2.3; sodium citrate, 0.65; sodium oxalate, 0.02; KH2PO4, 2.8; KCl, 1.6; NH4Cl, 1.0; urea, 25; and trypticase soy broth; pH 6.2; Soriano et al., 2009). AU was sterilized using a Millipore Membrane 0.45 μm pore size. To obtain OMV samples without bacterial cells, 100 mL of overnight batch cultures were centrifuged at 6000×g, for 15 min, at 4°C. The supernatants were filtered through 0.45 and 0.20 μm membrane filters, and ultracentrifuged at 100000×g, for 2 h, at 4°C using a fixed-angle rotor (90 Ti, Beckman). The pellets were resuspended in 300 μL of 50 mM HEPES, 0.85% NaCl (HEPES-NaCl buffer). For OMV purification, samples were adjusted to 1 mL of 45% (w/v) iodixanol (OptiPrep™, Sigma) in HEPES-NaCl, transferred to the bottom of ultracentrifuge tubes, and layered with iodixanol-HEPES-NaCl (2 mL of 40, 35, 30, 25, and 20%). The samples were ultracentrifuged at 100,000 × g, for 3 h, at 4°C using a swing rotor (sw 40 Ti, Beckman). Then, several 1 mL fractions were collected from each gradient. To confirm the fraction containing OMVs, the protein concentrations in each fraction were measured using the Bradford protein assay and UV–Vis absorbance at 280 nm. The fraction containing OMVs was ultracentrifuged at 100,000 × g, for 2 h, at 4°C and resuspended in HEPES-NaCl.
2.3 Charaterization of OMV for dynamic light scattering measurements and laser doppler electrophoresisThe hydrodynamic radius and polydispersity index (PdI) were acquired by the dynamic light scattering technique (DLS), and the zeta potential was measured by laser Doppler electrophoresis in a Zetasizer ZS (Malvern Instruments), with a He-Ne laser light source at 633 nm with a fixed scattering angle of 175°. The OMV samples were diluted with Milli-Q water and were measured on a DTS1070 cuvette.
2.4 Nanoparticle tracking analysisOMVs concentration was determined using the Nanoparticle Tracking Analysis (NTA). All the measurements were made in a Nanosight NS300 (Malvern Instruments) in triplicate at 25°C. The light dispersion was captured by a sCMOS camera. The samples were diluted 1/100 in filtered Milli-Q water. The obtained hydrodynamic radius, concentration, and standard deviation correspond to the arithmetic values for all the particles analyzed on the sample by the NTA software.
2.5 Transmission electron microscopyFor Transmission electron microscopy (TEM) analysis of OMVs, the samples were diluted and deposited onto a copper grid, stained with 1% phosphotungstic acid, dried at room temperature, and visualized at 10.00 kV using an Inspect F50 scanning transmission electron microscope (FEI Company, Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile). The OMVs size measurements were obtained as the average size from 10 microimages for each condition.
2.6 OMVs protein analysis by SDS-PAGEOMVs protein concentration was determined by densitometry analysis of Colloidal Coomassie stained gels. For that purpose, each sample was loaded into pre-cast gels (NuPAGE™ 4–12%, Bis-Tris, 1.0 mm, Mini Protein Gel, 10-well, Invitrogen), using LMW-SDS Marker Kit (Cytiva). Electrophoresis was run at 150 V. Gels were fixed with 50% ethanol and 10% acetic acid and stained overnight with colloidal Coomassie blue. After destaining with ultrapure water washing, gel images were digitalized with UMAX Power-Look 1,120 scanner and LabScan 5.0 software (GE Healthcare). Quantification was performed using the ImageQuant TL software (v8.1), 1D analysis module, and LMW-SDS Marker Kit (Cytiva) as standard.
2.7 Preparation of OMV proteins for mass spectrometry analysisProteomic analysis of OMVs obtained from E. coli and P. mirabilis grown in LB broth and AU was carried out using three independent biological replicates. Twenty-five μL of each sample were loaded into a 12.5% acrylamide gel and run until samples enter 1 cm into the resolving gel. The gel was fixed and stained as described above. Gel fragments were sliced with a scalpel and transferred to microcentrifuge tubes. Sample processing for mass spectrometry analysis was performed as described previously (Rossello et al., 2017; Gil et al., 2019). Briefly, the procedure consisted in sample reduction with 10 mM DTT at 56°C for 1 h with vigorous agitation; cysteine alkylation with 50 mM iodoacetamide at room temperature for 45 min with vigorous agitation and protected from light; in gel protein digestion with 0.1 μg/μL trypsin (sequence grade, Promega) in 50 mM ammonium bicarbonate, overnight at 37°C; peptide extraction by incubation with 60% acetonitrile/0.1% trifluoroacetic acid with vigorous agitation for 2 h at 30°C; vacuum drying concentration and resuspension in 0.1% trifluoroacetic acid. Samples were desalted using C18 micro-columns (C18 OMIX pipette tips, Agilent), eluted with 0.1% formic acid in ACN, vacuum dried, and resuspended in 0.1% formic acid. Peptide samples concentration were determined using a Denovix DS-11 FX+ spectrophotometer/fluorometer, at 215 nm. Final volume was adjusted in order to normalize all sample concentrations.
2.8 Nano-liquid chromatography and mass spectrometry analysisOMV protein samples were analyzed by nano-LC MS/MS using a shotgun strategy on a nano-HPLC, UltiMate 3,000 coupled on line to an Orbitrap Exploris 240 mass spectrometer through an Easy-Spray source. One ug of tryptic peptides was injected into an Acclaim PepMap™ 100 C18 nano-trap column (75 μm x 2 cm, 3 μm particle size), and separated using a 75 μm x 50 cm, Easy-Spray™ analytical C18 HPLC column (2 μm particle size, 100 Å pore size) at a constant flow rate of 200 nL/min and 40°C. The column was equilibrated at 1% of mobile phase B (A: 0.1% formic acid; B: 0.1% formic acid in acetonitrile) followed by an elution gradient from 1 to 35% B over 90 min and 35–99% B over 20 min. All nano-LC MS/MS equipment and supplies were from Thermo Scientific.
The mass spectrometer was operated in the positive mode. Ion spray voltage was set at 2.2 kV; capillary temperature at 250°C, and S-lens RF level at 50. Mass analysis was carried out in data dependent mode in two steps: acquisition of full MS scans in a range of m/z from 200 to 2000; followed by HCD fragmentation of the 20 most intense ions in each segment using a stepped normalized collision energy of 25, 30 and 35. Full MS scans were acquired at a resolution of 90,000 ppm at 200 m/z, and a maximum ion injection time of 100 ms. For MS/MS acquisition the resolution was 22,500 ppm, and maximum ion injection time: 50 ms. Precursor ions with charge states from 2 to 5 were included for fragmentation. Dynamic exclusion time was set at 30 s.
2.9 Protein identification and analysisDatabase generation, protein identification, and analyses were performed using the PatternLab V software (Santos et al., 2022). Protein sequences from uropathogenic E. coli CFT073 and P. mirabilis HI4320 were downloaded from Uniprot Consortium (in December, 2023) and protein sequences of the 127 most common mass spectrometry contaminants were used to generate target-decoy databases. Raw files were queried against each corresponding database using the Comet search engine (integrated in PatternLab V) applying the following parameters: fully specific trypsin as proteolytic enzyme with up to 2 missed cleavages allowed; methionine oxidation as variable modification and cysteine carbamidomethylation as fixed modification; 40 ppm tolerance from the measured precursor m/z.
Peptide spectrum matches were filtered using the Search Engine Processor (SEPro), setting the acceptable FDR criteria as follows: 3% at spectrum level, 2% at peptide level, and 1% at the protein level. PatternLab’s Approximately Area-Proportional Venn Diagram module was used to identify proteins exclusively detected in each sample set from each bacterium using a p-value ≤0.05. The Pairwise Comparison module was used to detect proteins present in both conditions but at significantly different relative abundance by means of extracted ion chromatogram (XIC) analysis. The following statistical criteria were applied: log2 fold change ≤1, and p-value ≤0.05.
In order to evaluate the surface-associated protein fraction in OMVs, the subcellular origin of identified proteins was analyzed in silico using the PSORTb v3.0 algorithm (Yu et al., 2010).
3 Results 3.1 Characterization of OMVs of Escherichia coli and Proteus mirabilisOMVs secreted by E. coli and P. mirabilis were isolated and then examined using scanning transmission electron microscopy (TEM) and dynamic light scattering (DLS). E. coli and P. mirabilis produced spherical OMV in both LB broth and AU with a size ranging from 85 to 260 nm, as shown in the TEM images (Figure 1). On the other hand, the sizes determined by DLS of P. mirabilis and E. coli OMVs obtained from cultures in LB were 267.6 ± 29.7 nm and 185 ± 25.9 nm, respectively. In AU, the sizes observed were 320.4 ± 32.2 and 257.6 ± 3.8 nm for P. mirabilis and E. coli, respectively. These measurements allowed us to observe that OMVs size recovered from bacteria grown in AU were larger compared to those grown in LB, for both strains. For bacteria grown in LB, the polydispersity factor was higher in P. mirabilis showing a value of 0.355, while in E. coli OMVs it was 0.263. In bacteria grown in AU, we also observed higher polydispersity factors in P. mirabilis OMVs compared to those obtained from E. coli, showing values of 0.344 and 0.214, respectively. Furthermore, these data indicated that all preparations presented low polydispersity (Table 1).
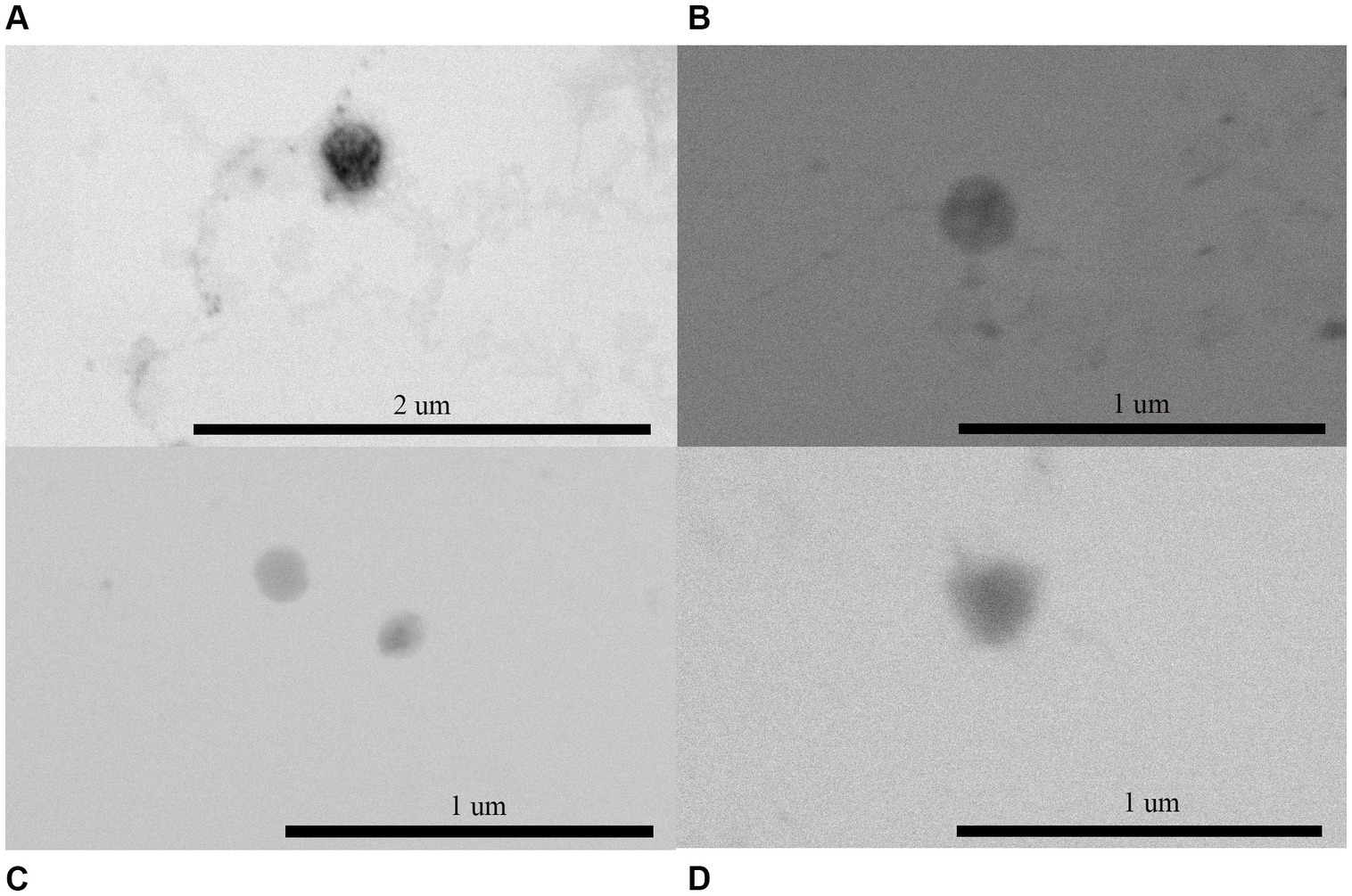
Figure 1. Visualization of OMVs isolated from (A) Escherichia coli in LB, (B) E. coli in AU, (C) Proteus mirabilis in LB broth, and (D) P. mirabilis in AU. OMVs were fixed and negatively stained and viewed under STEM (Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile) showing a spherical shape. The scale bar indicated in (A) 2 μm, and in (B–D) 1 μm.
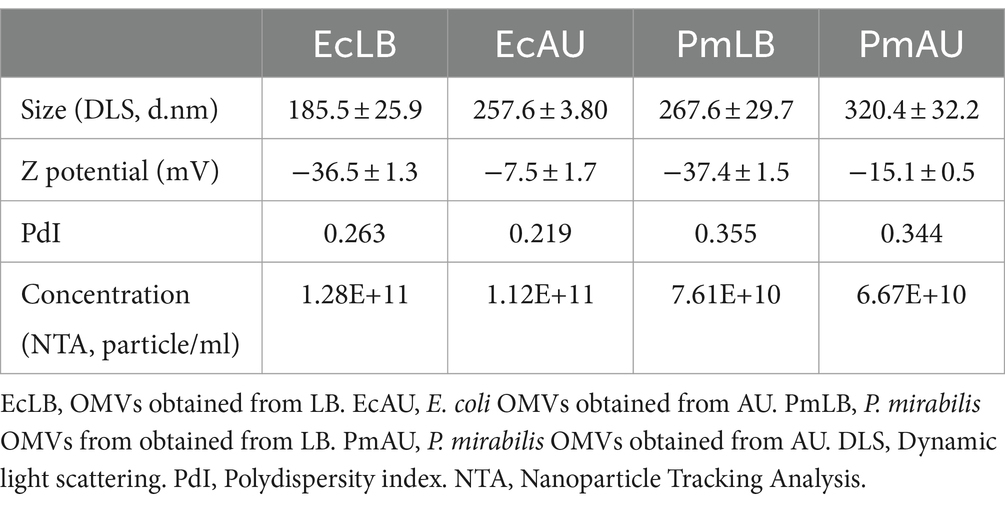
Table 1. Physicochemical characterization parameter of Escherichia coli and Proteus mirabilis OMVs.
The zeta potentials recorded were -37.4 ± 1.5 mV and -36.5 ± 1.3 mV in LB, and -15.5 ± 0.5 mV and -7.5 ± 1.7 mV in AU, for P. mirabilis and E. coli OMVs, respectively. These values suggest very minor differences in OMVs stability between the two growth conditions.
The OMVs concentration was measured using NTA, evidencing concentrations of approximately 1×1011 particles/ml and 7×1010 particles/ml for E. coli and P. mirabilis, respectively. This assay showed that the amount of particles for each bacteria were similar in LB and AU (Table 1).
To observe the OMVs protein profiles, the vesicles were subjected to SDS-PAGE stained with Coomassie blue (Figure 2). Equal amounts of OMVs were loaded in order to compare their protein content. Comparison of OMVs preparations showed different protein profiles and protein yields among strains and media conditions.
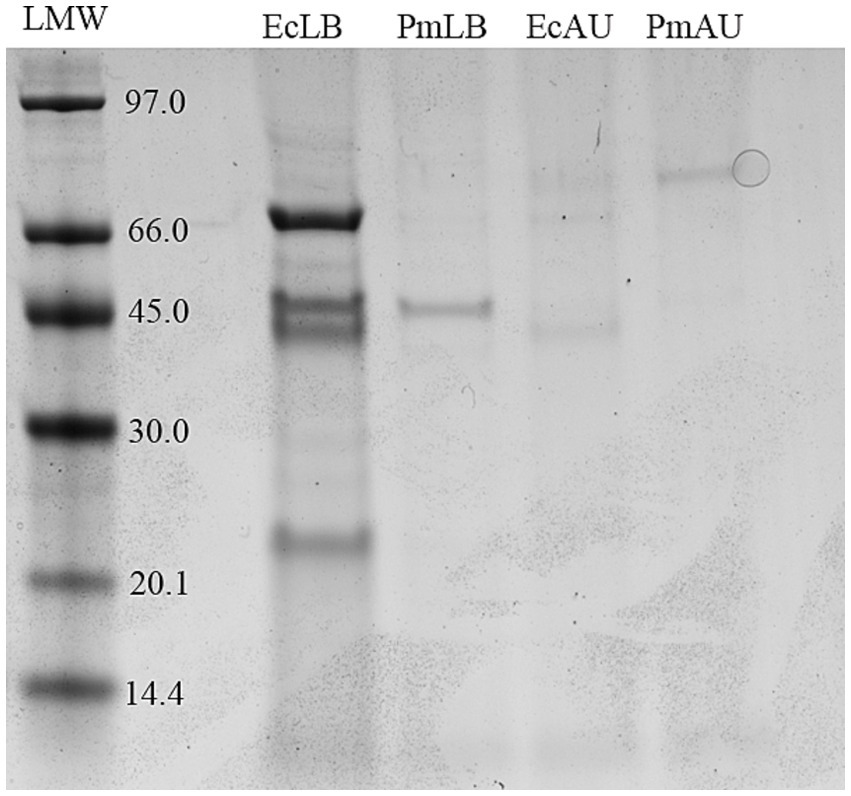
Figure 2. Protein profiles of OMV from E. coli in LB and AU (EcLB and EcAU), and P. mirabilis in LB and AU (PmLB and PmAU) run in 12% SDS-PAGE. Molecular weight standard is indicated on the left (kDa).
3.2 Proteomic characterization of Proteus mirabilis OMV in LB and AULC–MS/MS analysis identified 353 proteins of OMVs from P. mirabilis grown in LB broth (PmLB), and 103 proteins from P. mirabilis OMVs cultured in AU (PmAU), considering those proteins that were statistically detected in at least two of the three replicates.
The analysis of subcellular localization of the 353 identified proteins from P. mirabilis OMVs obtained in LB media showed the following classification: 105 (29.7%) cytoplasmic, 87 (24.6%) unknown, 66 (18.7%) inner membrane, 38 (10.8%) periplasmic, 38 (10.8%) outer membrane, and 19 (5.4%) extracellular (Figure 3).
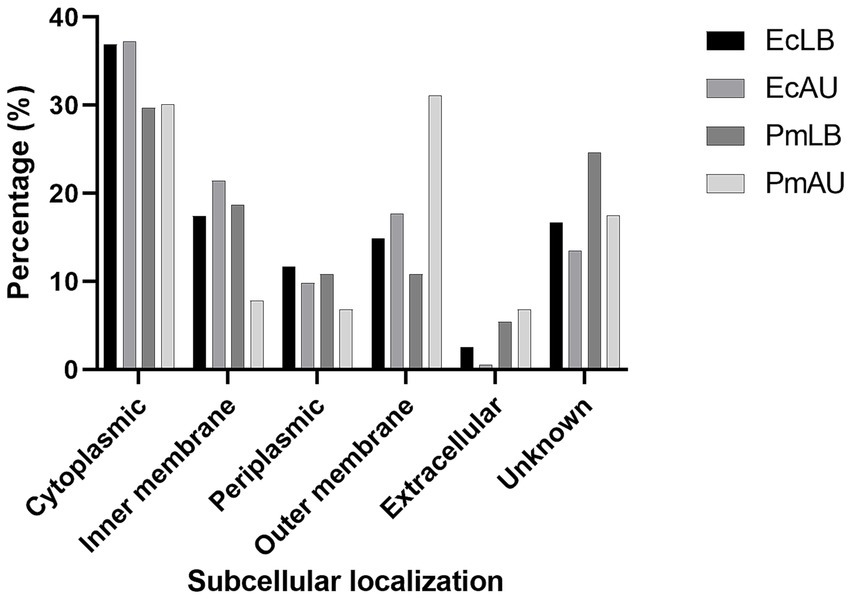
Figure 3. Distribution of subcellular localizations of vesicular proteins from E. coli and P. mirabilis in LB (EcLB and PmLB), and in AU (EcAU and PmAU), according to the PSORTb v3.0 algorithm.
On the other hand, the subcellular localization of the identified proteins from P. mirabilis OMVs cultured in AU showed the following distribution: 32 (31.1%) outer membrane, 31 (30.1%) cytoplasmic, 8 (7.8%) inner membrane, 7 (6.8%) periplasmic, and 7 (6.8%) extracellular. Eighteen identified proteins (17.5%) could not be classified into any of the above categories (Figure 3). Based on these analyses, we could observe that the percentage of inner membrane proteins was lower in AU than in LB OMVs. The opposite trend was found for the percentage of outer membrane proteins.
Among the most abundant proteins determined according to spectrum count in both culture conditions, we found some reported vesicle markers including OmpA, OmpF, OmpW, TolC, TolB, AcrA, TufB, Pal, and Lpp. Furthermore, several outer membrane proteins, such as TonB-dependent receptor (IreA), YaeT, MipA, NlpD, and FadL, were found to be highly abundant in this dataset. Also, the hemolysin HpmA was found (Supplementary Table 1).
3.3 Comparative analysis of Proteus mirabilis OMV in LB and AUProteins identified in P. mirabilis OMVs obtained from LB broth were compared to those from AU culture. These analyses allowed us to determine that 105 proteins were exclusively detected in LB broth and only 5 proteins in AU (p-value <0.05; Figure 4A; Supplementary Table 2).
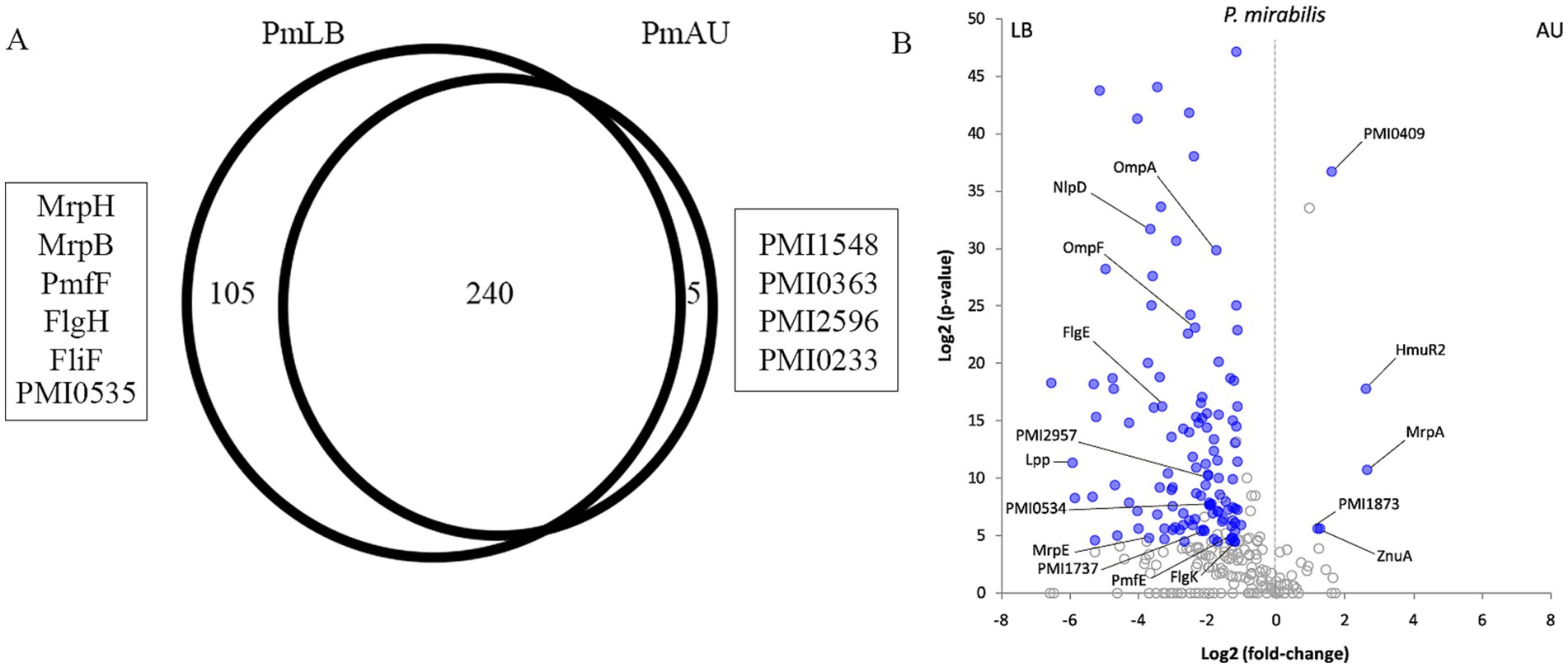
Figure 4. (A) Venn diagram of OMVs proteins overlapping between EcLB and EcAU, using the Venn diagram’s statistical module from the PatternLab V software. 271 proteins were shared between OMVs from both media, 47 proteins were exclusive of EcLB OMVs, and 14 proteins from EcAU OMVs. The proteins mentioned in the text are shown in the picture. (B) Differentially abundant proteins between E. coli OMVs obtained from AU and LB. The volcano plot shows the Log2 (p-value) on the y-axis and the Log2 (fold-change) on the x-axis. Proteins identified as common to both conditions are represented by a dot in the plot. Blue dots indicate proteins satisfying all statistical criteria and thus are considered as differentially abundant among culture conditions. Selected differential proteins discussed in the text are labeled. Information regarding all differential proteins are depicted in Supplementary Table 3.
Using the PatternLab V Pairwise comparison module, we identified 118 and 5 proteins significantly increased in PmLB and PmAU, respectively (p-value <0.05; Figure 4B; Supplementary Table 2). Taking into account that the protein dataset obtained from P. mirabilis OMVs grown in LB was considerably larger than the one obtained in the same bacterium but grown in AU, and considering that this difference may lead to some bias and potential false positive, we proceed to comparatively analyze proteomic profiles of E. coli grown on the same conditions. Therefore, we focused our discussion on proteins supported by the E. coli OMVs comparative analysis and corroborated by evidence reported in the literature.
Within the proteins that were detected only in PmLB OMVs we found several fimbrial proteins including MrpH, PmfF, MrpB, PMI0535, and the flagellar proteins FlgH and FliF. Also, many other members of this family proteins were increased in the OMVs from LB broth such as MrpE, PMI0534, PmfE, FlgE and FlgK. Within OMVs from P. mirabilis grown in LB broth, we could observe several virulence factors related to adhesion and motility. The only fimbrial protein that was increased in OMVs from AU was MrpA (Table 2).
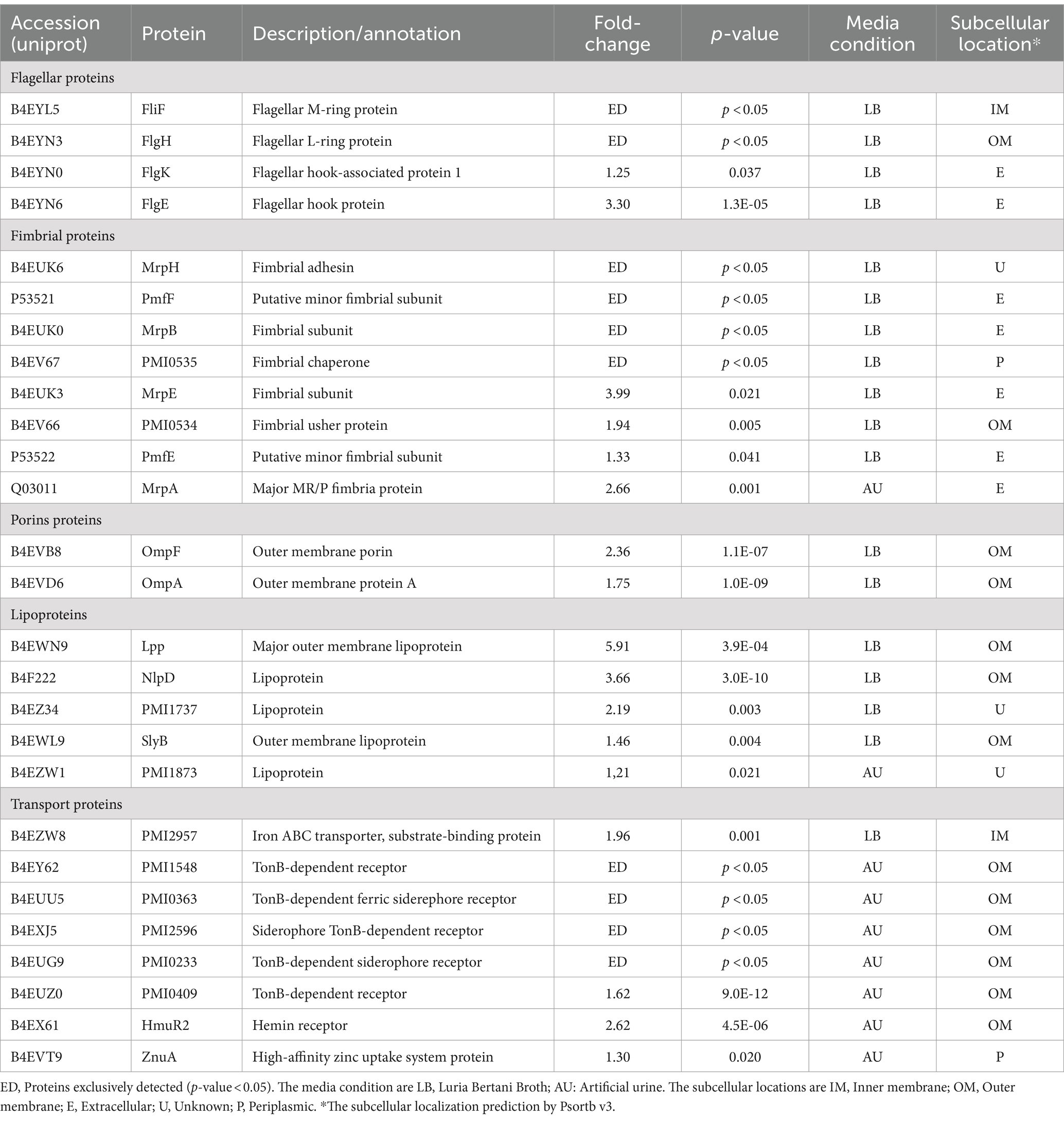
Table 2. Selected proteins identified in OMVs from P. mirabilis discussed in the text.
Several lipoproteins are shared between OMVs obtained in both culture conditions (LB broth and AU), but two of them (NlpD and SlyB) were increased in LB broth as shown in Table 2. The porin OmpA was more abundant in OMVs from PmLB as also in OMVs from EcLB.
The proteins present exclusively in OMVs from AU are most related with iron acquisition and belong to the TonB-dependent receptor family: PMI1448, PMI0363, PMI2596, and PMI0233. Also, PMI0409 was more abundant in AU. Other transporters showing significant increased relative abundance in OMVs from PmAU are ZnuA, related with zinc uptake, and HmuR2, an hemin receptor involved in Zinc and iron-acquisition.
3.4 Proteomic characterization of Escherichia coli OMV in LB and AULC–MS/MS analysis allowed the identification of 282 proteins from LB broth-grown E. coli OMVs and 215 proteins from AU-grown E. coli OMVs, considering those proteins that are statistically detected in at least two of the three replicates.
The analysis of subcellular localizations of the identified proteins from E. coli OMVs obtained in LB broth showed that the majority of them were from the bacterial envelope, including 49 (17.4%) from the inner membrane, 42 (14.9%) from the outer membrane, and 33 (11.7%) were classified as periplasmic proteins (Figure 3). On the other hand, 36.9% of total proteins (104) were classified as cytoplasmic, and 2.5% (7) extracellular. The subcellular localization of 46 proteins (16.6%) was classified as unknown (Figure 3).
A similar distribution of subcellular localization was observed in E. coli OMVs obtained in AU. Most of the proteins were from the bacterial envelope, including 46 (21.4%) from inner membrane, 38 (17.7%) from outer membrane, and 21 (9.8%) from the periplasm. Eighty of the identified proteins (37.2%) were cytoplasmic and only one was extracellular. Twenty-nine of identified proteins (13.5%) showed an unknown localization classification (Figure 3). From these analyses, it becomes apparent that, despite the similarities observed, there are slight differences in the percentage of cell envelope components, primarily in the inner and outer membranes in OMVs obtained from AU compared with those obtained from LB broth.
Among all the proteins identified in UPEC grown in both conditions, conserved outer membrane protein components were the most abundant as determined by spectrum count. Mainly, the outer membrane porin OmpA, OmpC, OmpF, OmpX, NmpC, and Maltoporine, exhibited the highest abundance. These porins were commonly observed as vesicle markers. Also, other outer membrane components considered as vesicles markers were found, including TolB, TolC, and AcrA. Additional proteins detected in OMVs from both media conditions, such as Lpp, Pal, Tsx, FhuA, and LamB, were previously reported in OMVs (Lee et al., 2007; Supplementary Table 2).
3.5 Quantitative comparative analysis of Escherichia coli OMV in LB and AUUsing the Venn diagram’s statistical module from the PatternLab V software, we could determine that 47 proteins were exclusively detected in LB and 14 in AU (p-value <0.05; Figure 5A; Supplementary Table 3).
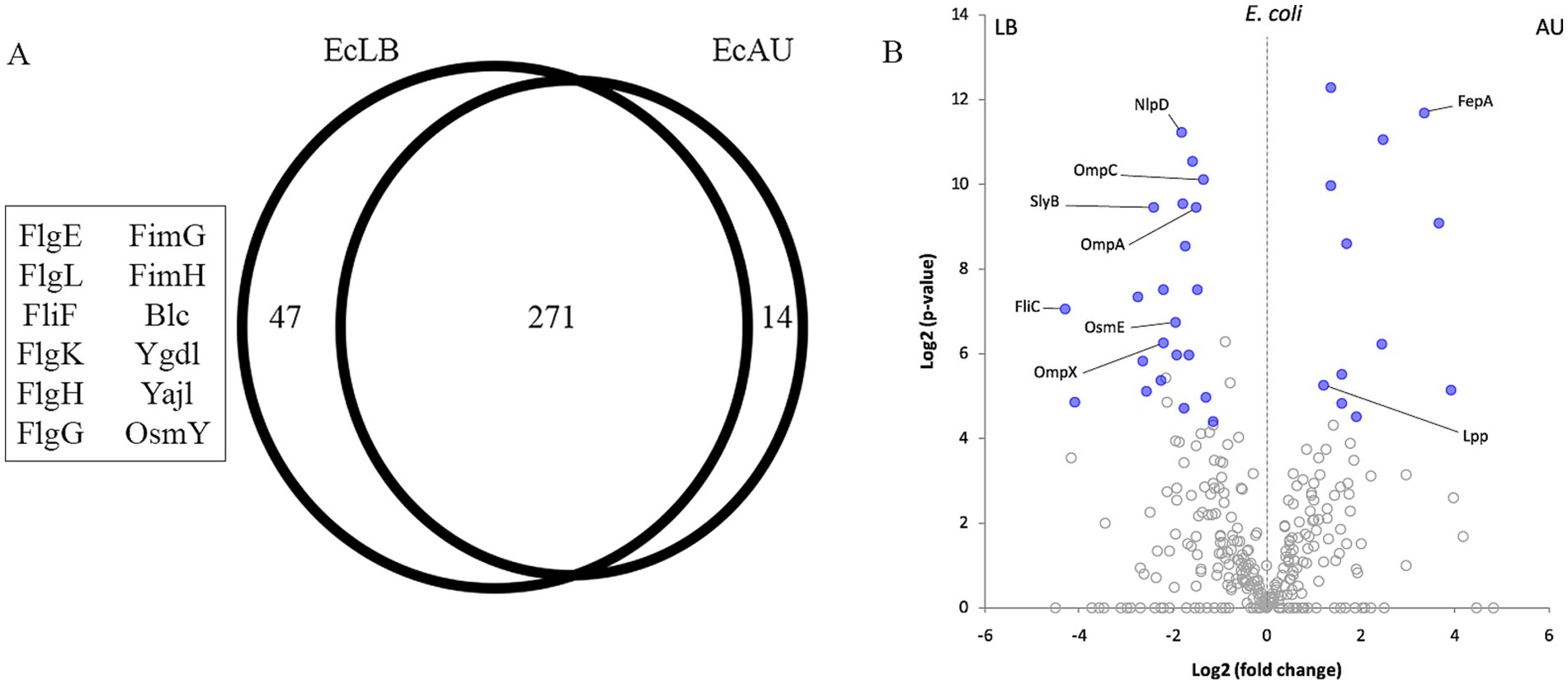
Figure 5. (A) Venn diagram of OMVs proteins overlapping between PmLB and PmAU, using the Venn diagram’s statistical module from the PatternLab V software. 240 proteins were shared between OMVs from both media, 105 proteins were exclusive of PmLB OMVs, and 5 proteins from PmAU OMVs. The proteins mentioned in the text are shown in the picture. (B) Differentially abundant proteins between P. mirabilis OMVs obtained from AU and LB. The volcano plot shows the Log2 (p-value) on the y-axis and the Log2 (fold-change) on the x-axis. Proteins identified as common to both conditions are represented by a dot in the plot. Blue dots indicate proteins satisfying all statistical criteria and thus are considered as differentially abundant between culture conditions. Selected differential proteins discussed in the text are labeled and information regarding all differential proteins are depicted in Supplementary Table 3.
In order to analyze proteins present in both conditions, OMVs from E. coli in LB broth (EcLB) and AU medium (EcAU), but exhibiting significant differences in their relative abundance, we employed the PatternLab V Pairwise Comparison module. Using this tool, we identified 22 and 12 proteins significantly increased in EcLB and EcAU, respectively (p-value <0.05; Figure 5B; Supplementary Table 3).
Within the proteins that were detected only in EcLB OMVs, we identified the fimbrial proteins FimH and FimG, and the flagellar proteins FlgE, FlgK, FlgG, FlgH, FlgL, and FliF. FliC was found in both conditions but increased in LB (Table 3). The lipoproteins Blc, YgdI, YajI and OsmY were detected as exclusive to EcLB OMVs. In addition, OsmE, SlyB, and NlpD were increased in LB, while Lpp showed a relative increased abundance in AU. On the other hand, although OMVs recovered from both cultured conditions express the same membrane porins, some of them were found significantly increased in the LB OMVs, such as OmpA, OmpC and OmpX (Table 3).

Table 3. Selected proteins identified in OMVs from UPEC discussed in the text.
Regarding the outer membrane proteins, two of them were found to be increased from EcAU OMVs: Ferrienterobactin receptor (FepA) and Colicin I receptor (CirA).
4 DiscussionOuter membrane vesicles (OMVs) are spherical particles released from the outer membrane of Gram-negative bacteria (Beveridge, 1999). The production of OMVs is not only a common physiological mechanism in bacteria but also serves as a response to environmental stress (Kulp and Kuehn, 2010). OMVs are associated with bacterial survival, playing key roles in nutrient acquisition, intra-and interspecific bacterial communication, biofilm formation, defense mechanisms, resistance, and pathogenesis (Kulp and Kuehn, 2010). The composition, function, and quantity of OMVs depend on the specific biogenesis pathways. Studies have shown that OMVs exhibit distinct structures and characteristics depending on their mode of production and the surrounding microenvironment (Kulp and Kuehn, 2010).
The primary mechanism through which Gram-negative bacteria produce OMVs is outer membrane blebbing. As a result, OMVs are enriched in outer membrane proteins, have specific lipid compositions, and differ in cargo molecule content compared to other extracellular vesicles (Orench-Rivera and Kuehn, 2016). In this study, we isolated, characterized, and compared the OMVs produced by Proteus mirabilis and Escherichia coli cultured in Luria-Bertani (LB) media and artificial urine (AU). Additionally, we conducted a quantitative and comparative proteomic analysis to determine and compare the protein profiles of OMVs derived from both pathogens grown in these two different media.
In a previous study, 619 unique proteins were identified in OMVs from 54 UPEC strains (Wurpel et al., 2015). Wurpel et al. further analyzed the proteome of OMVs from five UPEC reference strains (536, CFT073, F11, UMN026, and UTI89) EDTA-induced in human urine, identifying a distinct set of 173 non-redundant proteins (Wurpel et al., 2016). However, their study provides only an indirect evaluation of the outer membrane composition in UPEC. In contrast, our work evaluates the protein composition of naturally produced OMVs, offering a more direct insight into their biological role.
Outer membrane vesicles (OMVs) are nanoparticles ranging from 20 to 250 nm in diameter (Kulp and Kuehn, 2010), composed of a lipid membrane, lipopolysaccharides (LPS), and periplasmic content. OMVs contain periplasmic and outer membrane proteins, virulence factors, toxins, and genetic material (Pin et al., 2023). Their small size allows them to penetrate tissues typically inaccessible to bacteria (Kuehn and Kesty, 2005). Internalization pathways differ based on OMV size: smaller OMVs (20–100 nm) use caveolin-mediated endocytosis, medium-sized ones (20–250 nm) use clathrin-mediated endocytosis, and larger ones (90–450 nm) rely on macropinocytosis (Wang et al., 2023; O'Donoghue and Krachler, 2016).
In this study, we successfully purified OMVs from Proteus mirabilis and E. coli grown in LB media and artificial urine (AU). P. mirabilis OMVs were significantly larger, with an average diameter over 250 nm, while E. coli OMVs were smaller, with diameters under 250 nm. These size variations could influence how OMVs interact with eukaryotic cells. Specifically, the larger size of P. mirabilis OMVs may affect the mechanisms of internalization and subsequent immune responses.
Proteomic analysis revealed key OMV proteins, including OmpA, OmpC, and OmpF, common outer membrane protein markers. P. mirabilis OMVs showed higher relative abundance of Braun’s lipoprotein (Lpp) in LB-grown cultures, similar to E. coli OMVs in AU. Lpp is essential for outer membrane stability by linking it to the peptidoglycan layer (Mathelié-Guinlet et al., 2020). Variations in Lpp abundance and OMV size suggest that protein composition influences OMV structure and function in different environments.
Notably, around 50% of the proteins identified in OMVs were cytoplasmic, suggesting either random cytoplasmic protein incorporation or the production of other vesicle types, such as external-internal membrane vesicles (OIMVs) or cytoplasmic membrane vesicles (CMVs; Toyofuku et al., 2023). Our findings align with reports indicating significant cytoplasmic content in OMVs from Gram-negative bacteria (Kulp and Kuehn, 2010; Charpentier et al., 2023), providing further insight into the complexity of OMV biogenesis.
In contrast to the nutritionally-rich LB media, urine in the bladder represents a high-osmolarity, moderately oxygenated and, iron-limited environment that contains mostly amino acids and small peptides (Brooks and Keevil, 1997). To this must be added that the presence of a constant flow caused by urination poses a challenge to bacteria (Asscher et al., 1966; Stamey and Mihara, 1980). Artificial urine mimics the chemical composition of urine, but lacks the immune system components such as cells, chemokines, interleukins, and microorganisms from the urinary microbiota (Soriano et al., 2009). Comparing the proteomic profile of OMVs in LB and AU can provide evidence regarding the presence of proteins required by the uropathogens in the context of urinary tract infection.
4.1 Proteomic content in OMVs from Proteus mirabilis 2921Although the roles of OMVs have been studied extensively, little is known about their production, delivery, and involvement in P. mirabilis. This study represents the first report on the proteomic composition of OMVs from P. mirabilis under two different culture conditions: LB and artificial urine (AU). We used a P. mirabilis strain isolated from a patient with symptomatic UTI (Zunino et al., 2000), known for its high biofilm-forming capacity (Schlapp et al., 2011; Scavone et al., 2023).
Our findings revealed a significant difference in the number of proteins detected in LB compared to AU, showing higher numbers of proteins detected in LB, being this effect more evident in P. mirabilis than in E. coli. Despite the lower number of proteins detected in AU, those identified were biologically significant, mainly related to iron acquisition—a key response to the iron-depleted conditions in AU, similar to natural urine.
We identified several key OMV markers in P. mirabilis-derived OMVs, including OmpA, OmpF, OmpW, TolC, TolB, AcrA, TufB, Pal, and Lpp, confirming the reliability of our vesicle isolation protocol. Flagellar and fimbrial proteins, essential for P. mirabilis motility and colonization, were also detected. Notably, MR/P fimbriae components (MrpH, MrpB, MrpE, MrpF, and MrpG) were over-represented in OMVs from LB cultures, while the structural fimbrial subunit MrpA was more abundant in AU, aligning with their roles in urinary tract colonization.
We also identified UCA/NAF fimbriae proteins (PMI0535 and PMI0534) and several PMF fimbriae proteins (PmfA, PmfE, and PmfF), which are critical for bladder and kidney colonization (Zunino et al., 2003). Consistent with previous studies, OmpA and OmpF were more abundant in nutrient-rich LB media (D'Alessandro et al., 2011). Additionally, TonB-dependent receptors (PMI2596, IreA, PMI1548, PMI0363, PMI0233, PMI0409) were over-represented in AU, reflecting the bacterium’s response to iron-restricted conditions.
Among P. mirabilis virulence factors, only hemolysin (HpmA) was detected in OMVs from both media conditions. HmpB, which activates HmpA, was exclusively found in AU-derived OMVs, suggesting that OMVs could serve as a mechanism for hemolysin secretion.
4.2 Proteomic content in OMVs from Escherichia coli 144In this study, we analyzed the protein content of OMVs produced by a clinical E. coli strain grown in Luria-Bertani (LB) broth and artificial urine (AU; Robino et al., 2014). This UPEC strain is known for its ability to form biofilms and invade urothelial cells in the bladder, forming intracellular bacterial communities (González et al., 2017; González et al., 2020). However, unlike other UPEC strains, this isolate does not express typical toxins such as HlyA and CNF1, nor the virulence genes iutA, ibeA, PAI, and fyuA (González et al., 2020). As expected, we did not detect HlyA and CNF1 toxins in the OMVs from E. coli 144.
Pathogenic and commensal E. coli strains produce OMVs with differing effects. Pathogenic E. coli OMVs typically increase pro-inflammatory cytokines, while commensal E. coli OMVs have anti-inflammatory effects (Behrouzi et al., 2018). In this sense, it is interesting to highlight that OMV proteins such as OmpA, CirA, and FepA can trigger inflammatory responses from macrophages (Imamiya et al., 2023).
Adhesins play a crucial role in bacterial colonization by mediating adhesion to host tissues. FimH, for example, is an adhesin that enables UPEC to adhere to uroplakin molecules on urothelial cells, making it an important virulence factor (Behzadi and Behzadi, 2016). Blackburn et al. found that FimA was the dominant protein in OMVs from E. coli K12 and noted its co-regulation with FliC (Blackburn et al., 2021). In our study, we detected various fimbrial proteins, including FimG and FimH, with their abundance varying based on culture conditions. This suggests that OMVs may act as a delivery mechanism for adhesins, which can also activate various eukaryotic cell mechanisms.
Flagella and type 1 fimbriae are co-regulated and contribute to E. coli adhesion and biofilm formation (Blumer et al., 2005; Badea et al., 2009). Flagellum-mediated motility and chemotaxis help UPEC escape immune responses and spread within the urinary tract (Lane et al., 2005). Previous studies have shown downregulation of flagellation-related genes in vivo compared to LB growth (Snyder et al., 2004). In this work, FlgL and FlgK were only detected in OMVs from LB, while FliC was more abundant in LB OMVs. Consistently, both identified proteins, type 1 fimbriae (FimH) and flagellin (FliC), activate the immune system via TLR4 and TLR5 receptors (Mossman et al., 2008). Thus, their presence in OMVs is associated with pro-inflammatory cytokine induction.
In urine, the majority of upregulated genes and proteins are involved in iron acquisition, a crucial mechanism for bacteria adaptation to host environments (Alteri and Mobley, 2015). Iron-acquisition systems are vital virulence factors in UPEC (Alteri and Mobley, 2015). Previous studies identified iron-uptake proteins in E. coli OMVs, such as the catecholate siderophore receptor Fiu, iron uptake system component EfeO, and ferrichrome-iron receptor FhuA (Rajasekaran et al., 2010; Moeck et al., 1997; Grinter and Lithgow, 2019). OMVs may facilitate the collection of iron-bound siderophores through specific outer membrane receptors. Notably, we observed a significant increase in the relative abundance of the ferrienterobactin receptor FepA in OMVs from UPEC grown in AU. However, despite the high enrichment of TonB-dependent receptors in UPEC during growth in human urine (Alteri and Mobley, 2007), we did not detect TonB proteins in E. coli OMVs.
5 ConclusionBoth P. mirabilis and E. coli produce OMVs with distinct characteristics and functions. P. mirabilis OMVs are larger and enriched in proteins related to iron acquisition and motility, which are important for its virulence and adaptation to different environments. E. coli OMVs, on the other hand, are smaller and contain proteins involved in adhesion, motility, and iron uptake, but lack some typical toxins found in other uropathogenic strains.
The differences in OMV size, protein composition, and functional roles underscore the diversity in OMV-mediated strategies used by these bacteria for survival, adaptation, and pathogenesis in the urinary tract. Understanding the specific characteristics of OMVs in this context, under laboratory conditions mimicking urine, provides valuable insights into the mechanisms of urinary tract infections and could inform targeted therapeutic approaches.
Data availability statementThe mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052681.
Author contributionsMG: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. NN: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. EC: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. SS: Methodology, Writing – original draft, Writing – review & editing. JOM: Writing – original draft, Writing – review & editing. PZ: Writing – original draft, Writing – review & editing. LR: Conceptualization, Writing – original draft, Writing – review & editing. AL: Conceptualization, Methodology, Supervision, Writing - original draft, Writing -review & editing. PS: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Dirección Nacional de Innovación, Ciencia y Tecnología, Montevideo, Uruguay [I-FVF2017-106]; the Comisión Sectorial de Investigación Científica [CSIC iniciación-2019-511]; and the Comisión Académica de Posgrado. JOM received funding from FONDECYT Regular 1231154, PIA/ANID ACT240058, FONDAP 15130011, FONDEQUIP EQM160157.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1493859/full#supplementary-material
Footnotes ReferencesAguilera, L., Toloza, L., Gimenez, R., Odena, A., Oliveira, E., Aguilar, J., et al. (2014). Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics 14, 222–229. doi: 10.1002/pmic.201300328
PubMed Abstract | Crossref Full Text | Google Scholar
Alteri, C. J., and Mobley, H. L. (2007). Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 75, 2679–2688. doi: 10.1128/IAI.00076-06
PubMed Abstract | Crossref Full Text | Google Scholar
Alteri, C. J., and Mobley, H. L. (2015). Metabolism and Fitness of Urinary Tract Pathogens. Microbiol Spectr. doi: 10.1128/microbiolspec.mbp-0016-2015
Crossref Full Text | Google Scholar
Armbruster, C. E., and Mobley, H. L. (2012). Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 10, 743–754. doi: 10.1038/nrmicro2890
PubMed Abstract | Crossref Full Text | Google Scholar
Asscher, A. W., Sussman, M., Waters, W. E., Davis, R. H., and Chick, S. (1966). Urine as a medium for bacterial growth. Lancet 288, 1037–1041. doi: 10.1016/S0140-6736(66)92023-X
Crossref Full Text | Google Scholar
Badea, L., Beatson, S. A., Kaparakis, M., Ferrero, R. L., and Hartland, E. L. (2009). Secretion of flagellin by the LEE-encoded type III secretion system of enteropathogenic Escherichia coli. BMC Microbiol. 9, 1–10. doi: 10.1186/1471-2180-9-30
PubMed Abstract | Crossref Full Text | Google Scholar
Behrouzi, A., Vaziri, F., Rad, F. R., Amanzadeh, A., Fateh, A., Moshiri, A., et al. (2018). Comparative study of pathogenic and non-pathogenic Escherichia coli outer membrane vesicles and prediction of host-interactions with TLR signaling pathways. BMC. Res. Notes 11:539. doi: 10.1186/s13104-018-3648-3
PubMed Abstract | Crossref Full Text | Google Scholar
Blackburn, S. A., Shepherd, M., and Robinson, G. K. (2021). Reciprocal packaging of the main structural proteins of type 1 fimbriae and flagella in the outer membrane vesicles of “wild type” Escherichia coli strains. Ront Microbiol 12:557455. doi: 10.3389/fmicb.2021.557455
留言 (0)