For numerous years, conventional cytogenetics has been utilized to understand the nature of hematologic disorders (HD) and the behavior of cancer cells. These conventional techniques continue to be extensively employed in clinical genetics laboratories to this day (1). Karyotyping is used to define clonal chromosomal abnormalities that often occur in hematologic neoplasms, including number anomalies such as monosomies and trisomies, and also structural anomalies such as deletions, inversions, translocations, and others. Identifying recurrent anomalies enables the definition of a specific prognosis and facilitates the selection of appropriate treatment options (1). Abnormal karyotypes can be simple showing a single abnormality or present many alterations, and thus are named complex karyotypes (CK).
Whether in the context of simple karyotypes or CK, recurrent cytogenetic anomalies are identified. For example, chronic myeloid leukemia (CML) mainly results from the translocation between the long arms of chromosomes 9 and 22 leading to the BCR-ABL fusion (Philadelphia chromosome) (2, 3). Tyrosine kinase inhibitors, such as imatinib, constitute a cornerstone of CML management; however, a poor response to treatment is observed in patients with CK (4). Furthermore, in patients with chronic lymphoid leukemia (CLL), several chromosomal alterations are reported, including a deletion on the short arm of chromosome 17 [(del (17p)] that can impact therapeutic choices (5). Additionally, among recurrent cytogenetic anomalies, translocation between the long arms of chromosomes 15 and 17 [t(15,17)] resulting in PML-RARA fusion is a characteristic of acute promyelocytic leukemia (APL, AML-M3) (6). Many other recurrent cytogenetic anomalies are reported in the setting of different HD.
The first studies on CK were performed by Berger et al. in the 1980s who described CK in acute non-lymphocytic leukemia as presenting three or more different chromosomal abnormalities (CA) in the malignant clone (7). Over the years, CK were analyzed in different HD. Indeed, the significance of karyotyping has been underscored by Yunis et al. who reported that the presence of three or more CA in myelodysplastic syndromes (MDS) was associated with a poor prognosis (8). Concerning acute myeloid leukemia (AML), Mrozek et al. defined ‘typical’ CK as CK showing the following abnormalities of 5q, 7q and/or 17p, however, their absence denotes an “atypical” CK (9). Furthermore, AML patients with typical CK were older, had more TP53 mutations, and a shorter overall survival (OS) compared to AML patients with atypical CK.
Although there is no clear consensus on the exact definition of CK, the Francophone Group of Hematologic Cytogenetics (GFCH) described CK as abnormal karyotypes presenting three or more CA. Furthermore, GFCH defined subgroups according to the number of CA: low-CK with three CA, intermediate-CK with four, and highly-CK with five or more CA (10). The presence of CK at diagnosis holds a significant prognostic value in predicting response to treatment, relapse, and OS. However, interpreting CK remains challenging and varies among different hemopathies (10).
To our knowledge, there is a lack in studies that have exclusively investigated CK, focusing on their complexity, incidence, and associations. Based on our research, there is only one similar cytogenetic study that has been previously published. This study was carried out in Pakistan reporting data on 41 CK over a 6-years period (11). In the current study, we assessed the prevalence of CK in HD and compared their distribution among Lebanese patients. Additionally, we compared our results with those of the previously published study.
Materials and methodsA total of 5681 hematologic cases were referred to the Jacques Loiselet Center for Medical Genetics and Genomics (CGGM), previously named the Medical Genetics Unit (UGM), at Saint Joseph University in Beirut (USJ), a tertiary referral center in Lebanon, between January 2011 and May 2023. Among these, 255 cases presented CK.
For all samples, we employed conventional cytogenetic procedures. Karyotyping was conducted on bone marrow aspirate or peripheral blood. Two cell cultures were established in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. Mitogens were introduced to the cell cultures as deemed necessary based on the diagnosis or suspicion, such as IL2+DSP30 for mature B-cell neoplasms, and PHAm for T-cell neoplasms. Cell cultures were incubated at 37°C with 5% CO2. After 24h, 48h or 72h, cells were harvested in accordance with standard cytogenetic methods, using KCl hypotonic treatment and ethanol-acetic acid 3V:1V fixative. Then, cytogenetic pellets were spread on Superfrost slides, followed by RHG banding. Fifteen to 25 R-banded metaphases were analyzed according to the recommendations of the “international system of human cytogenetic nomenclature” (ISCN 2016 or 2021) (12), using the IKAROS software (Metasystems, Germany). Additionally, we used GFCH cut-off values to classify CK according to the number of CA. P values less than 0.05 were considered statistically significant.
ResultsIn our study, 255 CK were detected (4.48%). Among these, 151 (59.22%) were males and 104 (40.78%) were females, with a sex ratio of 1.5. The mean age was 59 years (median 65 years) ranging from 1 to 90 years; with 94.2% of adults.
Furthermore, we evaluated the recurrence of different CA detected in combination with CK. Our analyzes showed that hyperdiploidy was the most common CA detected, with a frequency of 22.41%, followed by 17p deletion (15.52%) and 7q deletion (13.79%) (Figure 1). Table 1 illustrates the classification of the most prevalent anomalies detected among the various HD.
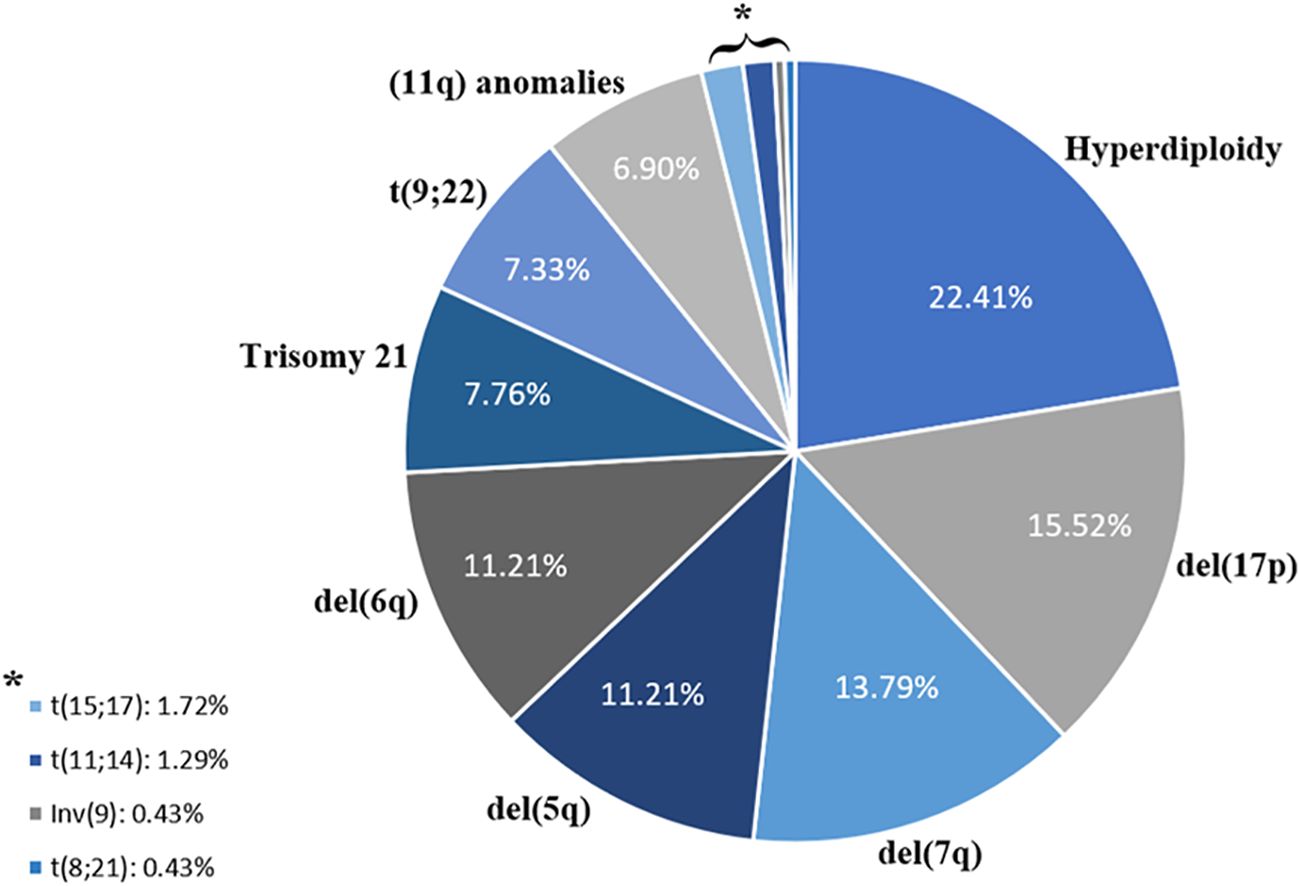
Figure 1. Distribution of chromosomal abnormalities associated with complex karyotypes.
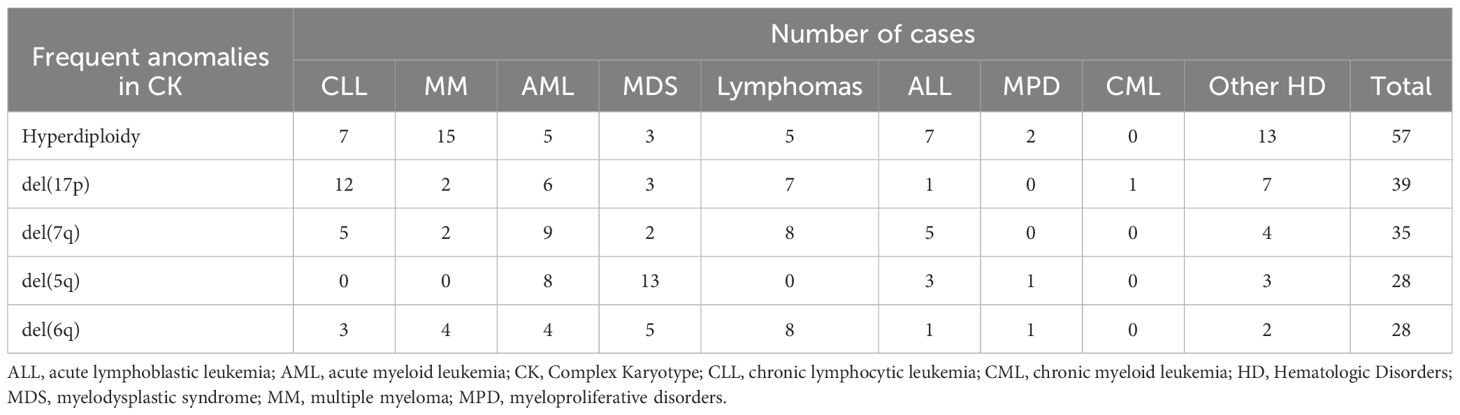
Table 1. Distribution of the frequent anomalies in CK among different HD.
Interestingly, and regardless of the sex distribution, we observed that most of the CK in our study (68.24%) had five or more CA and are therefore classified as highly complex (Table 2). Furthermore, CLL presented the highest CA rates among all other HD in our study (14.90%), followed by multiple myeloma (MM) (12.55%) and AML (11.76%) (Table 3). On a side note, the ‘other HD’ group included patients referred for isolated leucopenia or anemia, monoclonal gammopathy of undetermined significance (MGUS), among other HD.
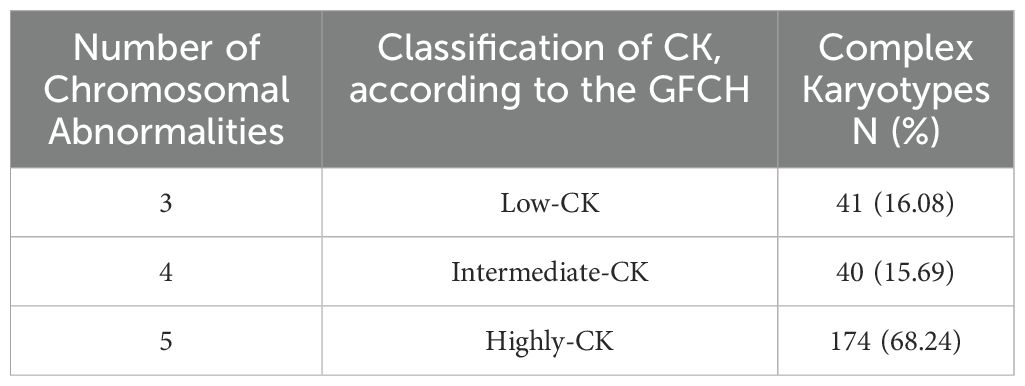
Table 2. Distribution of complex karyotypes according to the GFCH (Francophone Group of Hematologic Cytogenetics).
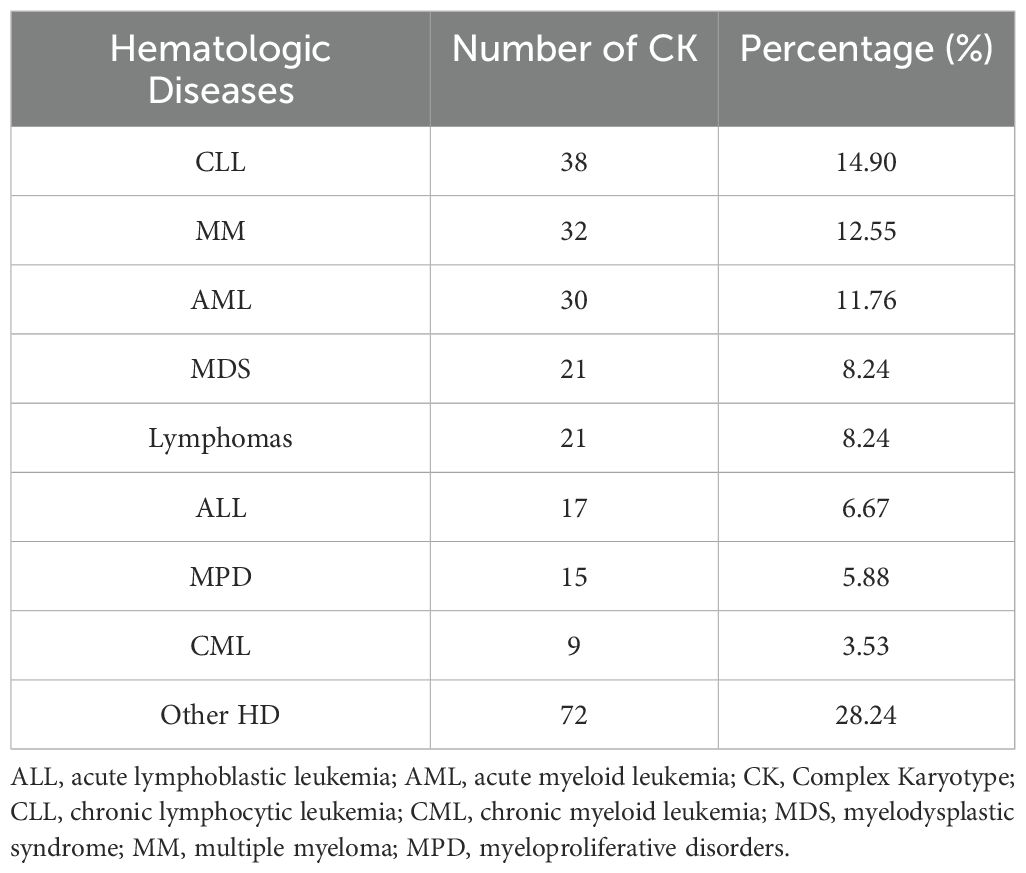
Table 3. Distribution of complex karyotypes among hematologic diseases.
On the other hand, our results did not show statistically significant differences in the distribution of CK between males and females, except for CML (p-value=0.01). In fact, a male predominance was observed in CML-CK (n=9), while zero females presented CK with CML in our study (Supplementary Table 1).
DiscussionIn the current study, we present, for the first time, comprehensive data from the Middle East concerning CK across a substantial series of hematologic neoplasms.
First, in comparison to the previously published study from Pakistan, the only similar study reporting on CK, we noticed that in our study we detected 4.48% of CK, slightly higher than the 3.4% reported by Waheed et al. (11). Furthermore, the male-to-female sex ratio in our study (1.5:1) differed significantly from the ratio of 4:1 reported in Pakistan. This divergence could be attributed to the small sample size in the Pakistani study (only 41 CK cases), and also to the different demographical characteristics among populations. Additionally, we noted an important difference in the mean age of CK patients: 59 years (median 65 years) in our study, versus 37 years (median 39 years) in the Pakistani study. This variation may be population-dependent and may be related to the increase in life expectancy in Lebanon, since life expectancy data in Lebanon align more closely with data collected from western populations such as the US (13).
Furthermore, while evaluating the association between recurrent CA and CK, our analyzes revealed that hyperdiploidy was the most common CA. This finding contrasts with the results reported by Waheed et al., where trisomy 21 was the most common CA followed by hyperdiploidy (11). This discrepancy is likely attributed to differences in demographic characteristics and the prevalence of various hematological disorders among populations. In this context, hyperdiploid karyotypes are significantly reported in several neoplasms, including (but not limited to) MM patients (14). Our study revealed an association between certain chromosome gains in cases of MM, notably identifying gains of chromosomes 3, 9, 11, 15, and 19 as the most frequently observed. Additionally, this CA is associated with a good response to treatment in children with Acute Lymphoblastic Leukemia (ALL) (15), but showed an unfavorable prognosis in cases of AML (10), depending on the associated cytogenetic abnormalities.
Regarding HD distribution, our findings diverge significantly from those reported in Pakistan, where the most prevalent diagnoses were MDS and AML (11). In our study, CLL emerged as the foremost diagnosis thus establishing itself as the most frequently encountered HD with CK.
In a brief review of the literature, we observed a decline in karyotyping requests for CLL cases over time, and that cytogenetic analyzes have predominantly shifted towards Fluorescence In Situ Hybridization (FISH) or microarray (16). The current study, in addition to our previous work on CLL, showed a low request for karyotyping in cases of CLL. Our data once again highlighted the importance of karyotyping in CLL, since CK regained its important prognostic value in CLL (17, 18), along with common abnormalities tested by molecular cytogenetics (deletion 17p, deletion 11q, deletion 13q and trisomy 12) (19, 20). In fact, Jondreville et al. (21) recommends a systematic karyotyping in patients with CLL before treatment because it can identify CK that are not detected by targeted techniques such as FISH. Additionally, several molecular biomarkers are important in CLL, such as the IGHV mutational status (22, 23).
The sample size of our study is important, but the main limitation of this study is the absence of survival and follow-up data from the patients. This information was not available for not all patients. However, based on the data that were accessible, most patients with CK in AML, ALL and MDS experienced poor outcomes and died due to the disease, adverse events or infections. In cases of CK-AML, Venetoclax was frequently used in combination therapies, yet a poor prognosis was still observed. T limitations of our study hindered our ability to draw conclusions regarding overall survival, treatment-free survival, or the worst prognosis linked to CK. However, our results still provide insightful cytogenomic information and can be considered as a basis for further investigations.
In conclusion, in the era of ‘molecular diagnostics’, it is clear that conventional cytogenetics still has room for the management and treatment of hematologic disorders. Despite significant advancements in the field, conventional karyotyping remains essential for detecting complex karyotypes. Emerging AI-driven technologies are poised to play a crucial role in the future (24). The current study is considered the first in the Middle East region and among a few published studies worldwide, with the largest sample size, highlighting the particularity of CK. To conclude, our study, which analyzes a representative sample size over a significant 12-year period, provides substantial credibility to our data. We believe these findings can serve as a valuable foundation for future clinical research focused on complex karyotypes, treatment strategies, and patient outcomes.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statementThis study was approved by the ethical committee at Saint Joseph university of Beirut. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements because the study was based on de-identified data.
Author contributionsSR: Data curation, Methodology, Visualization, Writing – original draft. FAA: Data curation, Methodology, Visualization, Writing – original draft. CA: Investigation, Methodology, Writing – original draft. ZEM: Investigation, Methodology, Writing – original draft. WS: Investigation, Methodology, Writing – original draft. AC: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a grant from the research council at Saint Joseph University of Beirut (Grant no. FM454).
AcknowledgmentsWe would like to acknowledge the contributions of former team members, Mrs Rima Korban and Dr Tony Yammine, for their valuable work.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1480793/full#supplementary-material
References1. Bain BJ. Overview. Best Pract Res Clin Haematology. (2001) 14:463–77.
3. Trela E, Glowacki S, Błasiak J. Therapy of chronic myeloid leukemia: twilight of the imatinib era? ISRN Oncol. (2014) 2014:1–9. doi: 10.1155/2014/596483
Crossref Full Text | Google Scholar
4. Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. (2020) 34:966–84. doi: 10.1038/s41375-020-0776-2
PubMed Abstract | Crossref Full Text | Google Scholar
5. Hallek M, Al-Sawaf O. Chronic lymphocytic leukemia: 2022 update on diagnostic and therapeutic procedures. Am J Hematology. (2021) 96:1679–705. doi: 10.1002/ajh.26367
Crossref Full Text | Google Scholar
7. Berger R, Bernheim A, Daniel MT, Valensi F, Flandrin G. Leucémies "induites". Aspects cytogénétique et cytologique. Comparaison avec des leucémies primitives [Induced leukemias. Cytogenetical and cytological aspects. Comparison with primitive leukemias (author's transl). Nouv Rev Fr Hematol (1978). (1981) 23:275–84.
PubMed Abstract | Google Scholar
8. Yunis JJ, Rydell RE, Oken MM, Arnesen MA, Mayer MG, Lobell M. Refined chromosome analysis as an independent prognostic indicator in de novo myelodysplastic syndromes. Blood. (1986) 67:1721–30. doi: 10.1182/blood.v67.6.1721.bloodjournal6761721
PubMed Abstract | Crossref Full Text | Google Scholar
9. Mrózek K, Eisfeld AK, Kohlschmidt J, Carroll AJ, Walker CJ, Nicolet D, et al. Complex karyotype in de novo acute myeloid leukemia: typical and atypical subtypes differ molecularly and clinically. Leukemia. (2019) 33:1620–34. doi: 10.1038/s41375-019-0390-3
PubMed Abstract | Crossref Full Text | Google Scholar
10. Nguyen-Khac F, Bidet A, Daudignon A, Lafage-Pochitaloff M, Ameye G, Bilhou-Nabéra C, et al. The complex karyotype in hematological Malignancies: a comprehensive overview by the Francophone Group of Hematological Cytogenetics (GFCH). Leukemia. (2022) 36:1451–66. doi: 10.1038/s41375-022-01561-w
PubMed Abstract | Crossref Full Text | Google Scholar
11. Waheed S, Hassan J, Naz M, Maqsood S, Abid M, Shan S, et al. Complex karyotype in hematological diseases: A 6-year single centre study from Pakistan. J Oncol. (2018) 2018:1–5. doi: 10.1155/2018/2019239
Crossref Full Text | Google Scholar
12. McGowan-Jordan J, Hastings R, Moore S. Re: international system for human cytogenetic or cytogenomic nomenclature (ISCN): some thoughts, by T. Liehr. Cytogenet Genome Res. (2021) 161:225–6. doi: 10.1159/000516655
PubMed Abstract | Crossref Full Text | Google Scholar
13. Krok-Schoen JL, Fisher JL, Stephens JA, Mims A, Ayyappan S, Woyach JA, et al. Incidence and survival of hematological cancers among adults ages ≥75 years. Cancer Med. (2018) 7:3425–33. doi: 10.1002/cam4.1461
PubMed Abstract | Crossref Full Text | Google Scholar
15. Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. (2012) 26:123–35. doi: 10.1016/j.blre.2012.01.001
PubMed Abstract | Crossref Full Text | Google Scholar
16. Parikh SA, Strati P, Tsang M, West CP, Shanafelt TD. Should IGHV status and FISH testing be performed in all CLL patients at diagnosis? A systematic review and meta-analysis. Blood. (2016) 127:1752–60. doi: 10.1182/blood-2015-10-620864
PubMed Abstract | Crossref Full Text | Google Scholar
17. Eichhorst B, Robak T, Montserrat E, Ghia P, Niemann CU, Kater AP, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2021) 32:23–33. doi: 10.1016/j.annonc.2020.09.019
PubMed Abstract | Crossref Full Text | Google Scholar
18. Ayoub G, Sinan H, Kourie HR, Kattan J, Nasr F, Karak FE, et al. Genetic markers of chronic lymphocytic leukemia: a retrospective study of 312 patients from a reference center in Lebanon. Future Oncol. (2023) 19:1991–2002. doi: 10.2217/fon-2023-0535
PubMed Abstract | Crossref Full Text | Google Scholar
19. Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. (2000) 343:1910–6. doi: 10.1056/nejm200012283432602
PubMed Abstract | Crossref Full Text | Google Scholar
20. Letestu R, Lévy V, Eclache V, Baran-Marszak F, Vaur D, Naguib D, et al. Prognosis of Binet stage A chronic lymphocytic leukemia patients: the strength of routine parameters. Blood. (2010) 116:4588–90. doi: 10.1182/blood-2010-06-288274
PubMed Abstract | Crossref Full Text | Google Scholar
21. Jondreville L, Krzisch D, Chapiro E, Nguyen-Khac F. The complex karyotype and chronic lymphocytic leukemia: prognostic value and diagnostic recommendations. Am J Hematol. (2020) 95:1361–7. doi: 10.1002/ajh.25956
PubMed Abstract | Crossref Full Text | Google Scholar
22. Delgado J, Doubek M, Baumann T, Kotaskova J, Molica S, Mozas P, et al. Chronic lymphocytic leukemia: A prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am J Hematol. (2017) 92:375–80. doi: 10.1002/ajh.24660
PubMed Abstract | Crossref Full Text | Google Scholar
23. Rammal S, Semaan W, Aprahamian N, Moussallem R, Chebly A. Spotlight on borderline-IGHV mutational status in chronic lymphocytic leukemia. Front Oncol. (2024) 14:1430225. doi: 10.3389/fonc.2024.1430225
留言 (0)