Notch receptors are heterodimeric transmembrane proteins that regulate cell fate, such as differentiation, proliferation, and apoptosis. Dysregulated Notch pathway signaling has been observed in glioblastomas, as well as in other human malignancies. Nerve growth factor (NGF) is essential for cell growth and differentiation in the nervous system. Recent reports suggest that NGF stimulates glioblastoma proliferation. However, the relationship between NGF and Notch1 in glioblastomas remains unknown. Therefore, we investigated expression of Notch1 in a glioblastoma cell line (U87-MG), and examined the relationship between NGF and Notch1 signaling.
MethodsWe evaluated expression of Notch1 in human glioblastomas and normal brain tissues by immunohistochemical staining. The effect of NGF on glioblastoma cell line (U87-MG) was evaluated by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. To evaluate the relationship between NGF and Notch1 signaling, Notch1 and Hes1 expression were evaluated by reverse transcription polymerase chain reaction (RT-PCR) and Western blot analysis, respectively. To confirm the effects of NGF on Notch1 signaling, Notch1 and Hes1 small interfering RNAs (siRNAs) were used.
ResultsIn immunohistochemistry, Notch1 expression was higher in glioblastoma than in normal brain tissue. MTT assay showed that NGF stimulates U87-MG cells in a dose-dependent manner. RT-PCR and Western blot analysis demonstrated that Notch1 and Hes1 expression were increased by NGF in a dose-dependent manner. After transfection with Notch1 and Hes1 siRNAs, there was no significant difference between controls and 100 nM NGF-β, which means that U87-MG cell proliferation was suppressed by Notch1 and Hes1 siRNAs.
ConclusionThese results indicate that NGF stimulates glioblastoma cell proliferation via Notch1 signaling through Hes 1.
Key Words: Glioblastoma · Nerve growth factor · Notch1.
INTRODUCTIONGlioblastoma (World Health Organization grade IV) is the most common type of glioma and the most common malignant primary brain tumor in adults. Glioblastomas usually develop in the cerebral hemisphere as a solitary tumor and may also occur in the cerebellum and the spinal cord. Even though the current standard of care includes maximal surgical resection, adjuvant chemotherapy and radiotherapy, the prognosis of glioblastoma is poor.
Notch signaling, a fundamental signaling system, regulates cell proliferation and differentiation through lateral inhibition and has been known to be involved with tumorigenesis [3,12]. Notch receptors are heterodimeric transmembrane proteins that regulate cell fate, such as differentiation, proliferation, and apoptosis in numerous tissues [39]. In mammals, there are four types of Notch receptors (Notch1, Notch2, Notch3, and Notch4) and five classic ligands (Delta-like1, Delta-like3, Delta-like4, Jagged1, and Jagged2) [1]. Dysregulated Notch pathway signaling has been identified in brain tumors, as well as in many other tumors including hematologic malignancies, cervical, lung, pancreatic, breast cancer, hepatocellular carcinomas, and in ovarian [2,4,7,17,19,22,23,26]. Notch expression and the correlation between tumor grades differ between previous reports and these differences have not been fully clarified. Of Notch receptors, Notch1 has been reported to be oncogenic and show positive correlation with glioma progression [20,38]. However, the tumor suppressive role of Notch1 has been identified in other reports [8,27]. Nerve growth factor (NGF) is a member of the neurotrophin family and is essential for cell growth and differentiation in both the peripheral and central nervous system [37]. NGF interacts with two receptors, such as TrkA and p75NTR, which have also been identified in glioblastoma cell lines [6,10,25]. NGF has been described as an inhibitor of tumor cell different proliferation, or mitogenesis [11,30-32]. Recent reports suggest that NGF stimulates glioblastoma proliferation [10,18,25,33].Although there is much evidence that Notch signaling pathway is involved in glioblastoma pathogenesis, the frequency of specific Notch receptor expression, the mechanisms underlying Notch activation, and whether NGF is involved with Notch1 signaling are still unknown. Therefore, we investigated expression of Notch1 in a glioblastoma cell line (U87-MG), and examined the relationship between NGF and Notch1 signaling.
MATERIALS AND METHODS Human tissue samplesHuman glioblastoma and normal brain tissues, including surgically resected tissue adjacent to infiltrating glioblastoma or traumatic brain tissue, were obtained from our hospital. Normal brain tissues were obtained at the area that has no visible tumor cells during tissue analyses by pathologist and during surgery for traumatic brain injury. This study was approved by the Hallym University Institutional Review Board (2016-I160).
Immunohistochemical stainingParaffin-embedded tissue sections were subjected to immunostaining using rabbit polyclonal to Notch1 antibodies (ab27526, Abcam, Cambridge, UK) at a 1 : 50 dilution. Primary antibody was diluted in phosphate-buffered saline with 5% normal blocking serum. Rabbit IgG antibody (PK-6101, Vector laboratories, Burlingame, CA, USA) was used as the secondary antibody. Immunohistochemistry was performed as described elsewhere. A streptavidin-biotin-peroxidase complex technique was used to reveal antibody-antigen reactions. Staining with 3,3’-diaminobenzidine was performed under a microscope for 1 minute (SK-4100, Vector laboratories). Slides were counterstained with hematoxylin (H-3401, Vector laboratories). After immunohistochemical controls were performed, normal brain tissue was used as a negative control and included omission of the primary antibody. The evaluation of immunohistochemical staining for Notch1 was performed by analyzing 10 different tumor fields and the mean percentage of tumor cells with positive staining was scored. For the qualitative assessment of immunohistochemical staining, the staining was divided as positive and negative.
For the quantitative assessment, staining has semiquantitatively been scored as : 1) negative - less than 10% of positive cells, 2) positive - immunoreactivity is more that 10% positive cancer cells. We did not count the percentage of positive cell numbers, but estimated the ratio in areas of the cancer cells.
Cell culture and cell proliferation assayThe U87-MG cells were cultured in minimum essential medium containing 10% fetal bovine serum, L-Glutamine (2 mM) and an antibiotic combination of 0.1 mg/mL streptomycin and 100 unit/mL penicillin (Gibco, Grand Island, NY, USA). The cells were incubated at 37°C in a humidified air atmosphere containing 5% CO2. Cells were placed in a 96-well culture plate at 1×104 cells/well in 200 μL culture medium. After 24 hours incubation at 37°C, cells were treated for 48 hours with drugs (human NGF-β) (N1408, Sigma-Aldrich, St. Louis, MO, USA) and Notch1 small interfering RNA (siRNA) (HSS107248, Invitrogen, Carlsbad, CA, USA). Briefly, the culture medium was replaced with 200 μL culture medium containing 0.5 mg/mL 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (M5655, Sigma-Aldrich) and incubated for 2 hours. After 2 hours, the supernatant was removed and 200 μL dimethyl sulfoxide was added to dissolve the formazan precimpitate at 37°C. After 30 minutes, absorbance was measured at 570 nm with an automated microplate reader (Multiskan GO, Thermo Fisher Scientific, Finland). All experiments were repeated at least eight times.
Western blotting of Notch1 and Hes1U87-MG cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium desoxycholate, 0.1% sodium dodecyl sulfate, 0.5 mM Tris, pH 8.0) on ice for 30 minutes, followed by centrifugation for 20 minutes at 4oC. Quantification was done by Bradford assay (Bio-Rad, Glattbrugg, Switzerland). In all, 50 microgram total cellular protein per lane was size fractionated on a 7% Tris-acetate gel (Invitrogen, Carlsbad, CA, USA) for Notch1 detection and transferred onto nitrocellulose (Schleicher and Schuell, Kassel, Germany). Equal loading and transfer efficiency was visually checked by Ponceau staining. Membranes were blocked overnight at 4oC temperature with 5% weight/volume (w/v) nonfat dry milk/Tris-buffered saline and Tween-20 (0.05% w/v). Membranes were incubated with rabbit polyclonal anti-Notch1 antibodies (H-131, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for the detection of the extracellular domain of Notch1. Experiments were repeated more than three times. The densities of the bands were measured by the software program (ImageJ 1.47v, NIH, Bethesda, MD, USA) and the density values were compared statistically.
Reverse transcriptionPCR (RT-PCR)Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using amfiRivert cDNA Synthesis Platinum Master Mix (R5600, GenDEPOT, Barker, TX, USA) according to the manufacturer’s instructions. Subsequently, cDNA was used for each PCR reaction with each primer pair. PCR products were separated on a 1 percent agarose gel containing ethidium bromide. Experiments were repeated more than five times. The densities of the bands were measured by the program (ImageJ 1.47v, NIH). The density values were analyzed statistically. Primers were as follows :
1) The Notch1 primer; sense : 5’-AGATCAACCTGGATGACTGTGCCA-3’, antisense : 5’-ACACGTAGCCACTGGTCATGTCTT-3’.
2) The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers; sense : 5-GCTCTCCAGAACATCATCCCTGCC-3; antisense : 5-CGTTGTCATACCAGGAAATGAGCTT-3.
3) The Hes1 primer; sense : 5’-AGATCAACCTGGATGACTGTGCCA-3’, antisense : 5’-ACACGTAGCCACTGGTCATGTCTT-3’.
Immunofluorescence stainingU87-MG cells were plated onto glass coverslips in 6-well culture dishes (100000 cells/2 mL medium/dish). After 24 hours incubation at 37°C, the cells were treated for 48 hours with drugs (NGF-β). The cells were rinsed with phosphate-buffered saline (PBS) and fixed in 4% formaldehyde in PBS for 10 minutes at room temperature, the fixed cells were incubated in 5% bovine serum albumin (BSA) in PBS for 2 hours. Cover slips were subsequently incubated 2 hours at room temperature with rabbit polyclonal antibodies against Notch1 (Abcam) (1 : 200 dilution). After washing, the slides were incubated in the dark at room temperature for 1 hour with fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit IgG secondary antibodies (Santa Cruz) (1 : 200 dilution). Slides were mounted with Fluorescent mounting medium (S3023, Dako Cytomation, Carpinteria, CA, USA) and immunofluorescence staining images were acquired using a laser-scanning confocal inverted microscope (LSM700, Carl Zeiss, Jena, Germany).
Statistical analysisNotch1 and Hes1 expression and clinico-pathological features were analyzed by paired t-test. p-values lower than 0.05 were accepted as statistically significant.
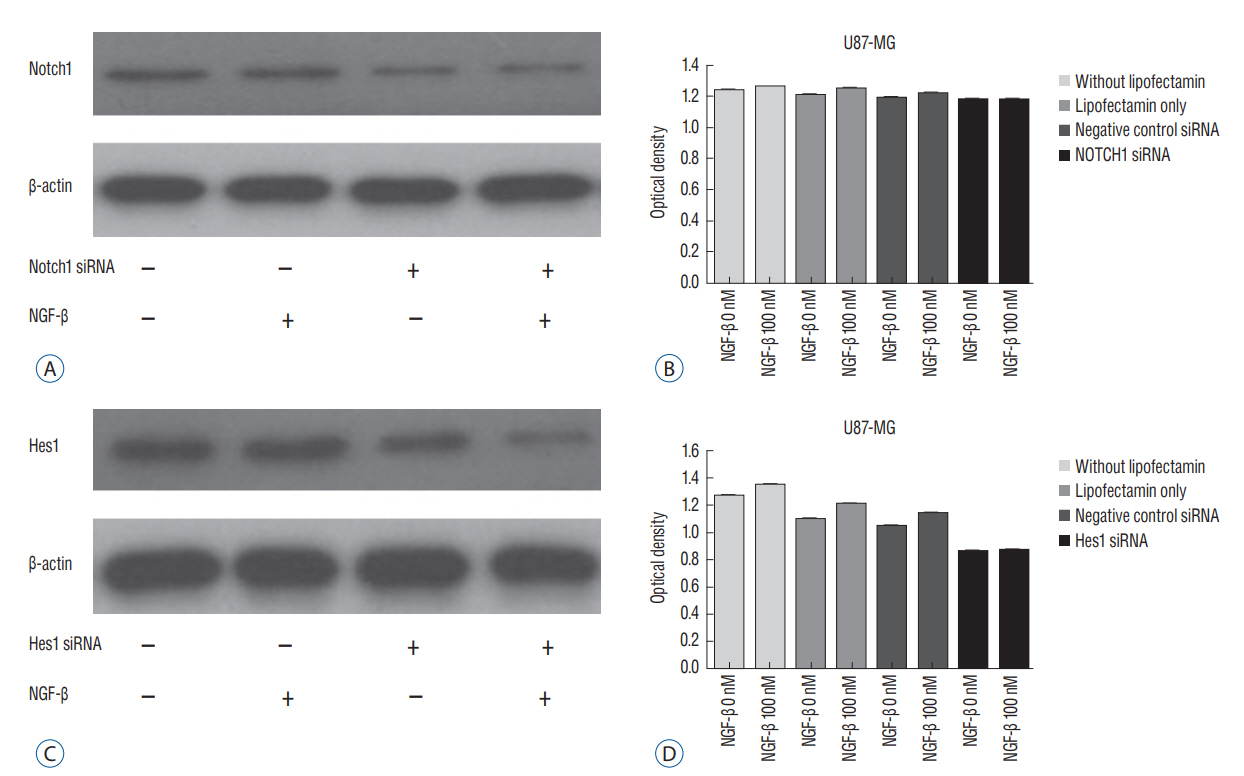
RESULTS Presence of Notch1 in primary human glioblastoma Immunohistochemistry was used to determine whether Notch1 expression is different in glioblastoma than in normal brain tissue. Notch1 expression was evaluated by immunohistochemistry in 10 human glioblastoma tissues and in six normal brain tissues, including surgically resected tissue adjacent to infiltrating glioblastoma or traumatic brain tissue. Based on quantitative scoring of Notch1 staining, a strong positive was identified in six cases (80%) and moderately positive in four cases. There was no significant staining in six normal brain tissues, suggesting no immunoreactivity of Notch1 in normal brain tissue (Fig. 1). Nerve growth factor and U87-MG proliferation The authors evaluated the effect of NGF-β in U87-MG cell line. After treatment of U87-MG cell lines with 0.1 nM, 1 nM, 10 nM, 50 nM, and 100 nM NGF-β, cell proliferation was measured by MTT assay. The mean values of the MTT assays were 1.430, 1.468, 1.512, 1.564, and 1.625 in the NGF-treated cell lines, respectively, and 1.380 in the non-NGF treated cell line (p=0.0054, 0.0005, 0.0001, 0.0001, and 0.0001) (Fig. 2). The findings suggested that NGF stimulates U87-MG cell line in a dose-dependent manner. Nerve growth factor induced expression of Notch1 and Hes1 After observing that NGF stimulates proliferation of the U87-MG cell line, Notch1 and Hes1 expression were evaluated by RTPCR and Western blot analysis, respectively, to evaluate the relationship between NGF and Notch1 signaling. In controls (0 nM NGF-β), the mean density of Notch1 was 100. The mean density of Notch1 in the RT-PCR was 143.02, 189.37, and 243.63 in 100 nM, 200 nM, and 500 nM NGF-β, respectively (Fig. 3A and B). There was a statistically significance at 500 nM (p=0.044), but no statistically significance at 100 nM or 200 nM (p=0.054, 0.054, respectively). The mean density of Notch1 in the Western blot analysis was 114.81, 116.05, and 137.93 in 100 nM, 200 nM, and 500 nM NGF-β, respectively (Fig. 3C and D). There was a statistically significance at 500 nM (p=0.037), but no statistically significance at 100 nM or 200 nM (p=0.059, 0.102, respectively). Hes1 expression was then evaluated. The mean density of Hes1 was 100 in controls (0 nM NGF-β). The mean density of Hes1 in the RT-PCR was 130.07 in 100 nM NGF-β, 136.92 in 200 nM NGF-β, and 146.67 in 500 nM NGF-β (p=0.271, 0.297, and 0.020, respectively) (Fig. 4A and B). The mean density of Hes1 in the Western blot analysis was 124.45 in 100 nM NGF-β, 125.28 in 200 nM NGF-β, and 142.03 in 500 nM NGF-β (p=0.109, 0.073, and 0.037, respectively) (Fig. 4C and D). These findings show that NGF has a dose-dependent effect on Notch1 and Hes1 expression in U87-MG cell line. Notch1 gene suppression by siRNA removes the NGF effect To confirm the effects of NGF on Notch1 signaling, siRNAs targeting Notch1 and Hes1 were used. The efficacy of siRNA for reducing the target was identified through quantitative PCR. Without lipofectamine, the mean values of Notch 1 were 1.239 in controls and 1.262 in 100 nM NGF-β samples (p=0.0003). In lipofectamine-only, the mean values were 1.201 in control and 1.253 in 100 nM NGF-β (p=0.0063). In negative controls with Notch1 siRNA, the mean values were 1.195 in control and 1.224 in 100 nM NGF-β (p=0.0013). After transfection with Notch1 siRNA, however, there was no significant difference between controls and 100 nM NGF-β (1.181 and 1.180, respectively) (p=0.887) (Fig. 5A and B). In the absence of lipofectamine, the mean values of Hes1 were 1.266 in the control and 1.346 in 100 nM NGF-β (p=0.0005). In lipofectamine-only samples, the mean values were 1.092 in the control and 1.201 in 100 nM NGF-β (p=0.0053). In the negative control of Hes1 siRNA, the mean values were 1.044 in the control and 1.135 in 100 nM NGF-β (p=0.0003). After transfection with Hes1 siRNA, there was no significant difference between controls and 100 nM NGF-β (0.857 and 0.869, respectively) (p=0.075) (Fig. 5C and D). These findings demonstrated that the effects of NGF on U87-MG cell line were suppressed by Notch1 siRNA. DISCUSSION Glioblastoma is the most common malignant brain tumor and the prognosis is poor. As knowledge has increased about the molecules involved in glioblastoma formation, various specific target therapies, such as epidermal growth factor, Akt, Hedgehog, mammalian target of rapamycin (mTOR), phosphoinositide 3-kinase, platelet-derived growth factor receptor, Raf, and transforming growth factor β (TGF-β) have been introduced [16]. The studies of the interactions between NGF and Notch signaling in glioblastoma may address the problems related to progress of malignancy and the difficulty of treatment. However, the relationship between Notch1 and NGF in glioblastoma remains unknown. In this study, the relationship between NGF and Notch1 signaling in U87-MG cell lines was evaluated and the results suggest that NGF stimulates glioblastoma proliferation through Notch1 receptor signaling. Notch signaling has been linked to the pathogenesis of various cancers [19]. Out of four Notch receptors, Notch1 has been reported to act as oncogenein many tumors, including pancreatic cancer, cervical cancer, renal cell carcinoma, and lymphomas, ovarian cancer and breast cancer [13,15,21,29,40]. However, it acts as a tumor suppressor in some tumors, such as murine skin tumors and non-small cell lung cancers [24,35]. Similar characteristics of Notch1 have been reported in gliomas. Xu et al. [38] reported that the expression of Notch1 showed positive correlation with glioma progression. In another report, high expression of Notch1 was shown to be an independent predictor of poor prognosis in glioma [20]. However, some authors reported an antiproliferative role of Notch1, which means a favorable prognosis with Notch1 expression in glioma [8,27,28]. These findings mean the action of Notch1 in gliomas and Notch1 signaling could be related to complex factors. A growing body of evidence suggests that Notch1 signaling is related to progression in gliomas [7,20]. However, the exact mechanism is still unclear. NGF is a member of the neurotrophin family, which includes brain-derived neurotrophic factor, neurotrophin 3, and neurotrophin 4/5, and it plays a critical role in cell growth and differentiation in both the peripheral and central nervous system [37]. NGF interacts with both the high affinity TrkA, and the common neurotrophin p75NTR receptors [6]. These receptors and NGF are produced during development, adult life, and aging of cells in various tissues such as the CNS, PNS, immune and inflammatory systems [34]. Recently, these two receptors have been identified in human glioblastoma cell lines (U87, U251, and U373) and there are several reports suggesting that glioblastoma growth could be enhanced by NGF [10,18,25,33]. Singer et al. [33], reported that glioblastoma growth can be induced by NGF acting by Trk A receptor phosphorylation. The exact role of Trk receptor in glioblastoma is, however, still unknown. We found that Notch1 expression was higher in glioblastoma tissue than in the adjacent or in normal brain tissue. Next, we evaluated the effect of NGF-β in U87-MG cell lines by MTT assay and showed that NGF stimulates U87-MG cell lines dosedependently. A similar finding was observed in previous reports. Oelmann et al. [25] reported that growth stimulation by NGF was dose-dependent in U87-MG but not significant in 87-HG-31. In another report, NGF exhibited promigratory and proliferatory activities in human glioma cell lines (LN229) [5]. To determine whether this proliferation by NGF was associated with Notch1 signal pathway, we evaluated the Notch1 and Hes1 expression and showed that Notch1 and Hes1 expression were increased by NGF dose-dependently. This results means NGF is related to up-regulation of Notch1 pathway in glioblastomas. Although there are no reports showing the exact relationship between the NGF and Notch1 pathway in glioblastomas, there are some reports suggesting NGF is related to the regulation of Notch1 pathway. Faux et al. [9] reported that growth factors such as fibroblast growth factor1 (FGF1) and FGF2 increased Notch1 expression and inhibited the neuronal differentiation of the neuroepithelial precursor (NEP) cells. However, some authors reported that NGF induced Notch1 downregulation in other cell types [14,36]. Subsequently, to confirm the NGF effect on the Notch1 signaling, siRNA was used to target Notch1 in this study. Compared to before transfection with Notch1 siRNA, there was no significant difference in U87-MG cell proliferation after transfection with Notch1 siRNA. This suggests that the NGF effect on U87-MG cell proliferation was suppressed by Notch1 siRNA, and that NGF might induce upregulation of Notch1 receptor. A recent study showed that Notch1 and Hes1 were also components of the NGF pathway. In an acute retinal injury model, Jian et al. [14] reported that transient Notch1 down-regulation and NGF up-regulation promoted de-differentiated Muller cells to differentiate into photoreceptors. Ström et al. [36] observed that endogenous Hes1 was related to the NGF pathway and that DNA binding of Hes1 was post-translationally inhibited during NGF signaling in PC12 cell lines. They also suggested that this posttranslational effect on Hes1 was conducted by the phosphorylation of the protein kinase C site on its DNA binding domain. In our study, however, NGF induced up-regulation Notch1 signaling and cell proliferation. The reason why our findings are not consistent with previous reports are as follows : 1) Notch1 signaling could be different depending on cell type and in each cell line, 2) decreased activity of kinases that phosphorylate Hes1, or increased activity of a phosphatase during NGF signaling in glioblastomas, and 3) the complexity of Notch1 function through other Notch family members or ligands. Which of these effects contributes to the response remains for future study. The reason for the inhibitory effect of Notch1 siRNA on cell proliferation in this study seems not so dramatic compared to the effect of hes1 siRNA may be as follows : 1) There may be more linkages between the Notch1 receptor and Hes1, 2) Second, inhibition of Hes1 may be more effective in blocking the effect of NGF in glioblastoma, and 3) The effect of hes1 inhibition in this cell may be more specific to glioblastoma cell proliferation.Some limitations could be considered in this study. 1) To identify for the relationship between NGF and Notch signaling, different kinds of glioblastoma cell lines are needed, 2) Although the results showed the NGF effect on the Notch signaling in glioblastoma, more molecular works looks necessary including r-secretase inhibition, Notch1 intracellular domain, other Hes families, and DNA binding proteins, 3) IHC staining showed clear positivity, but used the number of samples was too small, and 4) We did not study the downstream pathway of the NGF receptor including tyrosine kinases. It will be done in the next study.
CONCLUSIONThis study suggests that NGF stimulates glioblastoma cell proliferation in a dose-dependent manner through the Notch1 receptor, and that NGF also induces up-regulation of Notch1 signaling. These findings suggest that targeting NGF or Notch1 could be a therapeutic option for glioblastoma patients.
Fig. 1.Normal brain tissue (A) and negative control (B) show no staining, but human glioblastoma tissue (C) shows brown colored changes in the nucleus and cytoplasm by the streptavidin-biotin-peroxidase complex technique. Scale bar=200 μm.
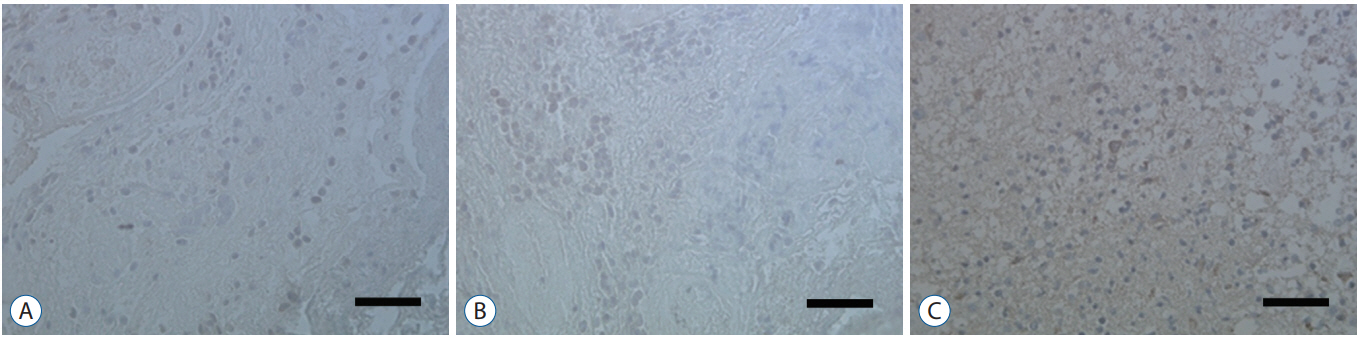 Fig. 2.
Fig. 2.
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay shows that nerve growth factor (NGF) stimulates U87-MG cell proliferation in a dose-dependent manner (p=0.005).
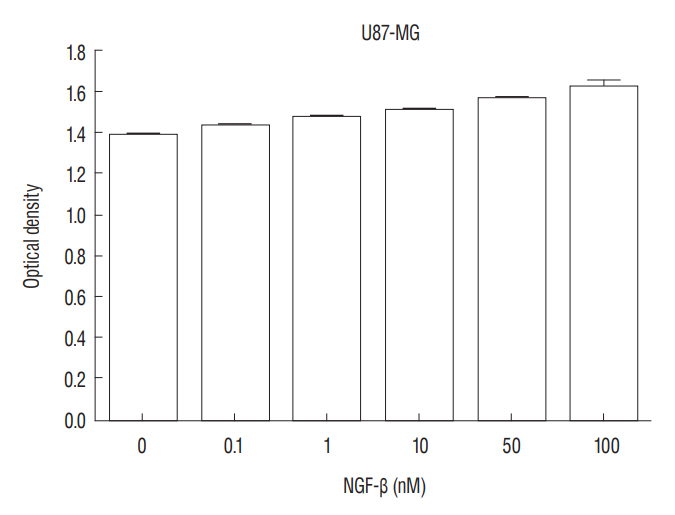 Fig. 3.
Fig. 3.
Expression of Notch1 in the reverse transcription polymerase chain reaction (RT-PCR) (A) and a dose-dependent effect of nerve growth factor (NGF) on Notch1 expression in the U87-MG cell line (p=0.044) (B); expression of Notch1 in the Western blot analysis (C) and a dose-dependent effect of nerve growth factor (NGF) on Notch1 expression in the U87-MG cell line (p=0.037) (D). GAPDH : glyceraldehyde-3-phosphate dehydrogenase.
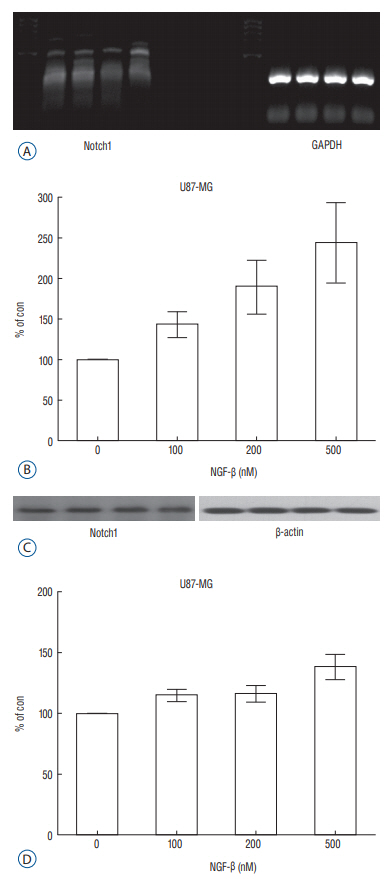 Fig. 4.
Fig. 4.
Expression of Hes1 in the reverse transcription polymerase chain reaction (RT-PCR) (A) and a dose-dependent effect of nerve growth factor (NGF) on Hes1 expression in the U87-MG cell line (p=0.020) (B); Expression of Hes1 in the Western blot analysis (C) and a dose-dependent effect of NGF on Hes1 expression in the U87-MG cell line (p=0.037) (D). GAPDH : glyceraldehyde3-phosphate dehydrogenase.
 Fig. 5.
Fig. 5.
Expression of Notch1 mall interfering RNA (siRNA) in the Western blot analysis (A). Notch1 siRNA suppresses the effects of nerve growth factor (NGF) on U87-MG cell proliferation. There was difference in without lipofectamin, in lipofectamin only, and in negative control siRNA (p<0.05), but no significant difference in Notch1 siRNA (p=0.887) (B). Expression of Hes1 siRNA in the Western blot analysis (C). Hes1 siRNA suppresses the effects of NGF on U87-MG cell proliferation. There was difference in without lipofectamin, in lipofectamin only, and in negative control siRNA (p<0.05), but no significant difference in Hes1 siRNA (p=0.075) (D).
 References
1. Artavanis-Tsakonas S, Rand MD, Lake RJ : Notch signaling: cell fate control and signal integration in development. Science 284 : 770-776, 1999
References
1. Artavanis-Tsakonas S, Rand MD, Lake RJ : Notch signaling: cell fate control and signal integration in development. Science 284 : 770-776, 1999




留言 (0)