The type VI secretion system (T6SS) is an apparatus composed of 13 structural proteins and several accessory proteins that deliver protein effectors into target cells by means of a contractile mechanism (Coulthurst, 2019; Cherrak et al., 2019). The T6SS needle is composed of an inner tube made of a stack of Hcp hexamer rings that is tipped by a trimer of VgrG and a proline-alanine–alanine-arginine repeat (PAAR) protein. This internal structure is surrounded by a contractile sheath of polymerized TssB/TssC subunits assembled in an extended, metastable conformation (Silverman et al., 2013; Cherrak et al., 2019). Contraction of the sheath propels the needle complex toward the target cell (Brackmann et al., 2017). T6SS effector proteins are classified as either cargo or specialized effectors. Cargo effectors are transported by non-covalent interaction with some core components (Coulthurst, 2019), while specialized effectors are VgrG, Hcp or PAAR proteins carrying additional domains (Durand et al., 2014; Whitney et al., 2014; Diniz and Coulthurst, 2015; Ma et al., 2017; Pissaridou et al., 2018).
T6SS effector proteins can target prokaryotic and/or eukaryotic cells (Coulthurst, 2019; Monjarás Feria and Valvano, 2020). Among the anti-bacterial effector proteins, some target the peptidic or glycosidic bonds of the peptidoglycan (Ma and Mekalanos, 2010; Russell et al., 2012; Srikannathasan et al., 2013; Whitney et al., 2013; Berni et al., 2019; Wood et al., 2019), or the FtsZ cell division ring (Ting et al., 2018). These anti-bacterial effectors are usually encoded in bi-cistronic elements with their cognate immunity proteins (E/I pairs) in order to avoid self-intoxication and killing of sibling cells (Russell et al., 2012). Other T6SS effectors target eukaryotic cells, such as those disrupting the actin or microtubule cytoskeleton networks (Monjarás Feria and Valvano, 2020), while trans-kingdom effectors target both bacterial and eukaryotic cells (Jiang et al., 2014). These effectors include those forming pores in membranes or targeting conserved molecules such as NAD+ and NADP+, and macromolecules such as DNA, RNA and phospholipids (Whitney et al., 2015; Tang et al., 2018; Ahmad et al., 2019). In many enteric pathogens (e.g., Salmonella, Shigella and Vibrio), the T6SS contributes to colonization of the intestinal tract of infected hosts (Sana et al., 2016; Chassaing and Cascales, 2018). On the other hand, strains of the gut commensal Bacteroides fragilis use their T6SSs for competition against other Bacteroidales species (Coyne and Comstock, 2019). Hence, the T6SS is a key player in bacterial warfare.
The Salmonella genus includes more than 2,600 serotypes distributed between species S. enterica and S. bongori (Issenhuth-Jeanjean et al., 2014), which differ in clinical signs and host range (Uzzau et al., 2000). In Salmonella, five T6SS gene clusters have been identified within Salmonella Pathogenicity Islands (SPIs) SPI-6, SPI-19, SPI-20, SPI-21, and SPI-22 (Blondel et al., 2009; Fookes et al., 2011; Bao et al., 2019). These T6SS gene clusters are distributed in 4 different evolutionary lineages: The SPI-6 T6SS gene cluster belongs to subtype i3, SPI-19 T6SS gene cluster to subtype i1, SPI-22 T6SS gene cluster to subtype i4a, and both SPI-20 and SPI-21 T6SS gene clusters to subtype i2 (Bao et al., 2019). Besides their distinct evolutionary origin, these five T6SS gene clusters are differentially distributed among distinct serotypes, subspecies, and species of Salmonella (Blondel et al., 2009; Bao et al., 2019).
In Salmonella, only a few studies have addressed the role played by the T6SSs in interbacterial and eukaryotic relationships, and most of our understanding regarding the contribution of T6SSs to Salmonella infection cycle, virulence and pathogenesis comes from studies of T6SSSPI-6 in S. Typhimurium and T6SSSPI-19 in S. Dublin (Mulder et al., 2012; Pezoa et al., 2013; Pezoa et al., 2014; Sana et al., 2016; Sibinelli-Sousa et al., 2022; Xian et al., 2020; Blondel et al., 2010; Hespanhol et al., 2022). Furthermore, knowledge of the presence and distribution of T6SS effector proteins is derived from studies using strains representing a limited number of serotypes (Russell et al., 2012; Benz et al., 2013; Sana et al., 2016; Whitney et al., 2013; Sibinelli-Sousa et al., 2020; Lorente-Cobo et al., 2022; Koskiniemi et al., 2014; Amaya et al., 2022; Jurėnas et al., 2022; Blondel et al., 2023). Consequently, information regarding Salmonella T6SS effector proteins is still scarce. Indeed, only 37 T6SS effectors and candidate effectors that target different bacterial molecules such as peptidoglycan, nucleic acids and bacterial ribosomes have been currently identified in a few serotypes (Blondel et al., 2009; Russell et al., 2012; Benz et al., 2013; Whitney et al., 2013; Koskiniemi et al., 2014; Sana et al., 2016; Ho et al., 2017; Sibinelli-Sousa et al., 2020; Amaya et al., 2022; Jurėnas et al., 2022; Lorente-Cobo et al., 2022; Hespanhol et al., 2022; Blondel et al., 2023). This is an important knowledge gap as the T6SS effector proteins are the ultimate mediators of the T6SS activity and thus, their identification and characterization are pivotal for a better understanding of Salmonella infectious cycle and in its contribution to environmental fitness and pathogenic potential.
Nowadays, there is increasing evidence that Salmonella enterica can persist in diverse environments such as aquatic ecosystems, maintaining a reservoir in surface waters and becoming a serious risk to public health and animal production systems. It is conceivable that the T6SS could mediate in part this persistence since it has been shown that S. Typhimurium requires the T6SSSPI-6 to survive intracellularly in environmental amoebas such as Dictyostelium discoideum (Riquelme et al., 2016). Interestingly, in Chile some serotypes such as S. Infantis, S. Newport and S. Typhimurium have been frequently isolated in surface waters during the last decade, imposing a significant threat to human and animal health since these serotypes usually carry an arsenal of antimicrobial resistance genes (Chen et al., 2024a,b). These Chilean isolates could be an untapped reservoir of new T6SS effector proteins. Importantly, Salmonella strains isolated from surface waters in Chile will shed light not only on the vast arsenal of T6SS effector repertoire but could also provide insight into geographic adaptation of Salmonella.
In this study, we performed bioinformatic and comparative genomic analyses of a dataset of 695 S. enterica genomes representing 44 serotypes isolated from different environmental sources in Chile, mostly surface waters. Our analysis revealed that most genomes only harbor the SPI-6 T6SS gene cluster, and that within its variable region 3 (VR3) we found four new candidate T6SS effectors with predicted nuclease activity. Noteworthy, many putative SPI-6 rearrangement hotspot (Rhs) effectors identified in this study harbor C-terminal extensions with unknown function. Overall, the diversity and distribution of T6SS effector proteins in Chilean Salmonella isolates suggest that different combinations of these proteins may contribute to the environmental fitness and pathogenic potential.
Materials and methods Environmental samples and Salmonella isolationWater samples were collected as part of a previous study (Toro et al., 2022) from sites in the Maipo, Mapocho, Claro and Lontué watersheds from the rivers themselves and connected tributaries, such as canals. Animal samples were collected as part of a previous study (Rivera et al., 2021) from industrial dairy farms, backyard systems and wild animals in the Región de Coquimbo, Región de Valparaíso, Región Metropolitana and Región del Libertador General Bernardo O’Higgins, Chile. A detailed description of sampling procedures, geographical location of samples and the procedure employed for Salmonella isolation from water an animal samples can be found elsewhere (Rivera et al., 2021; Toro et al., 2022).
Whole genome sequencing, assembly, and quality controlFor sequencing, each isolate was grown overnight at 37°C in tryptic soy broth and 1 mL of culture was used to purify DNA with the DNeasy Blood and Tissue Qiagen kit (Qiagen, CA, United States). Ratios of absorbance at 260 nm and 230 nm were obtained using a MaestroNano spectrophotometer (Maestro, Korea) and a QUBIT fluorimeter (Life Technologies, CA, United States). Libraries were prepared with the Illumina DNA Prep kit (Illumina, CA, United States) on the Sciclone G3 NGSx iQ Workstation (Perkin Elmer, MA, United States), and sequencing was performed on the Illumina NextSeq 2000 using the NextSeq 1000/2000 P2 reagents 300 cycles with the 150 paired-end chemistry (Illumina, CA, United States). Reads were examined for quality using FastQC (Galaxy version 0.69) (Wingett and Andrews, 2018) and trimmed using Trimmomatic (Galaxy version 0.36.4), with a minimum required quality of 20, averaging across 4 bases (Bolger et al., 2014). Processed reads were assembled using SPAdes (Galaxy version 3.11.1) with kmer sizes of 99 and 127, and careful correction (Bankevich et al., 2012). Assemblies were checked for quality using QUAST (Galaxy version 4.6.3) (Gurevich et al., 2013) and finally deposited in the NCBI Bioproject 560,080.
In silico serotyping was carried out using SeqSero (Galaxy version 2.0.1) (Zhang et al., 2015) and SISTR (Galaxy version 1.0.2) (Yoshida et al., 2016). Finally, a single-nucleotide polymorphism (SNP) analysis was performed to identify clonality among isolates from the same sample. Clones were defined as isolates with genomes having 20 or fewer SNPs, as described by Pightling et al. (2018). According to this criterion, genome sequences from non-clonal isolates obtained from the same sample were selected for subsequent analysis. Thus, the genome sequence dataset analyzed in this study includes 695 S. enterica genomes from 44 distinct serotypes (Supplementary Table S1).
Identification of T6SS gene clustersThe T6SS prediction tool from the Secret6 web server was used to identify T6SS gene clusters encoding the minimal 13 core components of a T6SS in each genome (Zhang et al., 2023). For selection of positive matches, a BLASTp 2.10.1+ identity threshold for T6SS prediction >30% and an E-value <0.0001 were used. These threshold values have been successfully used to identify T6SS gene clusters in Salmonella genomes (Amaya et al., 2022; Blondel et al., 2023).
Identification of candidate T6SS effectorsTo identify putative T6SS effectors encoded within the Salmonella genomes analyzed, each ORF encoded within the T6SS gene clusters identified was analyzed with the Bastion6 pipeline (Wang et al., 2018) excluding the 13 T6SS core components. ORFs presenting a Bastion6 score ≥ 0.7 were considered as candidate T6SS effectors. It is worth mentioning that a Bastion6 score ≥ 0.5 is routinely used as default setting for detection of T6SS effectors. However, we decided to use a score ≥ 0.7 to perform a more strict analysis. Each Bastion6 prediction was further analyzed using tools implemented in the Operon-Mapper web server (Taboada et al., 2018) to determine whether it was part of a single transcriptional unit that also encoded a putative immunity protein [i.e., a small protein with potential signal peptides (SignalP 6.0) and/or transmembrane domains (TMHMM 2.0)]. Conserved functional domains and motifs in the candidate T6SS effectors were identified using the PROSITE, NCBI-CDD, Motif-finder, and Pfam databases (Kanehisa et al., 2002; Sigrist et al., 2013; Finn et al., 2014; Lu et al., 2019) implemented in the GenomeNet search engine. An E-value cutoff score of 0.01 was used. In addition, for each putative effector and immunity protein identified, a biochemical functional prediction was performed by HMM homology searches using the HHpred HMM-HMM comparison tool (Zimmermann et al., 2017). Finally, a candidate T6SS effector was defined as “new” when it meets two criteria: (i) it includes at least one domain previously linked to antibacterial activity, and (ii) this domain has not been described as part of a T6SS effector in publicly available databases.
Hierarchical clustering analysis of the new T6SS effectorsFor hierarchical clustering analysis, a presence/absence matrix of each T6SS effector and candidate effector was constructed for each bacterial genome by means of BLASTn analyses and manual curation of the data (Supplementary Table S2). A 90% identity and 90% sequence coverage threshold was used to select positive matches, as done in previous analyses conducted by our group (Amaya et al., 2022; Blondel et al., 2023). The matrix generated was uploaded as a csv file to the online server MORPHEUS using default parameters (i.e., one minus Pearson’s correlation and average linkage method).
Phylogenetic analyses of Salmonella T6SS gene clustersTssC aminoacid sequences encoded in T6SS gene clusters from 605 Salmonella genomes were concatenated and aligned with ClustalW using the Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0 (Kumar et al., 2016). A phylogenetic tree was built from the alignments obtained from MEGA by performing a bootstrap test of phylogeny (1,000 replications) using the maximum-likelihood method with a Jones-Taylor-Thornton correction model.
Analysis of T6SS effectors distributionThe DNA sequence encoding each T6SS effector identified in this study was subjected to tBLASTx analyses to find orthologs in all Salmonella genome sequences deposited in the NCBI database (March, 2024) (Supplementary Tables S3, S4). For selection of positive matches, a 90% identity and 90% sequence coverage threshold was used. Conservation of sequences was determined by independent multiple sequence alignments using T-Coffee Expresso (Notredame et al., 2000), MAFFT (Katoh et al., 2017), and ESPript 3 (Robert and Gouet, 2014). Comparative genomic analyses of T6SS gene clusters were performed using Mauve version 2.3.1 (Darling et al., 2004) and EasyFig version 2.2.5 (Sullivan et al., 2011). Nucleotide sequences were analyzed using Artemis version 18 (Rutherford et al., 2000).
Results T6SS gene clusters are widely distributed among Chilean Salmonella isolatesPrevious analyses performed by our group have aimed in the identification of candidate T6SS effectors and cognate immunity proteins in Salmonella genomes deposited in public databases (Amaya et al., 2022; Blondel et al., 2023). In the present study, the analysis focused on genome sequences of Salmonella isolates obtained from different environmental sources in Chile, in order to shed light on the repertoire of T6SS candidate effectors present in Salmonella inhabiting our local geography. To this end, we analyzed a database of 695 high-quality sequenced Salmonella genomes from strains isolated from surface water and animal sources. Most isolates in this collection come from surface waters (674 isolates representing 34 serotypes), while 21 isolates representing only 8 serotypes were obtained from animal sources (14 in chicken, 3 in pigeon, 2 in pig and 2 in duck). Interestingly, the most frequently isolated serotypes were S. Infantis (n = 169), S. Agona (n = 71) and S. Newport (n = 11).
To identify T6SS gene clusters we used the T6SS prediction tool from the SecreT6 web server (see text footnote 2), which identified 622 putative T6SS gene clusters in 608 Salmonella genomes (Table 1; Supplementary Table S1). A more in-depth analysis revealed that these T6SS gene clusters correspond to those encoded in SPI-6, SPI-19 and SPI-21 (Table 1; Supplementary Figure S1). We could not identify T6SS gene clusters encoded in SPI-20 or SPI-22 in the genome of any isolate from our database. The SPI-6 T6SS gene cluster is widely distributed in 518 of the 695 genomes analyzed (74.5%), while the SPI-19 and SPI-21 T6SS gene clusters were only detected in 89 (12.8%) and 14 (2%) genomes, respectively (Table 1). Most isolates carried a unique T6SS gene cluster in SPI-6, SPI-19 or SPI-21, while a group of isolates belonging to serotype S. Livingstone harbors both SPI-6 and SPI-19 T6SS gene clusters. In contrast, no complete T6SS gene cluster was detected in isolates belonging to serotypes S. Enteritidis and S. Stanley.
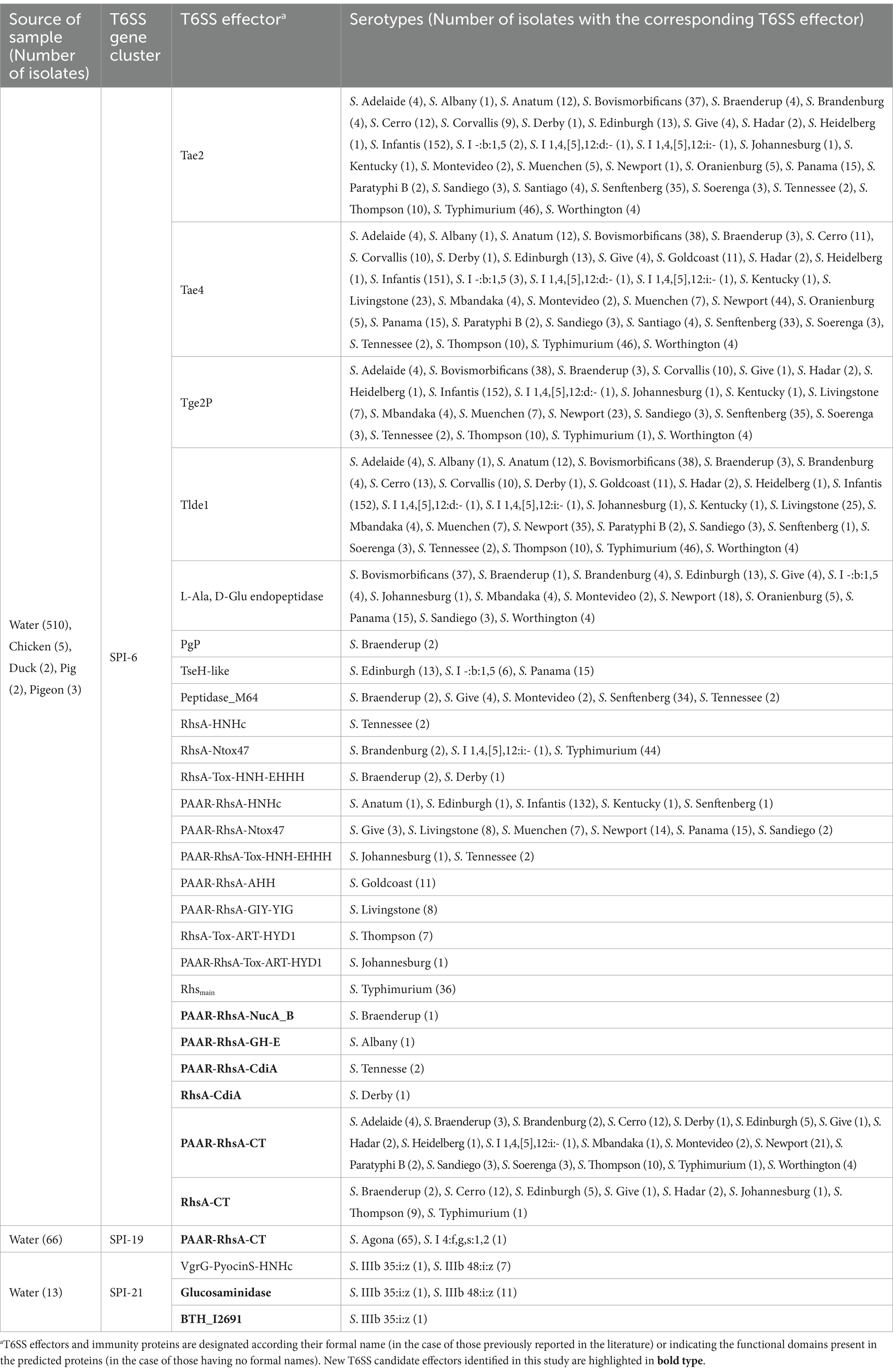
Table 1. T6SS effectors and cognate immunity proteins encoded in T6SS gene clusters in Chilean Salmonella isolates.
To identify high-confidence putative effectors encoded within every T6SS gene cluster detected, each ORF within these gene clusters was analyzed based on four criteria: (i) identification of candidate effectors through Bastion6 analysis (a bioinformatic tool that predicts T6SS effectors based on amino acid sequence, evolutionary information, and physicochemical properties); (ii) identification of putative immunity proteins by operon prediction (Operon-mapper; Taboada et al., 2018) and detection of signal peptides (SignalP 6.0) and transmembrane domains (TMHMM 2.0); (iii) identification of conserved functional domains associated with bona fide T6SS effectors (INTERPROSCAN, PROSITE, NCBI-CDD, MOTIF, and Pfam); and (iv) functional biochemical prediction using the HHpred HMM-HMM server. In addition, we further analyzed these T6SS gene clusters to identify potential unannotated ORFs that could encode putative effectors and cognate immunity proteins. Thus, our analysis revealed the presence of 6 new effector candidates encoded within the SPI-6 (4 effectors) and SPI-21 (2 effectors) T6SS gene clusters.
The VR3 within the SPI-6 T6SS gene cluster of isolates from surface waters harbor four candidate T6SS effector proteinsMost T6SS effector proteins identified in Salmonella are encoded within three variable regions (VR1-3) of SPI-6 (Blondel et al., 2023). We have previously shown that the VR3 of SPI-6, located downstream of the tssI gene, exhibits the greatest diversity of Salmonella T6SS effectors (Blondel et al., 2023). This is mainly due to the presence of a variable number of Rhs effector proteins that harbor C-terminal extensions encoding endonuclease domains, such as DNases, RNases, and deaminases, as well as ADP-ribosyltransferases (Blondel et al., 2023).
Our analysis identified 4 new putative effector proteins and cognate immunity proteins (Table 2; Figure 1) encoded in the VR3 of SPI-6 distributed in isolates of serotypes S. Braenderup, S. Albany, S. Tennessee and S. Derby. Three of these candidates are specialized Rhs effector proteins with predicted nuclease activity, including 2 DNases and 1 RNase, while only one is a cargo Rhs effector with putative RNase activity (Table 2). The first putative effector (FA1083_3621 in S. Braenderup FA1083) is a large 1,498 amino acid Rhs protein that harbors an N-terminal PAAR domain and a C-terminal Nuclease A/Nuclease B (NucA_B) domain with predicted DNase activity (Table 2; Figure 1). It should be noted that FA1083_3621 is predicted to be encoded in a bi-cistronic unit with FA1083_3620 (Table 2). This latter ORF encodes a 204 amino acid protein with a DUF6707 domain that may correspond to the cognate immunity protein of FA1083_3621. The second candidate effector (FA1443_1959 in S. Albany FA1443) with predicted DNase activity also corresponds to a 1,566 amino acid Rhs protein that harbors an N-terminal PAAR domain and the putative GH-E domain in its C-terminal end (Table 2; Figure 1). The GH-E domain is found in members of the HNH/ENDO VII superfamily nuclease with conserved glycine, histidine and glutamate residues. This putative effector was also predicted to be co-transcribed with its respective putative immunity protein gene that encodes a tetratricopeptide repeat (TPR)-containing protein (FA1443_1960 in S. Albany FA1443). The third candidate effector (FA1455_4074 in S. Tennessee FA1455) is a 1,560 amino acid Rhs protein with a predicted N-terminal PAAR domain and a C-terminal contact-dependent growth inhibition protein A (CdiA) domain with putative RNase activity (Table 2; Figure 1). The gene encoding this candidate effector is predicted to be part of a bi-cistronic unit with FA1455_4073, encoding its putative immunity protein (Table 2; Figure 1). Of note, FA1455_4073 harbors a multiple adhesin family I (MafI) domain that is frequently found in cognate immunity proteins of bacterial toxin systems (Zhang et al., 2012). The fourth new candidate effector identified in this study is a 372 amino acid Rhs protein with a predicted CdiA domain in its C-terminal end (FA1451_3438 in S. Derby FA1451) (Table 2; Figure 1). FA1451_3438 is predicted to be co-transcribed with FA1451_3439, encoding its cognate immunity protein (Table 2; Figure 1). FA1451_3439 harbors an anti-repressor A (AntA) domain usually found in phage anti-repressor proteins (Sandt et al., 2002). It is worth mentioning that the CdiA domain found in candidate effectors FA1455_4074 and FA1451_3438 has not been previously associated with any Rhs effector protein in Salmonella.
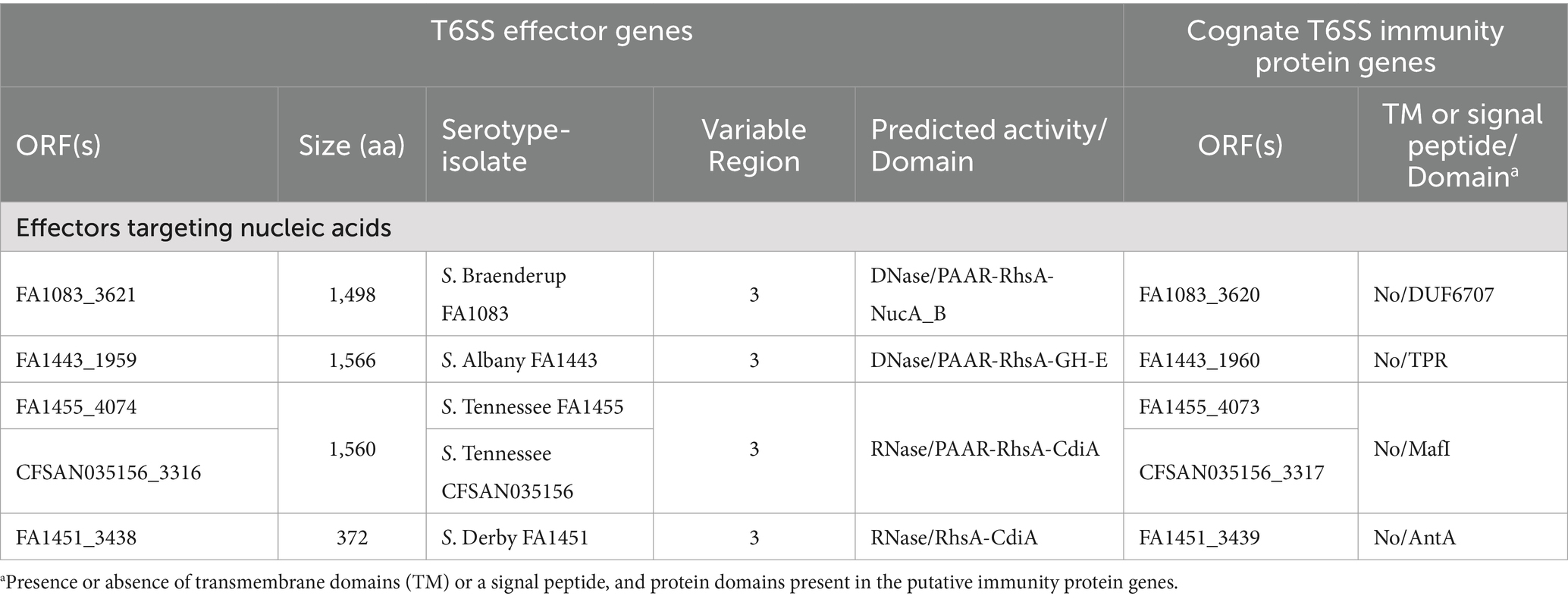
Table 2. New putative T6SS effectors and cognate immunity proteins encoded in the SPI-6 T6SS gene cluster of Chilean Salmonella isolates.
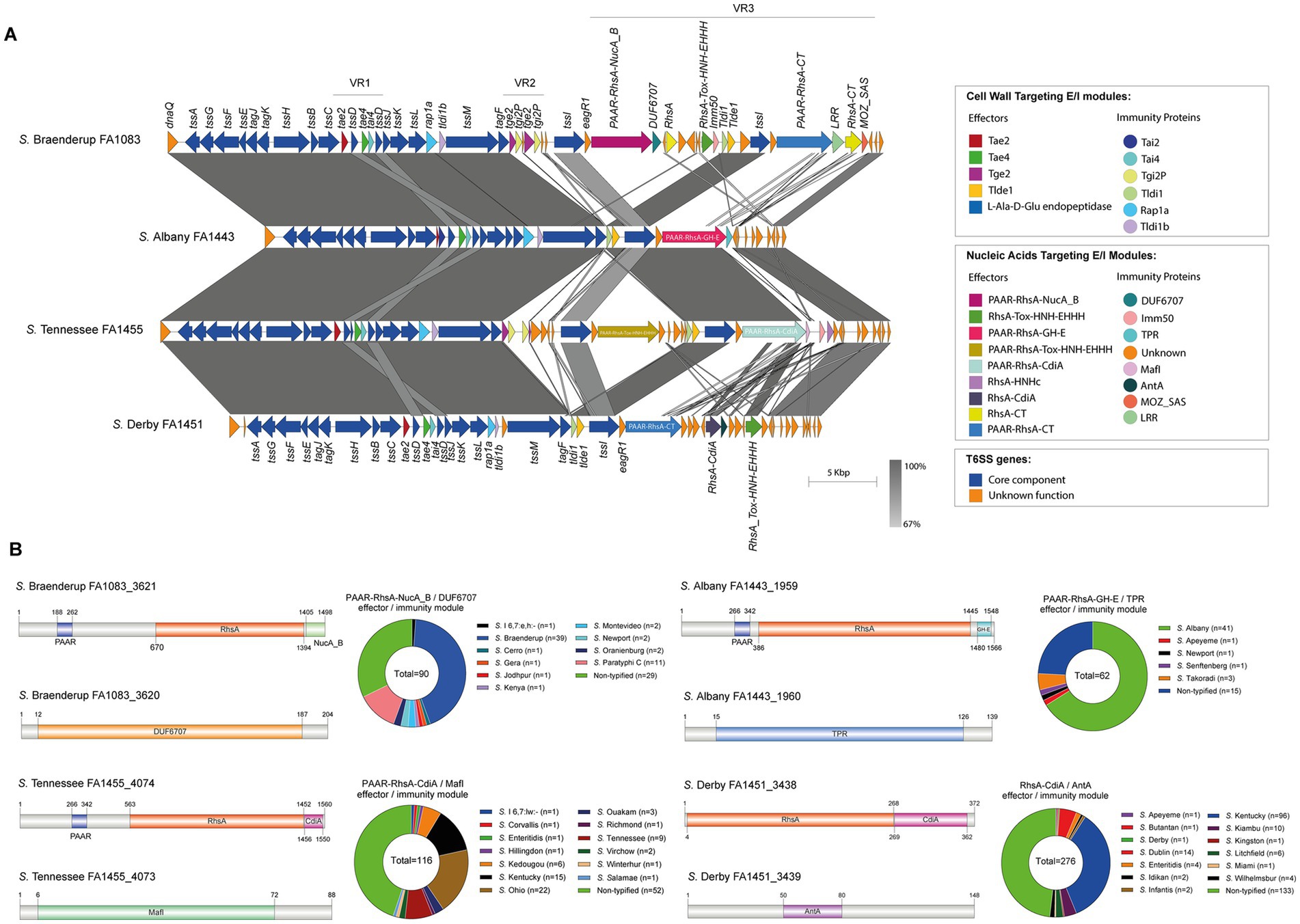
Figure 1. The SPI-6 T6SS gene cluster encodes new putative T6SS effector proteins. (A) Comparative genomic analysis of the SPI-6 T6SS cluster of S. Braenderup FA1083, S. Albany FA1443, S. Tennessee FA1455 and S. Derby FA1451. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). (B) Schematic representation and distribution among Salmonella genomes of each new effector and immunity protein identified. ORFs encoding new E/I modules are highlighted in different colors according to the predicted functions. Homologs for each component were identified by BLASTn analyses as described in Materials and Methods.
The genetic structure and repertoire of effector proteins encoded in the SPI-6 T6SS gene cluster vary considerably among Salmonella isolates of the same serotypeIt has been reported that the genetic structure of the T6SS gene clusters and the repertoire of effector proteins varies between different serotypes of Salmonella (Amaya et al., 2022; Blondel et al., 2023). Therefore we analyzed the genetic structure of SPI-6 and the distribution of previously identified effector proteins (Table 1; Supplementary Table S2). We identified 19 out of the 32 previously reported effectors encoded in the SPI-6 T6SS gene cluster. The three most frequently distributed T6SS effectors are encoded in VR1-2 of SPI-6. These effector proteins were Tae4 (34/36), Tae2 (32/36) and Tlde1 (29/36). In VR3, the region showing the greatest diversity of Salmonella T6SS effectors, the most prevalent effector proteins were PAAR-RhsA-Ntox47 (6/36) and PAAR-RhsA-HNHc (5/36).
Next, we performed a hierarchical clustering analysis to shed lights into the distribution of effectors and candidate effectors encoded in the SPI-6 T6SS gene cluster identified (Supplementary Table S1). As illustrated in Figure 2, the four bona fide effectors encoded within VR1-2 (Tae2, Tae4, Tge2 and Tlde1) were the most conserved across the genomes of isolates representing 29 to 34 Salmonella serotypes. However, some of these effectors are missing from the genomes of all isolates from a few Salmonella serotypes. In VR3, the most prevalent effector protein was PAAR-RhsA-Ntox47, while PAAR-RhsA-AHH, PAAR-RhsA-GIY-YIG, PAAR-RhsA-Tox-ART-HYD1, RhsA-Tox-ART-HYD1 and RhsA-HNHc were the least prevalent. It is worth mentioning that a greater diversity of VR3-encoded effectors is observed in those serotypes that lack some of the more conserved VR1-2-encoded effectors (Figure 2).
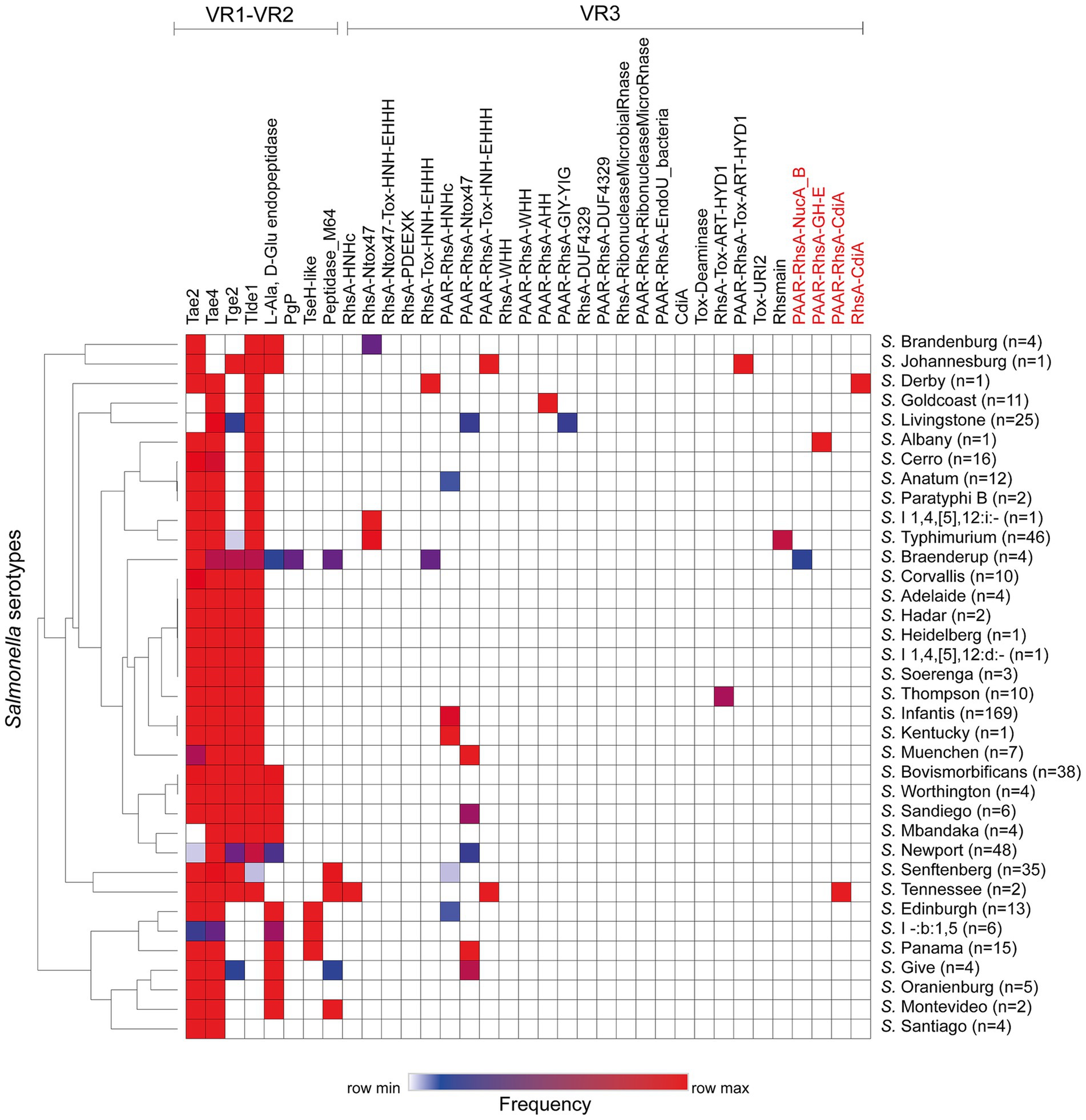
Figure 2. Prevalence of ORFs encoding T6SS effectors and candidate effectors in the SPI-6 T6SS gene cluster of Chilean Salmonella isolates. A hierarchical clustering analysis was conducted using MORPHEUS, as detailed in the Materials and Methods section. The color code in the heatmap indicates the frequency of a given ORF among all isolates of a particular Salmonella serotype. The names of new T6SS candidate effectors identified in this study are highlighted in red.
Analysis of genetic structure variation of the SPI-6 T6SS gene cluster between serotypes and between isolates of the same serotype revealed interesting observations. First, we identified a variable number of tssI-eagR-rhs gene modules encoded in VR3. A number of isolates from serotypes S. Braenderup, S. Kentucky, S. Sandiego and S. Tennessee harbor two tssI-eagR-rhs modules (Figure 3), while most isolates from serotypes carrying the SPI-6 T6SS gene cluster only harbor one tssI-eagR-rhs module (Figure 4). Remarkably, in S. Braenderup the genetic structure of SPI-6 differs between isolates CFSAN43223, FA0982 and FA1083. CFSAN43223 has only one tssI-eagR-rhs module, while FA0982 and FA1083 have two of these modules, as previously reported in S. Tennessee isolate CFSAN070645 (Blondel et al., 2023) (Figure 3; Supplementary Figure S2). Isolates FA0982 and FA1083 encode the RhsA-Tox-HNH-EHHH effector, as well as two other effectors harboring C-terminal ends with unknown function (PAAR-RhsA-CT and RhsA-CT). Additionally, isolate FA1083 encodes a new PAAR-RhsA-NucA_B effector with putative DNase activity, as described above (Figures 1, 3). It is important to note that isolate CFSAN43223 has an internal deletion within VR2 in comparison to isolates FA0982 and FA1083, and encodes only the Tlde1 effector. In contrast, isolates FA0982 and FA1083 encode two copies of the Tge2 effector in VR2 (Supplementary Figure S2). In S. Kentucky, our analysis of the single isolate present in the database (CFSAN035145) identified two tssI-eagR-rhs modules in VR3. These modules encode the PAAR-RhsA and PAAR-RhsA-HNHc effector proteins, respectively (Figure 3). Notably, the first tssI-eagR-rhs module has a high sequence identity with only one gene module previously reported in S. Tennessee CFSAN070645 (Blondel et al., 2023). Similarly, the second tssI-eagR-rhs module of S. Kentucky CFSAN035145 shows high sequence identity with the corresponding module encoded in VR3 of S. Typhimurium 14028s. Furthermore, S. Kentucky CFSAN035145 harbors an ORF with a predicted DUF4056 domain encoded in a bi-cistronic unit in VR2 never reported in Salmonella, which may constitute a new T6SS candidate effector (Figure 3). In S. Sandiego, the genetic structure of the SPI-6 T6SS gene cluster is conserved between isolates FA0894 and CFSAN105324, that harbor two tssI-eagR-rhs gene modules encoding a PAAR-RhsA-CT (C-terminal end with unknown function) and the PAAR-RhsA-Ntox47 effector proteins, respectively (Figure 3). A genomic comparative analysis of this latter effector with the corresponding T6SS effector in S. Typhimurium 14028s suggest that in isolates of serotype S. Sandiego the Rhsmain and RhsA-Ntox47 were at some point a single ORF that was later split due to the accumulation of nonsense mutations (Figure 3). Similar to S. Kentucky, the two tssI-eagR-rhs gene modules encoded in SPI-6 of S. Sandiego share high sequence identity with the corresponding gene modules encoded in S. Tennessee CFSAN070645 and S. Typhimurium 14028s, respectively (Figure 3). It is worth mentioning that Chilean S. Sandiego isolates harbor the Tae2 and Tae4 effector proteins encoded in VR1, as well as Tge2 and Tlde1 effectors encoded in VR2. Finally, in S. Tennessee, the genomic organization of the T6SS gene cluster encoded in SPI-6 is highly conserved not only among Chilean isolates but also among previously reported S. Tennessee isolates (Blondel et al., 2023) (Figure 3). Isolates of this serotype harbor two tssI-eagR-rhs gene modules encoding a PAAR-RhsA-Tox-HNH-EHHH and a PAAR-RhsA-CdiA T6SS effector proteins, respectively. Interestingly, unlike the other serotypes described above, these two tssI-eagR-rhs gene modules do not share any sequence identity with the corresponding module in S. Typhimurium 14028s. Altogether, these results suggest a distinct evolutionary origin of tssI-eagR-rhs gene modules within the SPI-6 T6SS gene cluster.
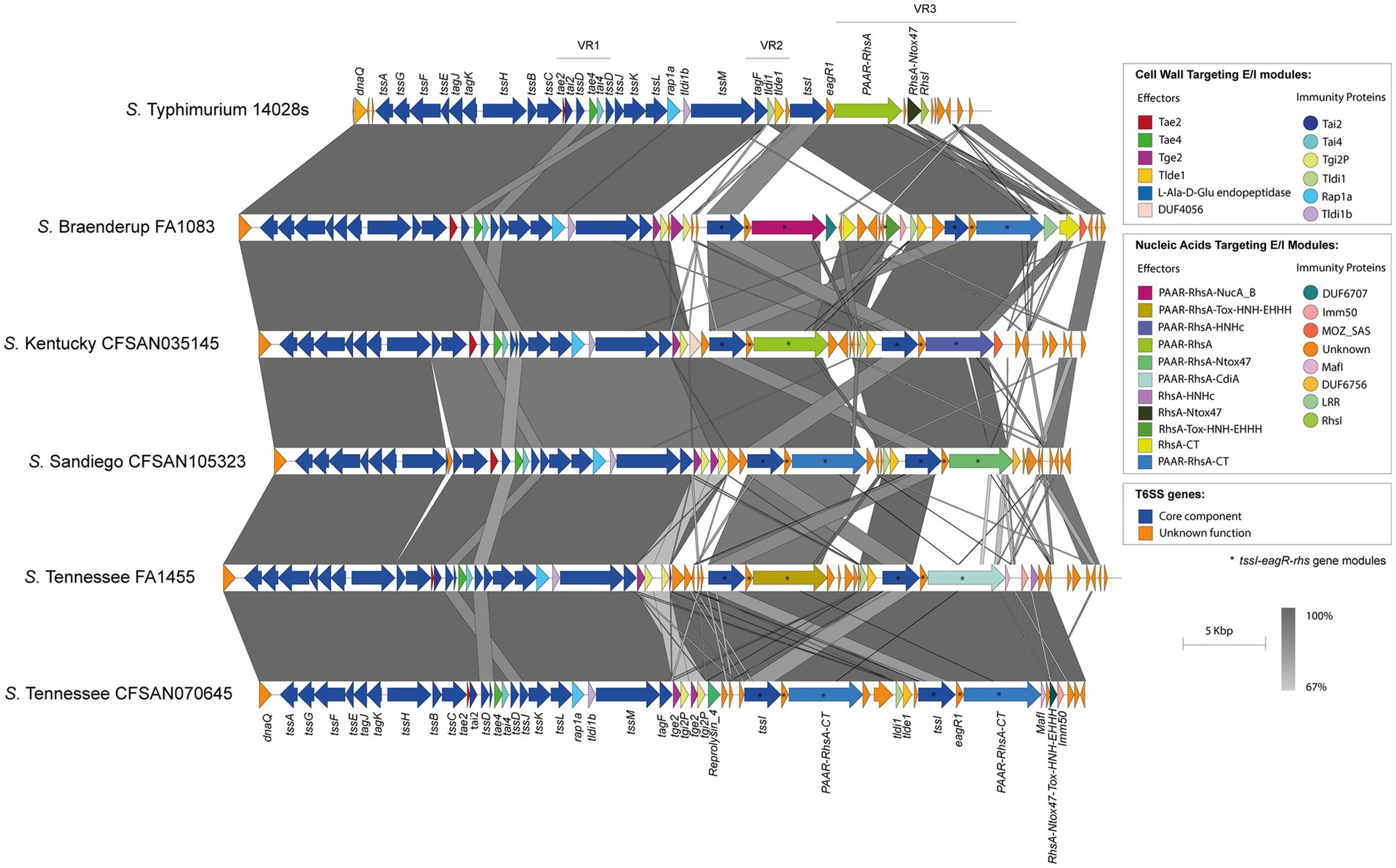
Figure 3. The SPI-6 T6SS gene cluster in a number of Chilean Salmonella isolates includes two tssI-eagR-rhs gene modules in VR3. Comparative genomic analysis of the SPI-6 T6SS cluster of S. Braenderup FA1083, S. Kentucky CFSAN035145, S. Sandiego CFSAN105323 and S. Tennessee FA1455 and CFSAN070645. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). ORFs encoding E/I modules are highlighted in different colors according to the confirmed or predicted functions. The tssI-eagR-rhs gene modules of the SPI-6 T6SS gene cluster are demarked by asterisks. Grayscale represents the percentage of identity between nucleotide sequences. The SPI-6 T6SS gene cluster from S. Typhimurium 14028s was used for comparative purposes.
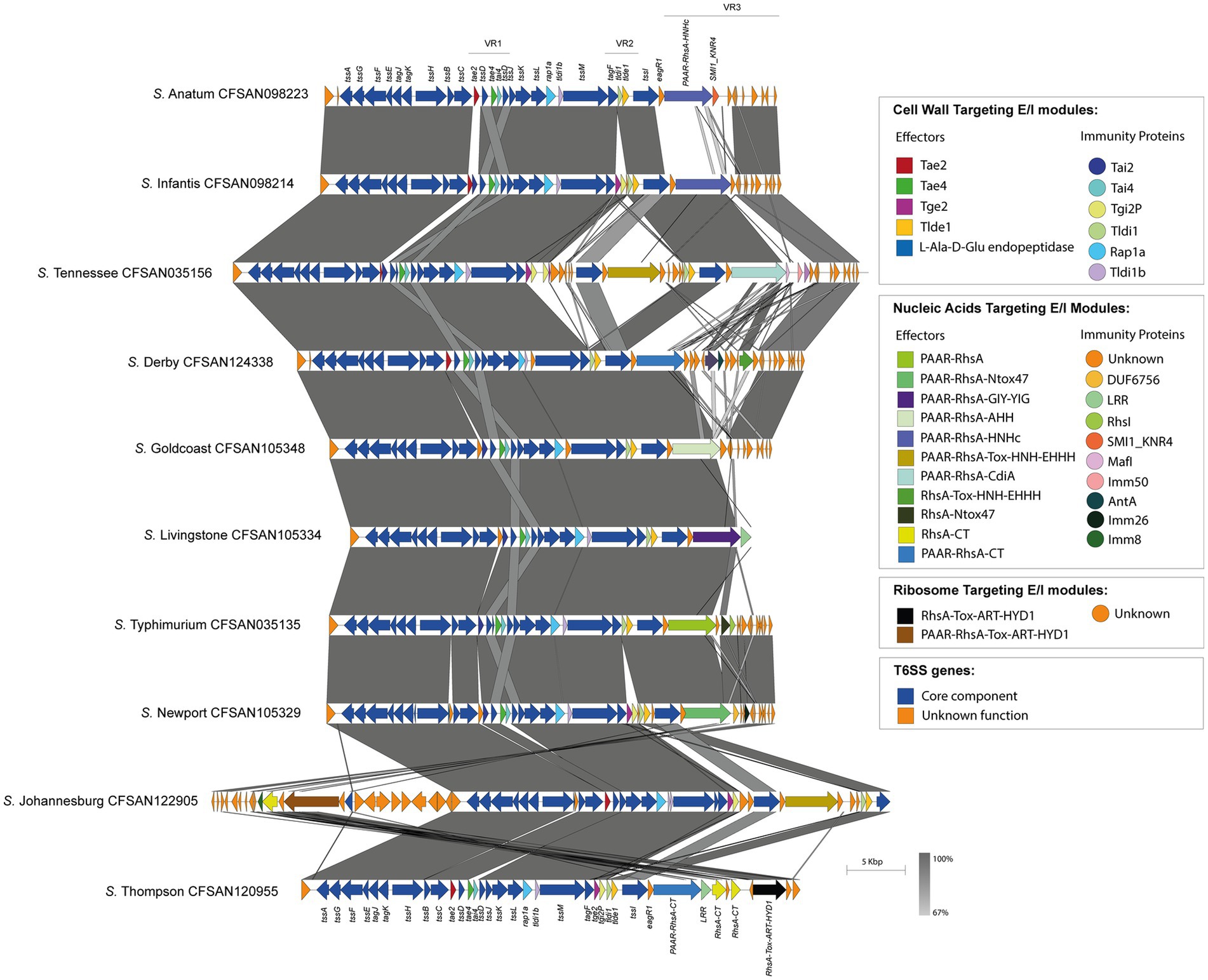
Figure 4. The T6SSSPI-6 effector repertoire varies among Chilean Salmonella isolates. Comparative genomic analysis of the SPI-6 T6SS cluster of selected Salmonella isolates representing different serotypes. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). ORFs encoding T6SS core components are shown in blue. ORFs encoding E/I modules are highlighted in different colors according to the confirmed or predicted functions. Grayscale represents the percentage of identity between nucleotide sequences.
On the other hand, the isolates belonging to the remaining 32 serotypes only contain one tssI-eagR-rhs gene module encoded in the SPI-6 T6SS gene cluster. In these isolates, the distribution of known and new candidate effectors varies considerably, even among representatives of the same serotype. This is the case of S. Livingstone, where two groups of isolates are distinguished. In the first group, the VR3 encodes the PAAR-RhsA-Ntox47 effector, while isolates in the second group harbor the PAAR-RhsA-GIY-YIG effector (Figure 5). In addition, the VR2 in the first group encodes the Tge2 and Tlde1 effector proteins, while in the second group only encodes Tlde1 (Figure 5; Supplementary Table S1). Remarkably, the first group only harbor the SPI-6 T6SS gene cluster while the second group also encodes the SPI-19 T6SS gene cluster. Furthermore, the genetic structure of the SPI-6 T6SS cluster in the first group differs more with the T6SS gene cluster of S. Typhimurium 14028s when compared to the second group (Figure 5).
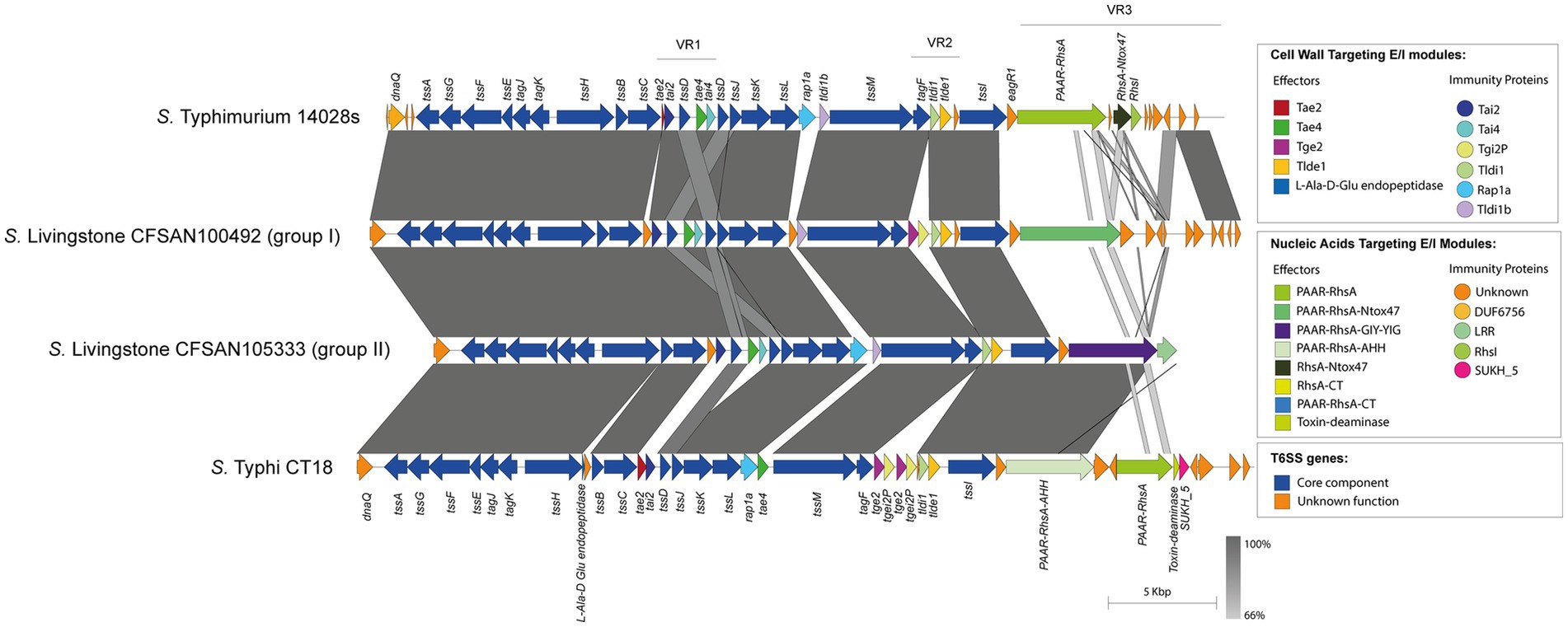
Figure 5. The genetic structure and repertoire of effector proteins encoded in the SPI-6 T6SS gene cluster vary among isolates of serotype S. Livingstone. Comparative genomic analysis of the SPI-6 T6SS cluster in S. Livingstone isolates. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). ORFs encoding E/I modules are highlighted in different colors according to the confirmed or predicted functions. SPI-6 T6SS gene clusters from S. Typhimurium 14028s and S. Typhi CT18 were used for comparative purposes.
In isolates of serotype S. Give, the SPI-6 T6SS gene cluster shows structural differences in VR2 and VR3. In VR2, the isolate CFSAN043231 encodes the Tge2 and Peptidase M64 effector proteins, while other isolates (CFSAN119452, CFSAN119453, and CFSAN119454) carry a bi-cistronic unit encoding proteins with unknown function (Supplementary Figure S3). The putative immunity protein encoding-gene of this bi-cistronic unit harbors a DUF4229 domain found in integral membrane proteins (Wang et al., 2023). Another intriguing structural difference exists in VR3, where isolates CFSAN119452, CFSAN119453, and CFSAN119454 encode a PAAR-RhsA-Ntox47 effector protein, while isolate CFSAN043231 encodes a PAAR-RhsA-CT and an RhsA-CT, both harboring C-terminal ends with unknown functions (Supplementary Figure S3). Notably, the putative immunity protein encoding-gene of the RhsA-CT candidate effector harbors the Imm9 domain, which is frequently found in cognate immunity proteins of bacterial toxin systems with RNase activity (Zhang et al., 2012). Thus, the presence of the Imm9 domain in the putative immunity protein-encoding gene suggests that the C-terminal end of the RhsA-CT candidate effector has RNase activity.
The genetic organization of the SPI-6 T6SS gene cluster in S. Newport varies between two groups of isolates. In the first group, the isolates encode the PAAR-RhsA-Ntox47 effector in VR3 and the Tge2 effector in VR2. Furthermore, in VR3, these isolates also contain an ORF with a predicted DUF6769 domain encoded in a bi-cistronic unit with an ORF harboring an Imm26 domain, which is typically found in cognate immunity proteins of bacterial toxin systems with RNase activity (Zhang et al., 2012). The presence of the Imm26 domain in this ORF suggests that the DUF6769-containing protein is a candidate effector with RNase activity. On the other hand, isolates in the second group encode the PAAR-RhsA-CT effector in VR3 and do not encode the Tge2 effector in VR2 (Supplementary Figure S4). Of note, there is no sequence identity between the Rhs elements of both groups of isolates, suggesting a different origin. In addition, the sequence of the C-terminal end of the PAAR-RhsA-CT effector encoded in these isolates shows high sequence similarity with the Rhs element of S. Typhi CT18 (Supplementary Figure S4).
Similar findings were also identified in S. Edinburgh, where two groups of isolates were distinguished. In VR3, isolates in the first group encode the PAAR-RhsA-HNHc effector protein, while isolates in the second group encode the PAAR-RhsA-CT and RhsA-CT effectors with C-terminal ends with unknown function (Supplementary Figure S5). Notably, S. Edinburgh is one of the three serotypes in which the TseH-like effector is predicted to be encoded in VR2 (Supplementary Figure S5; Supplementary Table S1).
Finally, the SPI-6 T6SS gene cluster in the remaining 32 serotypes is highly conserved among isolates within the same serotype. However, the T6SS effector repertoire and its distribution varies considerably among these 32 serotypes (Figure 4). Notably, in VR3 these serotypes encode several T6SS effector proteins with different anti-bacterial activities, including putative DNases such as PAAR-RhsA-HNHc (S. Anatum, S. Edinburgh, S. Infantis, S. Kentucky, S. Senftenberg), RhsA-HNHc (S. Tennessee), RhsA-Tox-HNH-EHHH (S. Braenderup, S. Derby), PAAR-RhsA-Tox-HNH-EHHH (S. Johannesburg, S. Tennessee), PAAR-RhsA-AHH (S. Goldcoast) and PAAR-RhsA-GIY-YIG (S. Livingstone); putative RNases such as RhsA-Ntox47 (S. Brandenburg, S. I 1,4,[5],12:i:-, S. Typhimurium), PAAR-RhsA-Ntox47 (S. Give, S. Livingstone, S. Muenchen, S. Newport, S. Panama, S. Sandiego) and DUF4329 (S. Anatum); and putative ADP-ribosyltransferases such as PAAR-RhsA-Tox-ART-HYD1 (S. Johannesburg), RhsA-Tox-ART-HYD1 (S. Thompson) and RhsAmain (S. Typhimurium). Notably, 19 out of these 32 serotypes encode PAAR-RhsA-CT and RhsA-CT effectors harboring C-terminal ends with unknown function (Table 1; Figure 4). For instance, S. Johannesburg isolate CFSAN 122905 encodes an RhsA-CT candidate effector, along with a putative immunity protein harboring an Imm8 domain, which is commonly found in immunity proteins of bacterial toxin systems with RNase activity (Zhang et al., 2012). This result suggests that the C-terminal end of the RhsA-CT candidate effector has RNase activity.
The SPI-19 Rhs effectors of Chilean Salmonella serotypes harbor C-terminal ends with protein domains of unknown functionThe SPI-19 encodes a T6SS gene cluster present in some of the most prevalent Salmonella serotypes worldwide, such as S. Dublin, S. Agona, S. Weltevreden and S. Gallinarum, among others. Despite its contribution to intestinal colonization, antibacterial activity and cytotoxicity against macrophages (Blondel et al., 2013; Blondel et al., 2010; Pezoa et al., 2013, 2014; Schroll et al., 2019; Xian et al., 2020) no effector protein of this T6SS has been experimentally validated and tested. This is an important knowledge gap as infections triggered by these serotypes cause major economic problems in animal production and public health issues.
Our analysis identified the SPI-19 T6SS gene cluster in isolates representing 4 out of the 42 serotypes encoding T6SS. Of note, the genetic structure of this T6SS gene cluster differs among isolates of these 4 serotypes (Figure 6). In S. Agona, there are two groups of isolates that encode a PAAR-RhsA-CT effector and differ in the putative cognate immunity protein. The first group encodes a putative immunity protein with a predicted TPR domain, while in the second group this protein harbors an Imm40 domain that is frequently found in cognate immunity proteins of bacterial toxin systems with RNase activity (Zhang et al., 2012) (Figure 6). Therefore, the presence of the Imm40 domain in the putative immunity protein-encoding gene suggests that the C-terminal end of the PAAR-RhA-CT candidate effector has RNase activity. Of note, a single S. Agona isolate (CFSAN100497) lacks the SPI-19 T6SS gene cluster and harbors that encoded in SPI-6, which encodes the effector RhsA-Ntox47. This SPI-6 T6SS gene cluster exhibits high homology to the corresponding cluster in S. Typhimurium 14028s (Supplementary Figure S6).
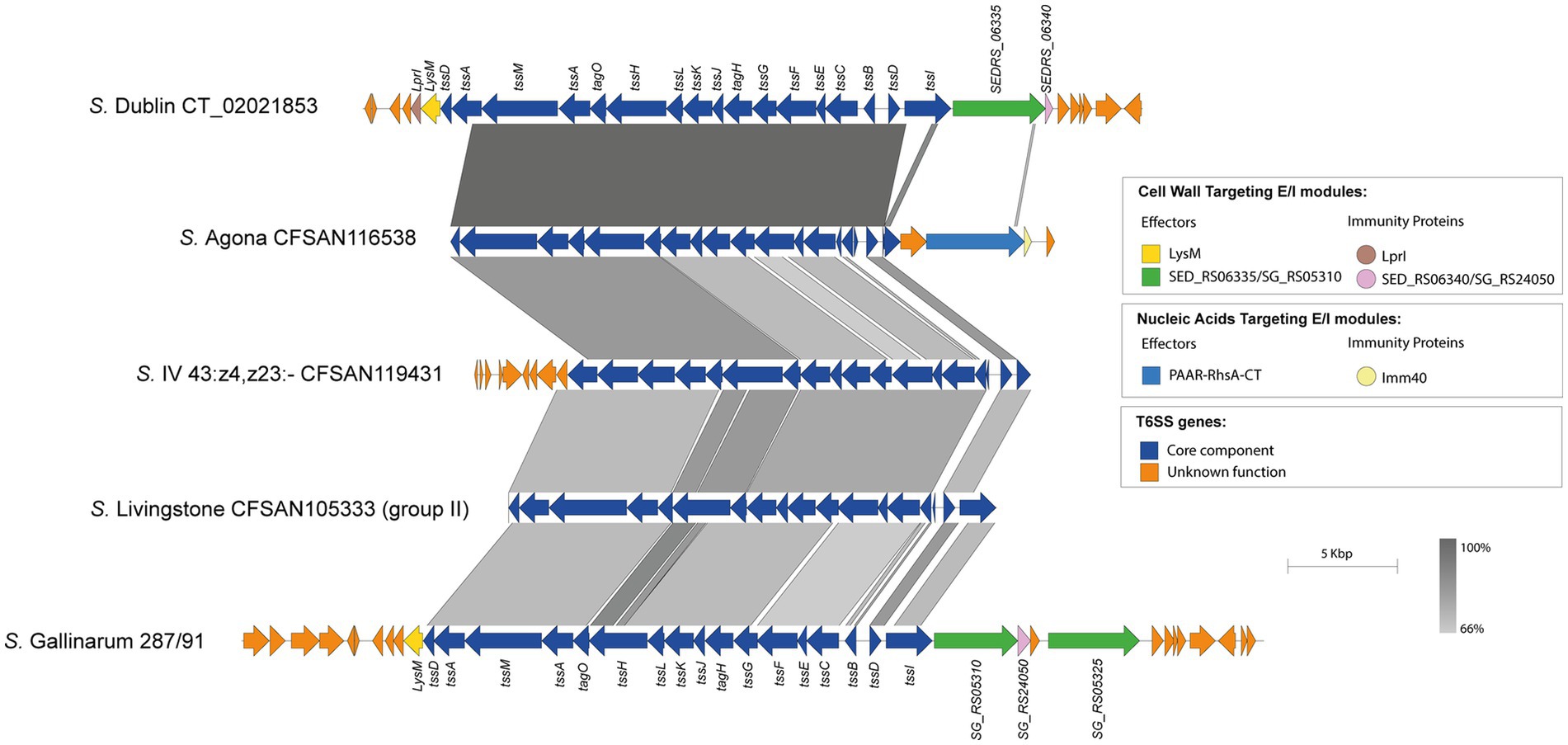
Figure 6. The SPI-19 T6SS gene cluster differs among Chilean Salmonella isolates and encodes putative T6SS Rhs effector proteins harboring C-terminal ends with domains of unknown function. Comparative genomic analysis of the SPI-19 T6SS gene cluster of S. Agona CFSAN116538, S. IV 43:z4,z23:- CFSAN119431 and S. Livingstone CFSAN105333. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). ORFs encoding T6SS core components are shown in blue. ORFs encoding E/I modules are highlighted in different colors according to the confirmed or predicted functions. SPI-19 T6SS gene clusters from S. Dublin CT_02021853 (top) and S. Gallinarum 287/91 (bottom) were used for comparative purposes. Grayscale represents the percentage of identity between nucleotide sequences.
In the case of the only isolate of serotype S. I 4:f,g,s:1,2 analyzed, the SPI-19 T6SS gene cluster exhibits high sequence conservation between the tssK and tssI core component genes with those encoded in the corresponding cluster of S. Dublin and S. Gallinarum (Figure 6). However, this serotype encodes a PAAR-RhsA-CT effector that has a different origin from the corresponding effector of S. Dublin and S. Gallinarum. Furthermore, the cognate immunity protein of this PAAR-RhsA-CT effector harbors an Imm40 domain (Zhang et al., 2012) (Figure 6), suggesting that the C-terminal end of PAAR-RhsA-CT has RNase activity.
Although we were not able to identify new effector candidates in the SPI-19 T6SS gene cluster of isolates belonging to serotypes S. IV 43:z4,z23:- and S. Livingstone, we found some features worth mentioning. In the case of serotype S. IV 43:z4,z23:-, the SPI-19 T6SS gene cluster is highly conserved among the 3 isolates analyzed. However, it shares lower degree of sequence identity with the corresponding gene cluster of S. Dublin and S. Gallinarum (Figure 6). The same was true for the group of 14 S. Livingstone isolates carrying both SPI-6 and SPI-19 T6SS gene clusters described above (Figure 6).
The SPI-21 T6SS gene cluster from S. enterica subspecies arizonae and diarizonae encodes two candidate effectorsTo date there is very limited information regarding the effector proteins encoded in the SPI-21 T6SS gene cluster. Only one candidate effector has been described in S. enterica subsp. arizonae serotype 62:z4,z23:- reference strain RSK2980, which corresponds to a specialized VgrG protein with a C-terminal extension including a pyocin domain (S Type) (Blondel et al., 2009; Ho et al., 2017). Indeed, our bioinformatic analysis identified the VgrG-PyocinS-HNHc effector in most isolates of S. enterica subsp. diarizonae serotypes 48:i:z and 35:i:z (S. IIIb 48:i:z and S. IIIb 35:i:z, respectively) analyzed (Table 1; Figure 7A). The predicted cognate immunity protein of this candidate effector includes a inhibitory immunity protein of colicin DNase and pyocins (Col_Imm_like) domain, frequently present in immunity proteins of bacterial toxin systems (Zhang et al., 2012) (Figure 7A). Noteworthy, the SPI-21 T6SS gene cluster in all isolates of S. enterica subsp. diarizonae analyzed encodes a new candidate effector including a glucosaminidase domain with predicted peptidoglycan hydrolase activity (Table 3; Figure 7B). The predicted cognate immunity protein carries the domain with no name (DWNN). Furthermore, the SPI-21 T6SS gene cluster in the only isolate of S. IIIb 35:i:z analyzed (CFSAN111176) encodes a second new candidate effector with a predicted BTH_I2691 domain (Table 3; Figure 7B). Of note, BTH_I2691 is a T6SS effector protein originally described in B. thailandensis (Russell et al., 2012), which exhibits structural homology to colicin Ia (Parret et al., 2003). This suggests that the BTH_I2691 candidate effector protein may have membrane pore-forming activity. Finally, the SPI-21 T6SS gene cluster in all isolates of S. enterica subsp. diarizonae analyzed exhibit a relatively low degree of sequence identity with the corresponding gene cluster in S. enterica subsp. arizonae RSK2980 (Figure 7A).
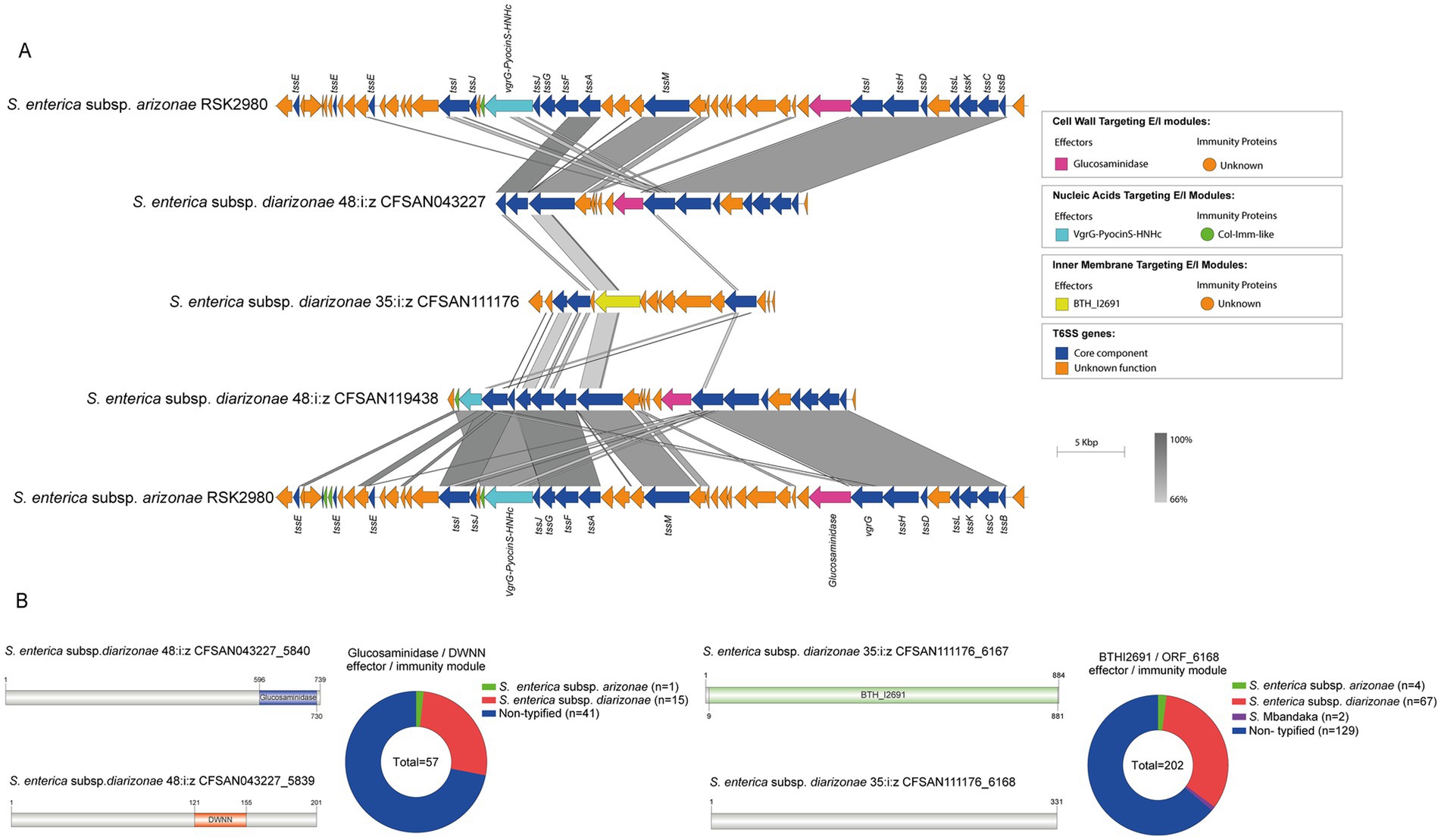
Figure 7. The SPI-21 T6SS gene cluster encodes new putative T6SS effector proteins. (A) Comparative genomic analysis of the SPI-21 T6SS cluster of S. enterica subsp. diarizonae 48:i:z CFSAN043227, S. enterica subsp. diarizonae 35:i:z CFSAN111176 and S. enterica subsp. diarizonae 48:i:z CFSAN119408. BLASTn sequence alignment was performed and visualized using EasyFig (Sullivan et al., 2011). SPI-21 T6SS gene cluster from S. enterica subsp. arizonae RSK2980 was used for comparative purposes. (B) Schematic representation and distribution among Salmonella genomes of each new effector and immunity protein identified. ORFs encoding new E/I modules are highlighted in different colors according to the predicted functions. Homologs for each component were identified by BLASTn analyses, as described in Materials and Methods.
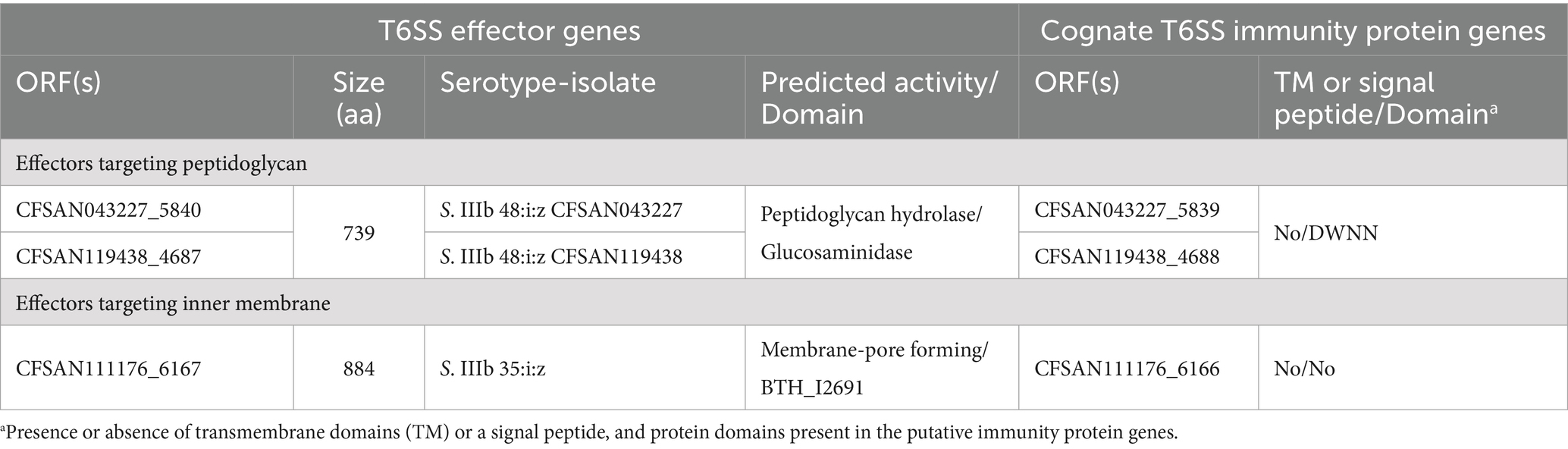
Table 3. New putative T6SS effectors and cognate immunity proteins encoded in the SPI-21 T6SS gene cluster of Chilean Salmonella isolates.
Global genome-wide distribution analysis of the new candidate effectors identified in SPI-6 and SPI-21 T6SS gene clustersThe identification of 6 new candidate T6SS effectors, harboring protein domains frequently found in bacterial toxin systems, prompted us to determine their distribution across Salmonella. To this end, the nucleotide sequence corresponding to the ORF encoding each candidate effector was used in tBLASTx searches in publicly available Salmonella genome sequences deposited in the NCBI database (March, 2024) and the distribution of each effector was determined. Our analysis revealed that the new candidate effectors are distributed in a limited number of serotypes (Figures 1B, 7B). Indeed, effectors PAAR-RhsA-NucA_B, PAAR-RhsA-CdiA and RhsA-CdiA (encoded in t
留言 (0)