Legionella, a bacterium that commonly colonizes aquatic environments, has emerged as a significant public health concern due to its ability to cause Legionnaires’ disease, a severe form of pneumonia (Graham et al., 2022). The pathogenicity of Legionella is multifaceted, involving complex interactions with the host immune system and metabolic systems (Luo et al., 2021; Yang et al., 2023). Among these, the metabolic perturbations triggered by Legionella infection are particularly noteworthy as they play a pivotal role in disease progression and pathogenesis.
The host’s metabolic homeostasis is a highly regulated process, ensuring the efficient conversion of nutrients into energy and the synthesis of macromolecules required for cellular growth and function. However, Legionella infection disrupts this homeostasis, leading to significant metabolic perturbations that favor bacterial survival and replication while compromising host cellular functions. These metabolic shifts are not limited to a single biochemical pathway but involve multiple interconnected systems. For instance, Legionella infection is known to perturb carbohydrate metabolism, altering glucose utilization, which in turn affects cellular energy production (Eisenreich and Heuner, 2016). Similarly, amino acid and protein metabolism are also disrupted, leading to changes in protein synthesis and degradation, further modifying cellular functions and immune defenses (Schunder et al., 2014; Belyi, 2020; Belyi et al., 2022). Furthermore, lipid metabolism is significantly affected by Legionella infection. Changes in lipid synthesis and breakdown can modify membrane composition and function, thereby influencing cellular signaling and the replicative niche of the bugs (Swart and Hilbi, 2020; Kowalczyk et al., 2021). These alterations contribute to the pathogenesis of Legionnaires’ disease and offer valuable insights into the complex host-pathogen interactions.
In this mini-review, we delve into the metabolic alterations triggered by Legionella infection, focusing on the specific biochemical pathways and mechanisms involved. We aim to provide a comprehensive understanding of how these metabolic perturbations contribute to disease progression and pathogenesis, and how they can be exploited for the development of novel therapeutic strategies. By doing so, we hope to gain deeper insights into the host-pathogen interactions that underlie Legionella infection and its associated diseases.
The type iv secretion system of LegionellaA distinctive feature of Legionella is its possession of the T4SS, a complex molecular machinery that enables intimate interactions with host cells (Durie et al., 2020; Bock et al., 2021). The T4SS serves as a crucial tool for Legionella to deliver effector proteins into the interior of host cells. These effector proteins play vital roles in the bacterium’s pathogenic process, manipulating various physiological activities of the host cell to aid in evasion of the immune system and successful reproduction (Song et al., 2021; Song et al., 2022; Fu et al., 2022b). Through this mechanism, Legionella establishes a favorable environment for its survival and multiplication within the host cell (Iyer and Das, 2021).
Specifically, the T4SS consists of multiple subunits that work synergistically to form a channel spanning the bacterial inner and outer membranes (Iyer and Das, 2021). This channel allows Legionella to directly transport its synthesized effector proteins into the interior of host cells. This direct delivery method enables the bacterium to quickly and effectively influence host cell functions, thereby achieving its pathogenic goals. The T4SS exhibits high specificity and selectivity, enabling precise targeting of effector proteins. Legionella can precisely select the effector proteins to be delivered and ensure their accurate targeting to specific locations within the host cell (Urbanus et al., 2016; Fu et al., 2022a, Fu et al., 2022b). This specificity ensures effective manipulation of the host cell without causing excessive damage. In conclusion, the T4SS of Legionella represents a complex and intricate molecular machinery that enables intimate interactions and manipulation of host cell functions. This mechanism not only enhances the bacterium’s pathogenic capabilities but also provides new insights into the understanding of the interaction between bacteria and host cells.
Role of Legionella Icm/Dot substrates in phosphoinositide metabolismLegionella effectors represent a sophisticated arsenal of virulence factors that target host phospholipid metabolism at multiple levels. By degrading, binding, or modifying specific phospholipids, these effectors disrupt membrane integrity, trafficking, and signaling, ultimately favoring Legionella’s infection and persistence within host cells. The integrated impact of these effectors is likely to be synergistic, as they target different aspects of phospholipid metabolism and membrane function. One such effector protein, MavQ, has recently been identified as a key player in the regulation of phosphatidylinositol (PtdIns) metabolism within the LCV (Hsieh et al., 2021; Li et al., 2021). MavQ, exhibits a unique enzymatic activity: it specifically phosphorylates PtdIns to generate phosphatidylinositol 3-phosphate (PtdIns3P) (Hsieh et al., 2021; Li et al., 2021). This activity is significant because PtdIns3P is an essential intermediate in the PtdIns phosphorylation cascade, leading to the production of more complex phosphoinositides like PtdIns4P (Li et al., 2021). Once PtdIns3P is generated by MavQ, it serves as a substrate for the subsequent actions of LepB (Dong et al., 2016). LepB, another Dot/Icm effector, exhibits phosphatidylinositol 4-kinase activity, catalyzing the phosphorylation of PtdIns3P to PtdIns3,4P (Dong et al., 2016). By acting downstream of LepB, the effector protein SidF functions as a phosphatidylinositol 3-phosphate phosphatase, converting PtdIns3,4P to PtdIns4P, which completes the biosynthetic pathway of PtdIns4P on the LCV membrane (Hsu et al., 2012; Dong et al., 2016).
Some other effector including SidP, LppA, VipD, PlcC, and LpdA, are Icm/Dot substrates delivered by Legionella pneumophila into host cells, where they play crucial roles in modulating phosphoinositide metabolism. SidP, a phosphatidylinositol 3-phosphatase, can convert PtdIns3,5P to PtdIns5P and PtdIns3P to PtdIns in vitro, although its activity in infected cells remains unclear (Toulabi et al., 2013). LppA, a CX5R motif PI phosphatase, hydrolyzes specific phosphoinositide species to produce PtdIns4P in vitro, but does not seem to significantly alter the PI pattern of Legionella-containing vacuoles (LCVs) in live cells (Weber et al., 2014). VipD, on the other hand, is a lipase that hydrolyzes PtdIns3P, and intriguingly, binds to Rab5 and Rab22 (Gaspar and Machner, 2014). This effector removes PtdIns(3)P from endosomal membranes, potentially promoting the evasion of the endocytic pathway by LCVs (Gaspar and Machner, 2014). PlcC, a metallophospholipase C, exhibits broad lipid hydrolyzing activity, including phosphatidylcholine (PC), phosphatidylglycerol (PG), and PtdIns (Aurass et al., 2013). Lastly, LpdA, a phospholipase D, hydrolyzes PG, PtdIns, PtdIns3P, and PtdIns4P to yield phosphatidic acid (PA) (Schroeder et al., 2015). Through these enzymatic activities, LpdA likely alters membrane properties and signaling cascades within LCVs (Schroeder et al., 2015). By modulating the PI composition and signaling cascades within LCVs, these effectors likely play important roles in Legionella’s pathogenesis, including the evasion of host defenses, modulation of membrane trafficking, and the establishment of a replicative niche within host cells. Further research is needed to fully elucidate the precise mechanisms and functional outcomes of these effectors within the context of Legionella infection.
In summary, the large cohort effectors play crucial roles in the regulation of PtdIns metabolism within the LCV. By targeting specific steps in the PtdIns phosphorylation cascade, these effectors generate PtdIns4P, which serves as an anchor for other Dot/Icm effectors (Figure 1). This process is essential for Legionella’s ability to establish and maintain infection within host cells. Further research into the mechanisms and functions of these effectors may provide new insights into Legionella’s pathogenesis and potential therapeutic targets for the treatment of Legionnaires’ disease.
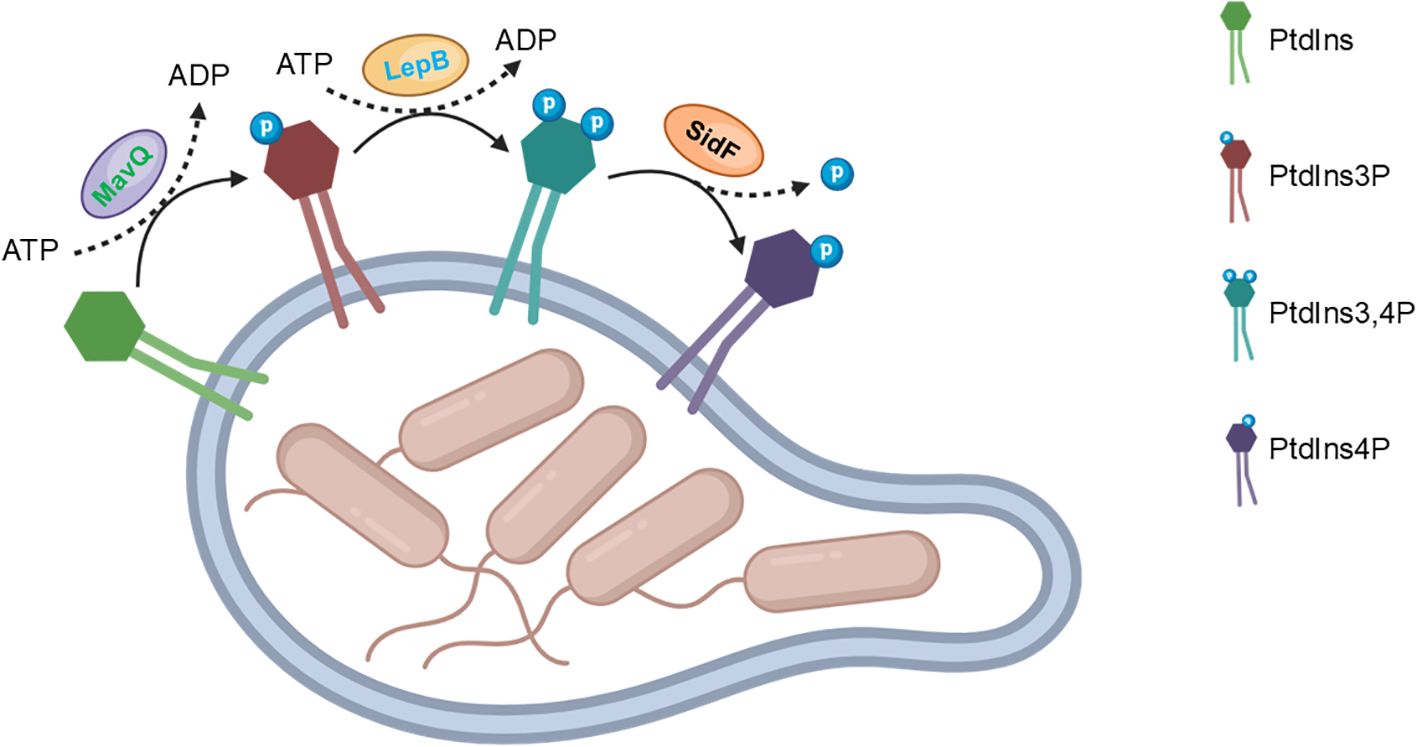
Figure 1. Legionella effectors synergistically convert PtdIns to PtdIns4P.
The impact of Legionella effectors on host protein synthesisThe impact of Legionella effectors on host cell protein synthesis is multifaceted and profoundly disrupts the normal cellular processes. Several effector proteins produced by Legionella, including Lgt1, Lgt2, Lgt3, SidI, LegK4, SidL, and RavX, have been identified to play a crucial role in inhibiting protein synthesis in infected host cells (Belyi et al., 2022). Among these, Lgt1-3 and SidI target elongation factors, the key components of the protein synthesis machinery (Shen et al., 2009; Sol et al., 2019). Lgt1-3 are members of the GT-A type glucosyltransferase family that mono-glucosylate elongation factor eEF1A (Belyi et al., 2006; Tzivelekidis et al., 2011). This glucosylation of eEF1A leads to its inactivation, thereby inhibiting protein synthesis (Belyi et al., 2006). Interestingly, SidI binds to both eEF1A and its guanine nucleotide exchange factor eEF1Bg, further disrupting their interaction and resulting in the inhibition of protein synthesis (Shen et al., 2009).
LegK4, another Legionella effector, functions as a protein kinase that phosphorylates Hsp70 proteins. Hsp70 proteins are involved in various cellular processes, including protein folding, translocation, and degradation (Moss et al., 2019). By phosphorylating Hsp70 proteins, LegK4 disrupts their function and ultimately inhibits protein synthesis (Moss et al., 2019). While SidL and RavX have not yet been assigned specific targets in the protein synthesis machinery, they are likely to affect protein synthesis indirectly by modulating other cellular processes (Subramanian et al., 2023). This suggests that Legionella effectors may have pleiotropic effects, targeting multiple cellular processes to promote infection.
The inhibition of protein synthesis by Legionella effectors has profound consequences for the host cell. It disrupts the normal functioning of the cell, impairs its ability to respond to stress, and ultimately leads to cell death. This allows Legionella to evade host immune defenses, replicate within the cell, and spread to other parts of the body. Further research into the mechanisms and functions of these effectors could provide novel insights into Legionella pathogenesis and potential therapeutic targets.
Conclusion and prospectiveMetabolic perturbations in Legionella infection provide deep insights into the complex host-pathogen interactions that underlie the pathogenesis of Legionnaires’ disease. The identification and characterization of Legionella effectors such as SidP, LppA, VipD, PlcC, and LpdA that target specific phosphoinositide species within host cells reveal a sophisticated mechanism of how the pathogen modulates membrane trafficking, alters signaling pathways, and establishes a favorable replicative niche. These effectors exhibit diverse enzymatic activities that perturb the host cell’s metabolic landscape, including the hydrolysis of phospholipids and PtdIns, altering membrane properties, and disrupting cellular signaling cascades. These perturbations enable Legionella to evade host immune defenses, hijack host cellular processes, and ultimately cause disease.
Future research in this area is poised to yield exciting new insights into Legionella pathogenesis and host-pathogen interactions. Understanding these interactions could provide novel targets for therapeutic intervention. The development of new diagnostic tools and therapeutic strategies based on the understanding of Legionella pathogenesis could significantly improve the management of Legionnaires’ disease. By investigating the synergistic actions of bacterial effectors, the broader impact of Legionella infection on host cell metabolism, and the interactions with host genetics, we are optimistic that continued research will lead to more effective treatments and better patient outcomes, reducing the morbidity and mortality associated with this infection.
Author contributionsZW: Conceptualization, Formal analysis, Writing – original draft. LS: Conceptualization, Funding acquisition, Writing – review & editing. JC: Conceptualization, Writing – review & editing. CL: Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported partly by The Bethune Project of Jilin University 2024B20 and the Science and Technology Development Project of Changchun City 23YQ10.
AcknowledgmentsThis article was polished for language and clarity with the assistance of ChatGPT 3.5.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1458276/full#supplementary-material
ReferencesAurass, P., Schlegel, M., Metwally, O., Harding, C. R., Schroeder, G. N., Frankel, G., et al. (2013). The Legionella pneumophila Dot/Icm-secreted effector PlcC/CegC1 together with PlcA and PlcB promotes virulence and belongs to a novel zinc metallophospholipase C family present in bacteria and fungi. J. Biol. Chem. 288, 11080–11092. doi: 10.1074/jbc.M112.426049
PubMed Abstract | Crossref Full Text | Google Scholar
Belyi, Y., Levanova, N., Schroeder, G. N. (2022). Glycosylating effectors of legionella pneumophila: finding the sweet spots for host cell subversion. Biomolecules 12, 255. doi: 10.3390/biom12020255
PubMed Abstract | Crossref Full Text | Google Scholar
Belyi, Y., Niggeweg, R., Opitz, B., Vogelsgesang, M., Hippenstiel, S., Wilm, M., et al. (2006). Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc. Natl. Acad. Sci. U.S.A. 103, 16953–16958. doi: 10.1073/pnas.0601562103
PubMed Abstract | Crossref Full Text | Google Scholar
Bock, D., Husler, D., Steiner, B., Medeiros, J. M., Welin, A., Radomska, K. A., et al. (2021). The polar legionella icm/dot T4SS establishes distinct contact sites with the pathogen vacuole membrane. mBio 12, e0218021. doi: 10.1128/mBio.02180-21
PubMed Abstract | Crossref Full Text | Google Scholar
Dong, N., Niu, M., Hu, L., Yao, Q., Zhou, R., Shao, F. (2016). Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat. Microbiol. 2, 16236. doi: 10.1038/nmicrobiol.2016.236
PubMed Abstract | Crossref Full Text | Google Scholar
Durie, C. L., Sheedlo, M. J., Chung, J. M., Byrne, B. G., Su, M., Knight, T., et al. (2020). Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex. Elife 9, e59530. doi: 10.7554/eLife.59530.sa2
PubMed Abstract | Crossref Full Text | Google Scholar
Fu, J., Li, P., Guan, H., Huang, D., Song, L., Ouyang, S., et al. (2022a). Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation. mLife 1, 51–65. doi: 10.1002/mlf2.12014
PubMed Abstract | Crossref Full Text | Google Scholar
Fu, J., Zhou, M., Gritsenko, M. A., Nakayasu, E. S., Song, L., Luo, Z. Q. (2022b). Legionella pneumophila modulates host energy metabolism by ADP-ribosylation of ADP/ATP translocases. Elife 11, e73611. doi: 10.7554/eLife.73611
PubMed Abstract | Crossref Full Text | Google Scholar
Gaspar, A. H., Machner, M. P. (2014). VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 4560–4565. doi: 10.1073/pnas.1316376111
PubMed Abstract | Crossref Full Text | Google Scholar
Graham, F. F., Finn, N., White, P., Hales, S., Baker, M. G. (2022). Global perspective of legionella infection in community-acquired pneumonia: A systematic review and meta-analysis of observational studies. Int. J. Environ. Res. Public Health 19, 1907. doi: 10.3390/ijerph19031907
PubMed Abstract | Crossref Full Text | Google Scholar
Hsieh, T. S., Lopez, V. A., Black, M. H., Osinski, A., Pawlowski, K., Tomchick, D. R., et al. (2021). Dynamic remodeling of host membranes by self-organizing bacterial effectors. Science 372, 935–941. doi: 10.1126/science.aay8118
PubMed Abstract | Crossref Full Text | Google Scholar
Hsu, F., Zhu, W., Brennan, L., Tao, L., Luo, Z. Q., Mao, Y. (2012). Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 109, 13567–13572. doi: 10.1073/pnas.1207903109
PubMed Abstract | Crossref Full Text | Google Scholar
Iyer, S., Das, C. (2021). The unity of opposites: Strategic interplay between bacterial effectors to regulate cellular homeostasis. J. Biol. Chem. 297, 101340. doi: 10.1016/j.jbc.2021.101340
PubMed Abstract | Crossref Full Text | Google Scholar
Li, G., Liu, H., Luo, Z. Q., Qiu, J. (2021). Modulation of phagosome phosphoinositide dynamics by a Legionella phosphoinositide 3-kinase. EMBO Rep. 22, e51163. doi: 10.15252/embr.202051163
PubMed Abstract | Crossref Full Text | Google Scholar
Luo, J., Wang, L., Song, L., Luo, Z. Q. (2021). Exploitation of the host ubiquitin system: means by legionella pneumophila. Front. Microbiol. 12, 790442. doi: 10.3389/fmicb.2021.790442
PubMed Abstract | Crossref Full Text | Google Scholar
Moss, S. M., Taylor, I. R., Ruggero, D., Gestwicki, J. E., Shokat, K. M., Mukherjee, S. (2019). A legionella pneumophila kinase phosphorylates the hsp70 chaperone family to inhibit eukaryotic protein synthesis. Cell Host Microbe 25, 454–462.e456. doi: 10.1016/j.chom.2019.01.006
PubMed Abstract | Crossref Full Text | Google Scholar
Schroeder, G. N., Aurass, P., Oates, C. V., Tate, E. W., Hartland, E. L., Flieger, A., et al. (2015). Legionella pneumophila effector lpdA is a palmitoylated phospholipase D virulence factor. Infect. Immun. 83, 3989–4002. doi: 10.1128/IAI.00785-15
PubMed Abstract | Crossref Full Text | Google Scholar
Schunder, E., Gillmaier, N., Kutzner, E., Eisenreich, W., Herrmann, V., Lautner, M., et al. (2014). Amino Acid Uptake and Metabolism of Legionella pneumophila Hosted by Acanthamoeba castellanii. J. Biol. Chem. 289, 21040–21054. doi: 10.1074/jbc.M114.570085
PubMed Abstract | Crossref Full Text | Google Scholar
Shen, X., Banga, S., Liu, Y., Xu, L., Gao, P., Shamovsky, I., et al. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 11, 911–926. doi: 10.1111/cmi.2009.11.issue-6
PubMed Abstract | Crossref Full Text | Google Scholar
Sol, A., Lipo, E., De Jesus-Diaz, D. A., Murphy, C., Devereux, M., Isberg, R. R. (2019). Legionella pneumophila translocated translation inhibitors are required for bacterial-induced host cell cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 116, 3221–3228. doi: 10.1073/pnas.1820093116
PubMed Abstract | Crossref Full Text | Google Scholar
Song, L., Luo, J., Wang, H., Huang, D., Tan, Y., Liu, Y., et al. (2022). Legionella pneumophila regulates host cell motility by targeting Phldb2 with a 14-3-3zeta-dependent protease effector. Elife 11, e73220. doi: 10.7554/eLife.73220.sa2
PubMed Abstract | Crossref Full Text | Google Scholar
Song, L., Xie, Y., Li, C., Wang, L., He, C., Zhang, Y., et al. (2021). The legionella effector sdjA is a bifunctional enzyme that distinctly regulates phosphoribosyl ubiquitination. mBio 12, e0231621. doi: 10.1128/mBio.02316-21
PubMed Abstract | Crossref Full Text | Google Scholar
Subramanian, A., Wang, L., Moss, T., Voorhies, M., Sangwan, S., Stevenson, E., et al. (2023). A Legionella toxin exhibits tRNA mimicry and glycosyl transferase activity to target the translation machinery and trigger a ribotoxic stress response. Nat. Cell Biol. 25, 1600–1615. doi: 10.1038/s41556-023-01248-z
PubMed Abstract | Crossref Full Text | Google Scholar
Toulabi, L., Wu, X., Cheng, Y., Mao, Y. (2013). Identification and structural characterization of a Legionella phosphoinositide phosphatase. J. Biol. Chem. 288, 24518–24527. doi: 10.1074/jbc.M113.474239
PubMed Abstract | Crossref Full Text | Google Scholar
Tzivelekidis, T., Jank, T., Pohl, C., Schlosser, A., Rospert, S., Knudsen, C. R., et al. (2011). Aminoacyl-tRNA-charged eukaryotic elongation factor 1A is the bona fide substrate for Legionella pneumophila effector glucosyltransferases. PLoS One 6, e29525. doi: 10.1371/journal.pone.0029525
PubMed Abstract | Crossref Full Text | Google Scholar
Urbanus, M. L., Quaile, A. T., Stogios, P. J., Morar, M., Rao, C., Di Leo, R., et al. (2016). Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12, 893. doi: 10.15252/msb.20167381
PubMed Abstract | Crossref Full Text | Google Scholar
Weber, S., Stirnimann, C. U., Wieser, M., Frey, D., Meier, R., Engelhardt, S., et al. (2014). A type IV translocated Legionella cysteine phytase counteracts intracellular growth restriction by phytate. J. Biol. Chem. 289, 34175–34188. doi: 10.1074/jbc.M114.592568
PubMed Abstract | Crossref Full Text | Google Scholar
Yang, Y., Mei, L., Chen, J., Chen, X., Wang, Z., Liu, L., et al. (2023). Legionella pneumophila-mediated host posttranslational modifications. J. Mol. Cell Biol. 15, mjad032. doi: 10.1093/jmcb/mjad032
留言 (0)