Exposure to stressful environmental conditions early in the life of an animal can result in changes of gene expression that can be manifested in adulthood (Fusco and Minelli, 2010; Xue and Leibler, 2018; Rodrigues and Beldade, 2020; Sommer, 2020). The nematode Caenorhabditis elegans responds to early-life stress conditions, such as starvation, high temperature, or crowding, through entry into an alternative life history trajectory (Hu, 2018). Upon exposure to these unfavorable growth conditions, L2d larvae enter a stress resistant, developmentally arrested, and non-feeding stage called dauer, at which they can remain for months. Dauer larvae exit as postdauers (PD) L4 larvae only when environmental conditions improve (Cassada and Russell, 1975). Animals that experience favorable conditions in early life continuously develop through four larval stages (L1-L4) before reaching reproductive adulthood (control adults, CON) (Sulston and Horvitz, 1977). We have previously shown that PD adults retain a cellular memory of passage through dauer that manifests as altered genome-wide gene expression, small RNA populations, chromatin states, and life history traits compared to control adult animals (Hall et al., 2010; 2013; Ow et al., 2018; Ow et al., 2021).
One target of this developmental programming is the osm-9 TRPV channel gene whose expression is downregulated in the ADL chemosensory neurons of PD adults (Sims et al., 2016). TRPV channels are members of a superfamily of evolutionarily conserved transient receptor potential (TRP) cation channels found in all eukaryotes and form homo- or heterocomplexes with other TRPs to regulate sensory perception (Kahn-Kirby and Bargmann, 2006; Nilius and Owsianik, 2011). In C. elegans, mutants of TRPV (vanilloid family) channel genes, such as those encoding for OSM-9 and its heteromeric TRPV channel partner member, OCR-2, exhibit defects in olfaction, nociception, and animal behavior. The osm-9 gene is expressed in the sensory cilia and in several sensory neurons including ADL (Colbert et al., 1997; Tobin et al., 2002; Kahn-Kirby and Bargmann, 2006; Jang et al., 2012). The reduced osm-9 mRNA expression in postdauer ADL neurons results in the abrogation of avoidance to ascr#3 (asc-ΔC9), which is an osm-9-dependent, ADL-mediated behavior (Sims et al., 2016). Ascr#3 is a dauer-inducing small signaling molecule with a nine-carbon α,β-unsaturated carboxylic acid side chain that is a component of pheromones produced by C. elegans that are crucial for olfactory behavior, aggregation, and mating (Butcher et al., 2007; Ludewig and Schroeder, 2018). Our early work indicated that TGF-ß signaling, chromatin remodeling, and endogenous small RNA interference (RNAi) pathways contribute to the mechanism by which osm-9 expression is reprogrammed in PD adults due to early-life environmental stress (Sims et al., 2016).
Endogenous small RNA pathways in animals are categorized into three major classes based on their biogenesis and their effector protein partners: PIWI-interacting RNA (piRNAs), microRNAs (miRNAs), and endogenous small interfering RNAs (endo-siRNAs). These small non-coding RNAs (snRNAs) range between ∼20 and 30 nucleotides (nt) long and are fully or partially anti-sense to their target transcripts (Billi et al., 2014; Ambros and Ruvkun, 2018; Bartel, 2018; Czech et al., 2018; Ketting and Cochella, 2021). In C. elegans, the biogenesis of endo-siRNAs starts with double-stranded RNA (dsRNA) being processed by a protein complex that includes the RNase III endoribonuclease DICER into two classes of 26 nt long primary siRNAs that have a 5′ guanosine (26Gs): ERGO-1 26G RNAs and 26G RNAs that associate with two paralogous Argonautes (AGOs), ALG-3 and ALG-4 (ALG-3/4) (Billi et al., 2014; Ketting and Cochella, 2021). The ERGO-1 26G RNAs primarily target transcripts produced in the oogenic germline, embryos, pseudogenes, and gene duplications while ALG-3/4 26G RNAs are derived from the spermatogenic germline (Han et al., 2009; Conine et al., 2010; Gent et al., 2010; Vasale et al., 2010). 26G RNAs stimulate the amplification of 22 nt long secondary siRNAs with a 5’ guanosine (22Gs) that are further subdivided into two categories, those that bind to the Worm specific AGO (WAGO) clade and those that bind to the CSR-1 AGO (Guang et al., 2008; Claycomb et al., 2009; Gu et al., 2009; Burton et al., 2011; Ashe et al., 2012; Buckley et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). The WAGO-bound 22G RNAs effect the silencing of target transcripts while CSR-1-bound 22G RNAs do not silence their targets but rather promote the expression of germline genes and proper chromosome segregation (Yigit et al., 2006; Claycomb et al., 2009).
Out of 26 AGO proteins encoded in C. elegans, 19 have been attributed as functional, most of which remain uncharacterized or partially characterized (Ketting and Cochella, 2021; Seroussi et al., 2023). While most characterized AGOs function primarily in the germ line, only four AGOs are reported to be expressed in neurons: ERGO-1, NRDE-3, and the miRNA-specific ALG-1 and ALG-2 (Seroussi et al., 2023). We previously showed that NRDE-3 and its nuclear complex members are required for the downregulation of osm-9 expression in PD ADL neurons and the resulting abrogation of the avoidance response to ascr#3 (Sims et al., 2016). NRDE (nuclear RNAi defective)-3/WAGO-12 is the effector protein of the somatic nuclear RNAi pathway and associates with 22G RNAs (Guang et al., 2008; 2010; Gent et al., 2010; Burton et al., 2011). Once bound to 22G RNAs, cytoplasmic NRDE-3 translocates to the nucleus where it is directed to target nascent pre-mRNA and recruits NRDE-2 to inhibit RNA polymerase II transcription elongation (Guang et al., 2008). NRDE-3 and NRDE-2 then recruit NRDE-1 to the nascent pre-mRNA to potentiate its association with chromatin and the trimethylation of Histone 3 Lysine 9 (H3K9me3) at the targeted locus (Guang et al., 2010; Burkhart et al., 2011). The small RNA-directed chromatin modification, but not the association with pre-mRNA, by NRDE-1 depends on a fourth nuclear RNA pathway factor, NRDE-4 (Guang et al., 2008; 2010; Burkhart et al., 2011). Similarly, NRDE-1, NRDE-2, and NRDE-4 also participate in the germline nuclear RNAi pathway and transgenerational epigenetic inheritance (TEI) directed by the AGO HRDE-1/WAGO-9 (Ashe et al., 2012; Buckely et al., 2012; Luteijn et al., 2012).
Compared to the prominence of endo-siRNAs in germline function, maintenance, and integrity, our knowledge of their role in the C. elegans nervous system is limited (Ow and Hall, 2021). Several reports have emerged in recent years describing their critical neural function. Mutations in genes encoding for CSR-1 and a component of the Mutator foci, a perinuclear structure where production of siRNA occurs, MUT-16 (Phillips et al., 2012), disrupt dauer formation due the exposure to starvation, crowding, and high temperature (Bharadwaj and Hall, 2017). This dauer defective (daf-d) phenotype was rescued by the expression of mut-16 using a pan-neuronal promoter. In addition, MUT-16 and CSR-1 are required for the expression of the gpa-1, gpa-3 and gpc-1 G proteins whose mutations are associated with the daf-d phenotype, suggesting that endogenous RNAi pathways play a role in the phenotypic plasticity of an animal in response to the environment (Bharadwaj and Hall, 2017). C. elegans’ attraction towards the odorant benzaldehyde was found to be heritable for at least two generations in the absence of the original odorant trigger through neuronal endo-siRNAs transmitted through the germ line. Animals expressing a co-factor necessary for the biogenesis of 26G RNAs, RDE-4 (Duchaine et al., 2006; Lee et al., 2006; Vasale et al., 2010), from a neuronal-specific promoter, led to the identification of neuronal small RNAs that affected the expression of germline mRNAs. Some of these germline gene expression changes perdured for at least three generations and were dependent on the germline-specific nuclear RNAi pathway AGO HRDE-1 silencing the saeg-2 gene encoding a component of the SAEG-1/SAEG-2 histone deacetylase complex in response to benzaldehyde (Posner et al., 2019). Loss of NRDE-3 results in defective olfactory adaptation behavior to the odorant butanone mediated by the AWC olfactory sensory neurons. Accordingly, an increase in 22G RNAs antisense to the odr-1 guanylyl cyclase gene required for AWC function was found, suggesting that a NRDE-3-mediated reduction in odr-1 expression in AWC is responsible for C. elegans’ olfactory adaptation to butanone (Juang et al., 2013).
While much is known about NRDE-3 in regulating somatic nuclear small RNA pathway triggered by exogenous substrates (Guang et al., 2008; 2010; Gent et al., 2010; Burkhart et al., 2011; Burton et al., 2011), its role in endogenous gene expression is yet to be well-characterized. To further investigate the role of NRDE-3 in regulating the developmental reprogramming of gene expression in ADL neurons due to early-life crowding stress, we profiled the mRNA transcriptome of control and postdauer ADL in wild-type and nrde-3 mutant adults. We found that hundreds of genes typically associated with the germ line, such as those encoding for major sperm protein (MSP) gene family, are also differentially expressed in ADL neurons in a NRDE-3 dependent manner. Mutation in the PP1 phosphatase, GSP-3, which is required for MSP function in sperm, significantly decreases avoidance behavior in response to nociceptive stimuli and the chemotaxis behavior towards attractive stimuli. We demonstrate that germline RNAi AGO, WAGO-4, and miRNA AGO, ALG-1, are expressed in ADL and modulate ascr#3 avoidance. Furthermore, we show that AGO WAGO-11, previously classified as a pseudogene, is expressed in ADL and is required for its function. Together, our results highlight a nexus between neuronal and reproductive networks in recalibrating the life history and the neuroplasticity of an animal in response to early-life environmental experience.
2 Materials and methods2.1 C. elegans strains and husbandryWorms were grown at 20°C in Nematode Growth Medium (NGM) seeded with E. coli OP50 (Stiernagle, 2006) unless otherwise stated. Cultivation of worms used for FACS were grown on NGM plates (100 mm petri dishes) seeded with 20× concentrated E. coli OP50 at 20°C. PDPhe adults were obtained as previously reported (Ow et al., 2018). All worm strains used in this study are listed in Supplementary Table S1. All un-backcrossed mutant strains were backcrossed 4–6 times into our laboratory N2 Bristol wild-type strain before use.
2.2 Transgenic strainsSEH306 pdrEx80 (sre-1p::gfp; unc-122p::dsRed) was made by injecting N2 adults with a plasmid containing 3 kb of the sre-1 promoter fused upstream of gfp (sre-1p::gfp) and a unc-122p::dsRed reporter as the co-injection marker. The sre-1p::gfp; unc-122p::dsRed extrachromosomal array was integrated using a UV crosslinker and backcrossed 6x with our N2 laboratory strain to create SEH318 pdrIs9 (sre-1p::gfp; unc-122p::dsRed). SEH318 was crossed into YY158 nrde-3(gg66) X to build SEH321 nrde-3(gg66) X; pdrIs9 (sre-1p::gfp; unc-122p::dsRed). All mutant strains expressing pdrIs9 (sre-1p::gfp; unc-122p::dsRed) were built by genetically crossing SEH318 into each mutant strain and confirmed by PCR or Sanger sequencing (Supplementary Table S1).
2.3 Adult neuronal isolationIsolation of adult ADL neurons of SEH318 pdrIs9 (sre-1p::gfp; unc-122p::dsRed) CON and PDPhe adults and SEH321 nrde-3(gg66) X; pdrIs9 (sre-1p::gfp; unc-122p::dsRed) CON and PDPhe adults were done as described by (Kaletsky et al., 2016). Briefly, staged 1-day old adult animals grown at 20°C on 100 mm NGM plates seeded with 20x concentrated OP50 were harvested and washed with M9 buffer until the supernatant was clear. 750 μL of Neuronal Isolation Lysis Buffer (200 mM DTT; 0.25% SDS; 20 mM HEPES pH 8.0; 3% sucrose) was added to a packed pellet of worms (up to 250 μL) and rotated at room temperature for 6.5 min. The worm mixture was then washed with M9 and 500 μL of a freshly made pronase (Sigma P6911) solution (20 mg/mL) was added and vigorous pipetted for 100 times (∼40 s) in 2 min intervals until ADL head neurons dissociated from the worm body as monitored using fluorescence microscopy. Once worm heads disintegrated (∼14–16 min), the pronase reaction was stopped with 250 μL of 1x PBS buffer supplemented with 2% Fetal Bovine Serum (Fisher Scientific 10082139) and iced. The worm mixture was then filtered through 5 μM filter, stored on ice, and immediately transported to the cell sorting facility (∼15 min interval) where it was sorted upon arrival.
2.4 Fluorescence activated cell sorting (FACS)ADL chemosensory neurons were collected from a neuronal population obtained from 1-day old adults using with a Becton Dickinson FACS Aria III Cell Sorter. Neurons collected from 1-day old wild-type N2 adults were used to set the FACS gate. The average total FACS event for a strain and condition ranged between 128,229 and 846,079. FACS events were collected directly into TRIzol (Life Technologies) and immediately frozen in dry ice. All frozen samples were stored at −80°C until ready for processing.
2.5 RNA extraction, mRNA-Seq library preparation, and data analysesTotal RNA extraction of FACS events was done using NEB Monarch Total RNA Miniprep (New England Biolabs [NEB] T2010). Four biological independent RNA-Seq libraries for a strain and a condition were prepared using NEBNext® Poly(A) mRNA Magnetic Isolation Module (NEB E7490S) and NEBNext® Ultra™ II RNA Library Prep with Sample Purification Beads (NEB E7775S) following the recommendations of the manufacturer. Indexing of libraries was done using NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 1) (NEB E7335S) and NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 2) (NEB E7500S) according to the manufacturer’s instructions. Data analysis was performed on the CLC Genomics Workbench (Qiagen) with differential expression assessed using EdgeR (Robinson, McCarthy, and Smyth, 2010) as previously described (Ow et al., 2018). Raw data can be accessed through NCBI GEO GSE268801.
2.6 Gene ontology (GO) term analysesGO terms enrichment was determined using Gorilla (Eden et al., 2009).
2.7 Statistical significanceStatistical significance was determined using GraphPad Prism version 9.5 (San Diego, CA, United States). The significance of hypergeometric distributions was determined using a hypergeometric p-value software calculator (https://systems.crump.ucla.edu/hypergeometric/index.php).
2.8 Bioinformatics analysis of the PD motifTo identify genes whose upstream regulatory sequence contain the DAF-3/SMAD binding site and the conserved sequence of the PD motif, a custom BLAST database including 500 bp upstream of ATG translational start sites for all genes in the C. elegans genome was created using NCBI. This database was used to search for genes whose 5′UTR contained “GTCTA” or the “CT[AT]TAA[AT]TTN(0,3)[AC]AN(0,2)TTTTG[CT]CATAA[TA]C[TC]” pattern, where “N” could be any nucleotide and the numbers signify the range of possible number of nucleotides. The overlap of the genes in each list were genes that contained the full PD motif. The python script used for this analysis can be found at https://github.com/mnishiguchi7/Hall_Lab/blob/main/script.
2.9 Single molecule fluorescence in situ hybridization (smFISH)One-day old adults expressing the sre-1p::gfp transgene were used for smFISH (Ji and Oudenaarden, 2012) and carried out as previously described (Sims et al., 2016). Custom Stellaris™ smFISH probe sets consisting of 27 probes for msp-19 (Quasar® 570) and 32 probes for gfp (Quasar® 670) were designed using the Stellaris™ probe designer website (LGC Biosearch Technologies). Probe sequences will be made available upon request. Images were acquired on a Leica DM5500 B microscope equipped with a Leica CTR5500 electronic box with a Hamamatsu Digital Camera C10600 ORCA R2 using maximally projected images of z-stacks as optimized by the Leica LAS AF 3.1.0 software followed by quantification on ImageJ. SmFISH experiments were done using two independent biological samples.
2.10 ImmunofluorescenceOne-day old adults expressing the sre-1p::gfp transgene were used for immunofluorescence using the method reported by Serrano-Saiz et al. (2017). In brief, 1-day old adults were fixed overnight with 1x PBS supplemented with 4% paraformaldehyde (Thermo Fisher AA433689M) followed by washing in PT Buffer (1x PBS, 0.5% Triton X-100). Fixed worms were reduced for ∼18–20 h at 37°C with 5% ß-mercaptoethanol in 1% Triton X-100 and 0.1 M Tris-HCl pH 7.5 followed by washing with TT Buffer (1% Triton X-100, 0.1 M Tris-HCl pH 7.5) and CTT Buffer (1 mM CaCl2, 1% Triton X-100, 0.1 M Tris-HCl pH 7.5). Worms were pelleted and shaken at room temperature for ∼30 min in CTT Buffer supplemented with 1 mg/ml of collagenase Type IV (Millipore Sigma C5138) until they fragmented (20%–50%). Fragmented worm samples were rinsed with PT Buffer and incubated with 1x PBS supplemented with 1 mg/mL of NaBH4 (Millipore Sigma 71320) for 1 h at 4°C with rotation. Samples were washed with PT Buffer and blocked in 0.2% gelatin solution [1x PBS, 0.5% Triton X-100, 0.2% gelatin (Millipore Sigma G7041)] supplemented with 2% donkey serum (Jackson Immunoresearch 102644-006) at room temperature for 2 h followed by an overnight rotation with mouse anti-MSP (Developmental Studies Hybridoma Bank, University of Iowa, United States; 4A5) (1:5) or chicken anti-GFP (Thermo Fisher A10262) (1:500) at 4°C in 0.2% gelatin solution supplemented with 0.5% donkey serum. Next day, samples were rinsed with PT Buffer followed by rotation in PT Buffer for 2 h at room temperature. Samples were incubated with goat anti-mouse Alexa Fluor 568 (Thermo Fisher A11031) (1:100) or goat anti-chicken Alexa Fluor Plus 647 (Thermo Fisher A32933) (1:1000) and rotated in the dark overnight at 4°C in 0.2% gelatin solution supplemented with 0.5% donkey serum. Next day, samples were washed with PT Buffer followed by rotation in PT Buffer for 1.5 h at room temperature in the dark. The DNA counterstain DAPI (Thermo Fisher D1306) was added and rotated for an additional 30 min followed by rinsing with PT Buffer. Worm samples were pelleted and VectaShield H1000 antifade mounting medium (Vector Laboratories) was added. Samples were imaged on a Leica DM5500 B microscope fitted with a Leica CTR5500 electronic box and a Hamamatsu Digital Camera C10600 ORCA R2 with maximally projected images of z-stacks optimized by the Leica LAS AF 3.1.0 software followed by quantification on ImageJ (NIH). Immunofluorescence was done using two to three independent biological replicates.
2.11 Expression of osm-9p::gfpImaging of osm-9p::gfp expression in ADL was assessed as previously done (Sims et al., 2016) except that animals were dye filled with either DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt, Thermo Fisher D7757) (for 2–4 h) or DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, Sigma Aldrich 42364) (for 30 min) (Wallace et al., 2016).
2.12 Octanol avoidance assaysAvoidance assays to the odorant 1-octanol (TCI O0036) by 1-day old adults was done as detailed by Troemel et al. (1997). In short, square petri dishes (Fisher Scientific FB0875711A) containing 2% BD Difco Bacto agar (Fisher Scientific DF0140154) in 5 mM potassium phosphate, 1 mM CaCl2, and 1 mM MgSO4 was divided into six equally sized sectors (A-F) where the two most distal sectors (A and F) from the center served as the sites for the deposition of 1-octanol and the control (100% ethanol) with 1 μL 1 M NaN3 anesthetic pipetted equidistantly near the plate’s edge. One-day adults grown at 20°C in NGM plates seeded with OP50 were collected and washed with S-basal buffer (0.1 M NaCl, 0.05 M potassium phosphate pH 6.0) until the supernatant was clear followed by one rinse in dH2O. Worms (∼200) were placed onto the center (between sectors C and D) of the petri dish equidistantly to sectors A and F where 1 μL of 100% ethanol (sector F) and 1-octanol (sector A) was immediately pipetted. Excess liquid was rapidly removed using a kimwipe without disturbing the animals or puncturing the agar. Assays were placed an undisturbed location at room temperature for 1 h after which worms were assessed for their reaction to octanol using the following formula: Avoidance Index = (A + B)-(E + F)/N; where A, B, E and F are the number of animals in these sectors and N is the total number of animals in all the six sectors.
2.13 Ascr#3 avoidance assaysAvoidance of ascr#3 was determined off-food using 1-day old adults grown at 20°C on NGM plates seeded with OP50 and performed essentially as previously done (Sims et al., 2016) except that forward-moving adults were first tested for their reaction to a drop of benign M13 buffer (30 mM Tris-HCl pH 7.0, 100 mM NaCl, 10 mM KCl) deposited near their tail and migrating up to their nose. 10–12 adults that were indifferent upon encountering M13 buffer were then tested for their behavior when presented with 100 nM ascr#3 and 1 M glycerol. A backward-moving motion consisting of 1.5 body lengths in 4 s was considered an avoidance response. Only animals that were tested in the same timeframe were subjected to statistical comparisons to the wild-type N2 control.
2.14 Chemotaxis assaysChemotaxis assays for the benzaldehyde and diacetyl attractants were performed on 1-day adults as described in (Bargmann et al., 1993; Hart, 2006). Assays plates consisting of 2% BD Difco Bacto agar (Fisher Scientific DF0140154) in 5 mM potassium phosphate, pH 6.0, 1 mM CaCl2, and 1 mM MgSO4 were prepared on 100 mm round petri dishes. Two marks (one for the attractant and one for the 100% ethanol control), 180o degrees opposite each other, were indicated on the bottom of the plate near its edge where 1 μL of 1 M NaN3 anesthetic was deposited. Worms grown at 20°C on OP50-seeded NGM plates were collected and washed with S-buffer (0.1 M NaCl, 0.05 M potassium phosphate pH 6.0) until the supernatant was cleared of their bacteria food followed by one wash with dH2O. Pipette worms (∼200) onto the center of the assay plate, equidistantly from the marks and slightly off center. Excess liquid was quickly soaked off using a kimwipe without disturbing the animals or puncturing the agar. Immediately place 1 μL of the attractant and the ethanol on their respective marks, assay plate was closed with the lid and placed in an undisturbed location at room temperature for 1 h. Worms that were within a 0.5 cm radius of the spot where the attractant and the ethanol were deposited were then counted and the chemotaxis index was calculated with the following formula: Chemotaxis Index (CI) = (number worms at the attractant after 1 h - number worms at control ethanol after 1 h)/total # of worms. Chemotaxis indexes of +1.0 to −1.0 indicated perfect attraction or repulsion, respectively.
2.15 Brood size assaysTen L4 larvae were placed individually onto 35 mm NGM plates seeded with OP50 and transferred daily until egg laying ceased at 20°C. Brood sizes were determined by counting their live progeny as previously described (Ow et al., 2018; Ow et al., 2021).
3 Results3.1 Wild-type adult ADL neurons express germline-enriched genesTo identify additional genes expressed in ADL neurons that may be regulated by passage through dauer, we used fluorescence activated cell sorting (FACS) followed by RNA-Seq to characterize the transcriptome of ADL chemosensory neurons in 1-day old control (CON) and postdauer adults that have experienced dauer resulting from crowding (PDPhe). This experiment was performed using a strain that expressed an integrated extra-chromosomal array of sre-1p::gfp to facilitate isolation of ADL neurons (Supplementary Figure S1). Using an expression cutoff of 10 RPKM (reads per kilobase of transcript per million reads), we identified 5908 and 6375 genes that are expressed in control and PDPhe ADL neurons, respectively (Supplementary Table S2). We compared our set of expressed genes with the ADL transcriptome profiled by the CeNGEN consortium (https://cengen.org) (7217 genes) that generated expression profiles of the 302 neurons comprising the C. elegans L4 larval nervous system in hermaphrodites (Taylor et al., 2021). The ADL CeNGEN dataset overlapped with 62% (3652 genes; p = 1.26e-514) and 65% (4129 genes; p = 6.30e-719) compared to our adult control and PDPhe transcriptomes, respectively (Supplementary Table S3). Although there are significant similarities between our gene lists and the CeNGEN consortium, the difference in transcriptomes may be ascribed to procedural variances or developmental stages assayed. We also compared a previously described pan-neuronal transcriptome of 8436 genes collected from 1-day old adults with our dataset (Kaletsky et al., 2018). We found an 81% (4779 genes; p = 9.75e-1161) and 82% overlap (5240 genes; p = 3.71e-1396) with our control and PDPhe gene lists, respectively (Supplementary Table S4). Based on these similarities, we conclude that our gene lists are an accurate representation of the ADL transcriptome.
We next identified the set of differentially expressed (DE) genes between control and PDPhe ADL neurons. Using an FDR <0.05 cutoff, we identified 116 and 595 genes that were up- and downregulated, respectively, between the two populations (Figure 1A; Supplementary Table S5). From here on, these genes will be called “WT DE Up” or “WT DE Down” to refer to wild-type PDPhe adult genes that are differentially up- or downregulated, respectively, compared to their control counterparts. The osm-9 gene was not present in this group due to its low expression levels, despite being previously validated as significantly downregulated in ADL PDPhe neurons (Sims et al., 2016), suggesting that the genes we identified have robust gene expression changes following the dauer experience. GO term analyses of WT DE Down genes uncovered significant associations with functions related to collagen and cuticle development, endoplasmic reticulum (ER) stress, immune and stress response (Figure 1B; Supplementary Table S6), whereas WT DE Up genes were associated with lipid transport and function, stimuli response, oxidation-reduction process, and metabolic processes (Figure 1C; Supplementary Table S7). The WT DE Up genes include the G protein-coupled receptor (GPCR) genes srh-281, sri-32, srw-89, srz-47, srh-204, srsx-7, srsx-6, and srw-23, as well as insulin-like genes ins-12 and irld-15 (Supplementary Table S5). A comparison with CeNGEN showed that six out of eight of GPCRs and both insulin-like genes were expressed in ADL: sri-32, srw-89, srz-47, srh-204, srsx-7, srsx-6, ins-12, and irld-15 (Taylor et al., 2021). Notably, srh-281, the most highly expressed GPCR gene in our PDPhe gene list is solely expressed in ADL (Vidal et al., 2018), further highlighting the validity our ADL transcriptome (Supplementary Tables S3, S5).
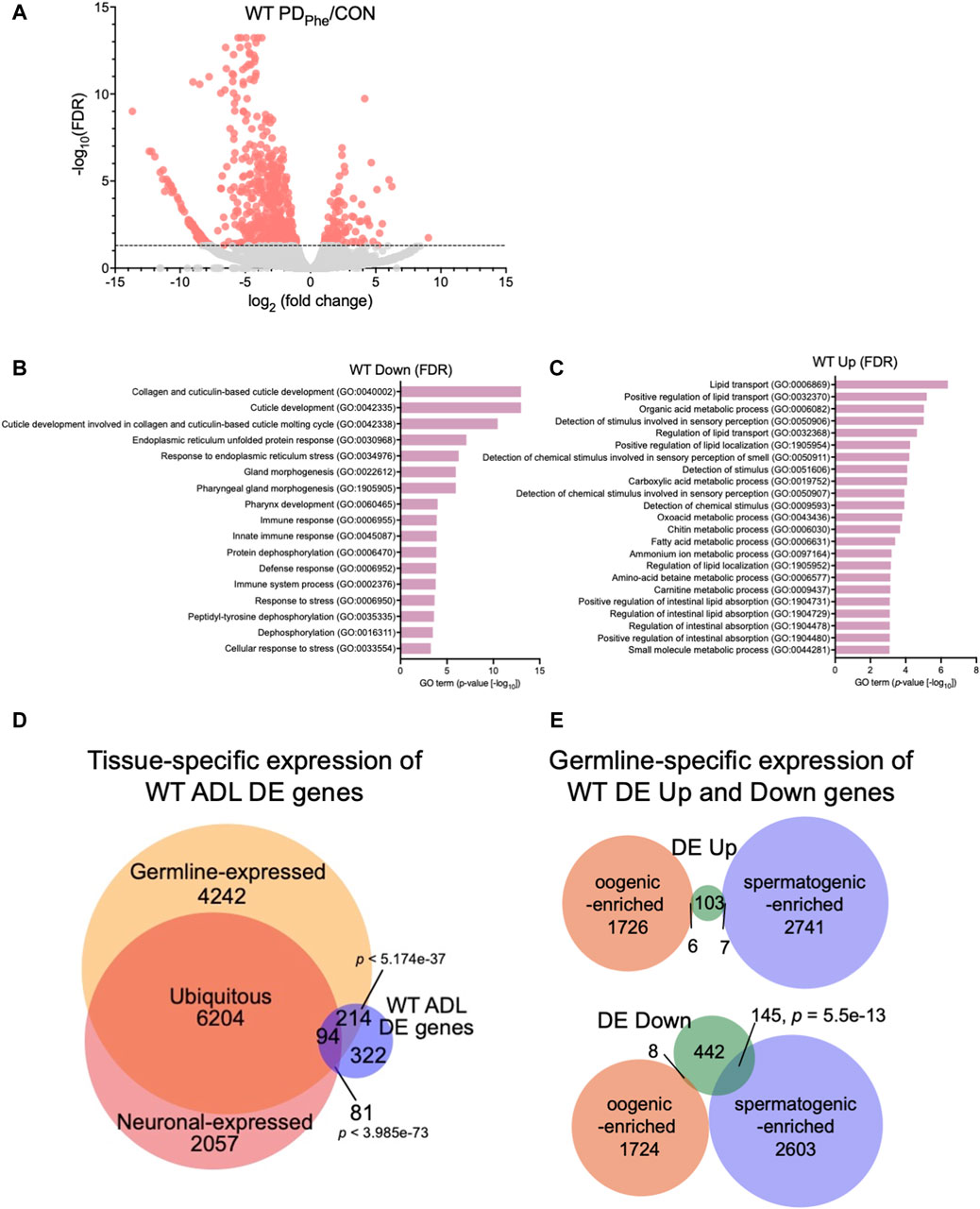
Figure 1. Wild-type adult ADL neurons express germline genes. (A) Volcano plot representing WT CON and PDPhe adult ADL RNA-Seq data. Dotted line indicates FDR cutoff of p < 0.05; salmon dots represent mRNAs that are either significantly upregulated or downregulated in PDPhe compared to CON. (B,C) GO terms of genes that are most significantly (B) decreased or (C) increased based on FDR < 0.05 in WT PDPhe compared to WT CON. (D) Venn diagram comparing all differentially expressed genes in WT PDPhe ADL neurons compared to WT CON ADL neurons (WT ADL DE) to germline (Ortiz et al., 2014) and neuronal expressed gene lists (Kaletsky et al., 2018). (E) Venn diagrams comparing spermatogenic- and oogenic-enriched (Ortiz et al., 2014) gene lists with wild-type differentially expressed up- (DE Up) and downregulated (DE Down) genes. p-values for Venn diagrams were determined using a hypergeometric distribution.
Comparison of the overlap between the 711 WT DE genes with the pan-neuronal transcriptome from 1-day old adults (Kaletsky et al., 2018) found that only 24.6% of the differential expressed genes in ADL were previously shown to be expressed in neurons (175 genes; p = 3.99e-73) (Figure 1D; Supplementary Table S8). The limited overlap between these two gene sets indicates that the vast majority of differentially expressed genes in ADL neurons are not the previously identified genes that function in neuronal plasticity. However, comparison of the ADL DE gene list with a germline-specific gene list (Ortiz et al., 2014), showed that a significant proportion of 43% of ADL DE genes (308 genes; p = 5.17e-37) overlapped with genes known to be germline-expressed (Figure 1D; Supplementary Table S9). To further distinguish between the classes of germline genes that are in common between our ADL-derived gene list and germline-enriched genes, we compared our list of upregulated and downregulated genes with previously identified spermatogenic- and oogenic-enriched genes (Ortiz et al., 2014). For upregulated genes in ADL, 7 genes were in the spermatogenic-enriched gene list (2.3-fold under enriched; p = 0.008) while 6 genes were in the oogenic-enriched gene list (1.7-fold under enriched; p = 0.12) (Figure 1E; Supplementary Table S10). For the downregulated genes, 145 spermatogenic genes (1.8-fold enriched; p = 5.5e-13) and 8 oogenic genes (6.4-fold under enriched; p = 1e-14) overlapped with both gene lists (Figure 1E; Supplementary Table S10). The 145 downregulated spermatogenic enriched genes included genes encoding for two major sperm protein domain-containing genes (msd-3/-4) and 21 (msp-3/-10/-19/-31/-36/-38/-45/-49/-50/-51/-55/-56/-64/-65/-77/-78/-79/-81/-113/-152) of the 48 predicted major sperm protein genes expressed in C. elegans (http://www.wormbase.org, release version WS287) (Figure 1E; Supplementary Table S10). These comparisons indicate that ADL neurons express hundreds of genes that have been previously considered to be exclusively expressed in the germ line. While the number of oogenic genes expressed in ADL is greater that spermatogenic genes, the spermatogenic genes, such as MSPs, are specifically targeted for downregulation in postdauer ADL neurons similar to osm-9 (Figures 1D,E; Supplementary Tables S9, S10). Collectively, these results indicate that a subset of germline genes are expressed in ADL neurons and are targeted for differential expression based early-life experiences.
3.2 NRDE-3 is required for differential expression of major sperm protein genes in ADLWe showed previously that osm-9 downregulation in PDPhe ADL neurons requires components of the somatic nuclear RNAi pathway, including NRDE-3, NRDE-1, and NRDE-4 (Sims et al., 2016). To examine whether the somatic nuclear RNAi complex plays a broader role in programming an animal’s mRNA transcriptome in response to early-life environmental experience, we set out to identify additional targets of NRDE-3 AGO by performing RNA-Seq of control and PDPhe ADL neurons collected from the nrde-3(gg66) mutant strain. Using an expression cutoff of RPKM >10, we identified 6001 and 6377 genes that are expressed in the ADL neurons of control and PDPhenrde-3 adults, respectively (Supplementary Table S11). Comparison of the transcriptomes from wild-type and nrde-3 control ADL neurons resulted in a 77.7% similarity (5206 genes) (p = 5.17e-3045) (Supplementary Table S12). Similarly, the transcriptomes of the wild-type and nrde-3 PDPhe neurons shared an 87.5% similarity (5951 genes) (p = 4.07e-3939) (Supplementary Table S12). Differential expression analysis of ADL genes in the nrde-3 mutants revealed 105 genes that are downregulated and have functions in signal transduction, G protein-coupled receptor signaling, dauer entry, and detection of stimuli (Figures 2A,B; Supplementary Table S13). In addition, we found 96 genes that are upregulated in nrde-3 PDPhe ADL neurons compared to controls with functions in endoplasmic reticulum stress response pathways, organic molecule transport, and histidine metabolism (Figures 2A,C; Supplementary Table S14).
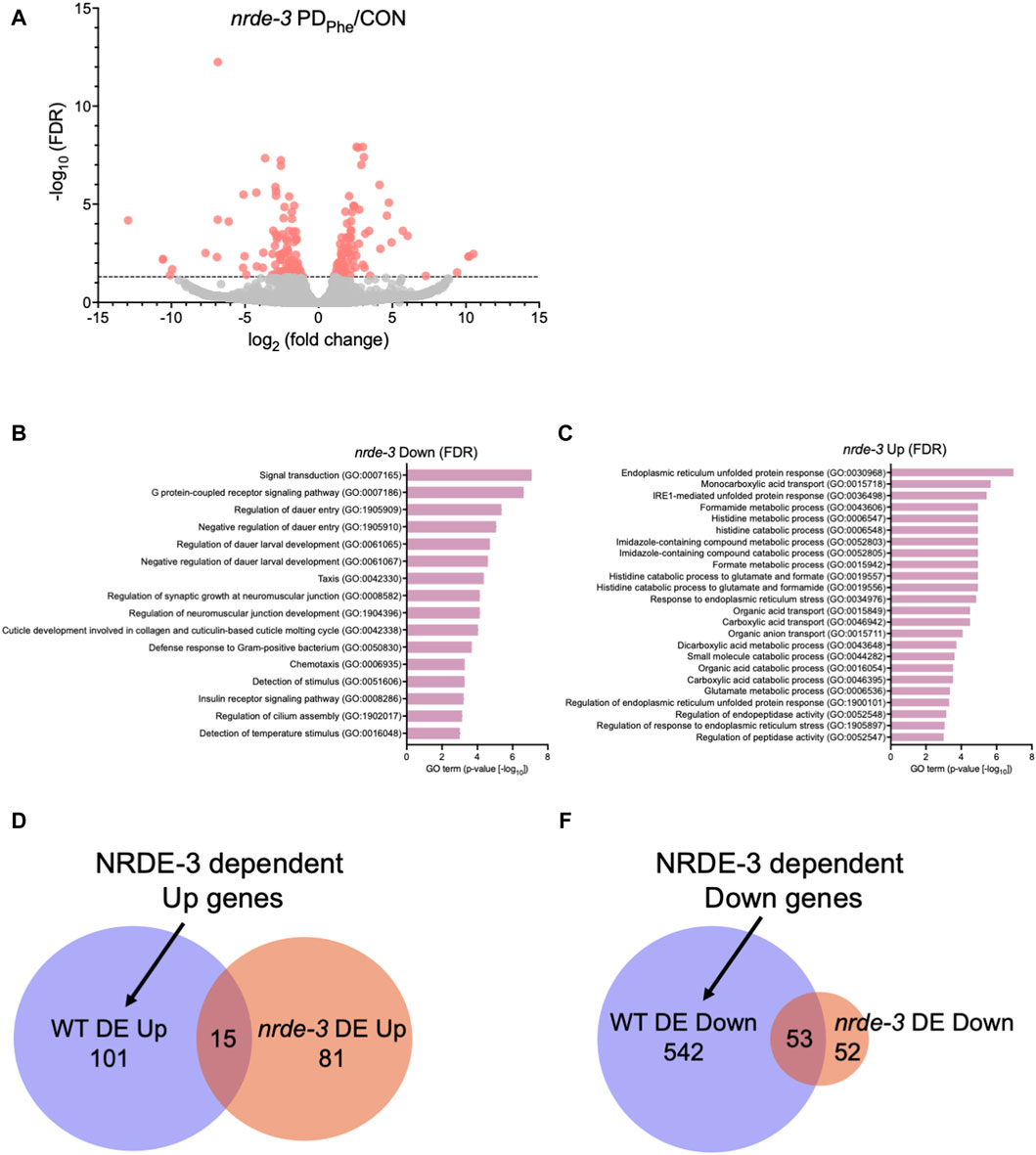
Figure 2. The NRDE-3 somatic nuclear RNAi pathway is required for differential expression of ADL genes. (A) Volcano plot representing nrde-3(gg66) CON and PDPhe adult ADL RNA-Seq data. Dotted line indicates FDR cutoff of p < 0.05; red dots represent mRNAs that are either significantly up- or downregulated in PDPhe compared to CON. (B,C) GO terms of genes that are most significantly (B) decreased or (C) increased based on FDR <0.05 in nrde-3 PDPhe compared to nrde-3 CON. (D,E) Venn diagrams comparing WT and nrde-3 gene lists for (D) DE Up and (E) DE Down genes.
Next, we identified which genes were dependent on NRDE-3, either directly or indirectly, for their differential expression in wild-type ADL neurons. First, we compared the 116 genes upregulated in wild-type PDPhe ADL neurons with the 96 genes that are upregulated in nrde-3 PDPhe ADL neurons (Supplementary Tables S5, S15). We found only 15 genes overlapping between the two gene lists, indicating that 101 genes require NRDE-3 for their upregulation in PDPhe ADL neurons in wild-type adults (Figure 2D; Supplementary Table S16). Amongst this gene set were 33 (32.7%) secreted protein genes, 26 (25.7%) transmembrane domain genes, 10 (9.9%) serpentine receptor genes involved in chemoreception, and 10 (9.9%) lipid metabolism genes (Suh and Hutter, 2012; Zhang et al., 2013). The lipid metabolism genes included the Rieske-like oxygenase daf-36 important for cholesterol metabolism, Δ9 desaturase fat-7 involved in lipid metabolism, and six vitellogenins, vit-1/-2/-3/-4/-5/-6, that have known functions to transport lipids into the germ line (Ow et al., 2021). We then compared the 595 downregulated ADL genes in wild-type PDPhe adults to the 105 genes that are differentially downregulated in nrde-3 mutants. We found 53 genes overlapping between the two gene lists, indicating that 542 downregulated genes in wild type ADL neurons were dependent on NRDE-3 function for their differential expression (Figure 2E; Supplementary Tables S5, S15, S17). Notably, sperm-expressed genes were members of this gene set, including 20 msp genes and the two PP1 phosphatases responsible for sperm development and motility, gsp-3 and gsp-4 (Wu et al., 2012). Additional genes in this group include 81 collagen genes (14.9%), 273 genes (50%) predicted to encode secreted proteins, 77 genes (14.2%) encoding transmembrane domain proteins, and 58 endoplasmic reticulum (ER) genes (10.7%) (Suh and Hutter, 2012), the latter of which include those in ER unfolded protein response and are amongst the most significantly upregulated genes present in nrde-3 PDStv adults. Also included is the odr-7 gene, required for the function of the AWA chemosensory neurons in a germline-dependent neural circuitry response to the odorant diacetyl during development (Sengupta et al., 1994; Fujiwara et al., 2016). Together, these results show that NRDE-3 plays a significant role in the regulation of genes in ADL neurons, particularly after passage through dauer due to crowding. The NRDE-3-dependent differentially expressed genes included those with functions related to fertility, reproduction, odor response, and fat metabolism, which are phenotypic traits that we have previously observed to be significantly altered in adults that experienced dauer (Sims et al., 2016; Ow et al., 2018; Ow et al., 2021).
3.3 A conserved upstream regulatory sequence is enriched for genes with NRDE-3 dependent expression changesIn C. elegans, neuronal gene expression is modular in that the presence and arrangement of transcription factor binding sites in the upstream regulatory sequences can specify in which neurons that gene is transcribed (Stefanakis et al., 2015; Reilly et al., 2020; 2022; Hobert, 2021; Leyva-Díaz and Hobert, 2022). We next asked if ADL-expressed germline genes have the hallmarks of neuronal gene expression in their upstream regulatory sequences. C. elegans expresses over 40 MSP genes, and 20 of these genes exhibited differential expression in our ADL gene set (Supplementary Table S10). Previously, an ADL-enhancer element consisting of the CASCTG E-box motif (S=C or G) was found by analyzing promoters of ADL-expressed chemoreceptor genes (McCarroll et al., 2005). A similar motif of CACGTG was identified as overrepresented in msp promoter regions (McCarroll et al., 2005). While the modified E-box CACGTG motifs were present in 7 of the MSP genes expressed in ADL (msp-19, -36, -55, -56, -64, -142, -152), we also identified 1 to 3 copies of the ADL specific CASCTG E-box motif in the 5’ UTR sequences of 11 MSP genes (msp-10, -19, -31, -38, -45, -50, -51, -55, -77, -81, -113) (Figure 3A). These observations underscore the possibility that msp expression in ADL could be dictated at the transcriptional level for ADL specification.
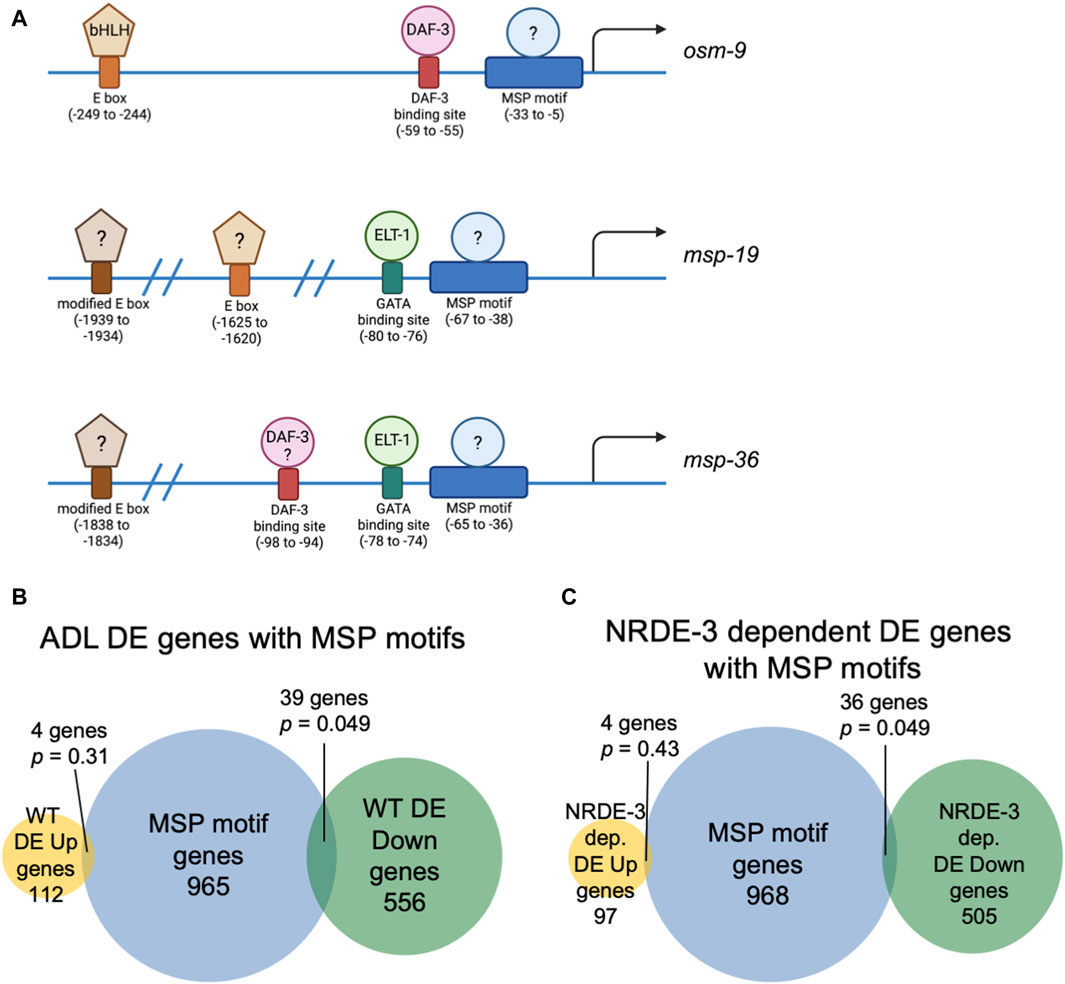
Figure 3. Genes with NRDE-3-dependent expression changes are enriched for a conserved sequence in their upstream regulatory regions. (A) Diagrams of upstream regulatory regions of osm-9, msp-19, and msp-36 genes. Names of sequence motifs, location relative to translational start sites, and known interactors are indicated. (B,C) Venn diagrams comparing the set of genes containing an MSP motif in their 5′ UTRs with (B) all WT and (C) NRDE-3 dependent DE Up and DE Down gene lists. p-values were determined using a hypergeometric distribution.
We next asked whether the differential regulation of gene expression in ADL is also dependent upon upstream regulatory sequences. We previously identified a cis-acting motif located in the osm-9 promoter, referred to as the “PD motif”, that was required for its transcriptional silencing in ADL of PDPhe adults (Sims et al., 2016). This motif is composed of a DAF-3/SMAD binding site (GTCTA) and a ∼30 base pair conserved sequence (CT[A/T]TAA[A/T]TTN0-3[A/C]AN0-2TTTTG[C/T]CATAA[T/A]C[T/C]) that is responsive to TGF-ß signaling (Sims et al., 2016; Pekar et al., 2017). While >6000 genes possess the DAF-3 binding site in their upstream regulatory regions, 1008 genes contain the conserved sequence, and 116 genes the full PD motif (Supplementary Table S18). Interestingly, nine genes, in addition to osm-9, that were downregulated in WT postdauer ADL neurons compared to controls contain a full PD motif: msd-3, msp-36, msp-51, msp-65, msp-81, msp-79, col-133, B0205.14, and T16G1.2 (Supplementary Tables S5, S18). Since five of these genes were MSP genes, we next examined the overlap of differentially expressed genes in ADL and the list of genes containing just the conserved sequence in their upstream regulatory regions. For all genes differentially expressed in ADL, we found an overlap of 4 and 39 genes for the up- and downregulated gene lists, respectively (Figure 3B). A comparison of genes that exhibit NRDE-3-dependent differential expression in ADL with the list of genes with conserved sequences revealed a similar overlap of 4 and 36 genes for the up- and downregulated gene set, respectively (Figure 3C; Supplementary Tables S16–S18). The downregulated genes with conserved sequences included 19 MSP genes (msp-3, -10, -19, -31, -36, -38, -45, -49, -50, -51, -55, -56, -65, -77, -78, -79, -81, -113, -152). Future work will determine if the downregulation of MSP genes in postdauer ADL neurons may be attributed to binding of an unidentified factor to the conserved sequence in their promoter (Figure 3A). Given the enrichment of the conserved sequence in MSP gene regulatory sequences, we will henceforth refer to this ∼30 base pair conserved sequence element in the PD motif as the “MSP motif”.
3.4 Major sperm proteins are expressed in ADL neuronsSince the expression of spermatogenic genes in wild-type ADL neurons was unexpected, we sought to validate the expression of msp genes in ADL. First, we performed single molecule fluorescence in situ hybridization (smFISH) to detect the presence of msp mRNAs in wild-type and nrde-3 control adults carrying the sre-1p::gfp transgene expressed in ADL neurons (Supplementary Figure S1). Our RNA-Seq results showed that wild-type control adults express msp-19 at an 11-fold higher level than in nrde-3 control ADL neurons (RPKM = 129.91 vs. RPKM = 11.61, respectively) (Supplementary Table S19); thus, we chose msp-19 as the template for smFISH probes design. However, because of the high sequence homology of the msp genes (Kasimatis and Phillips, 2018), it is likely that most, if not all, msp mRNAs were detected using our probes. Using smFISH, we detected msp-19 mRNAs in wild-type control ADL neurons (Figures 4A,B; Supplementary Figure S2). Consistent with our RNA-Seq results, the msp-19 signal was significantly decreased in nrde-3 mutants, validating that NRDE-3 is required for the expression of msp mRNA in control ADL neurons (Figures 4A,B; Supplementary Figure S2). As a positive control, we also performed smFISH with probes detecting gfp mRNA, and we observed no significant differences in gfp fluorescence intensity in ADL neurons of wild-type and nrde-3 control adults (Figure 4A; Supplementary Figure S2).
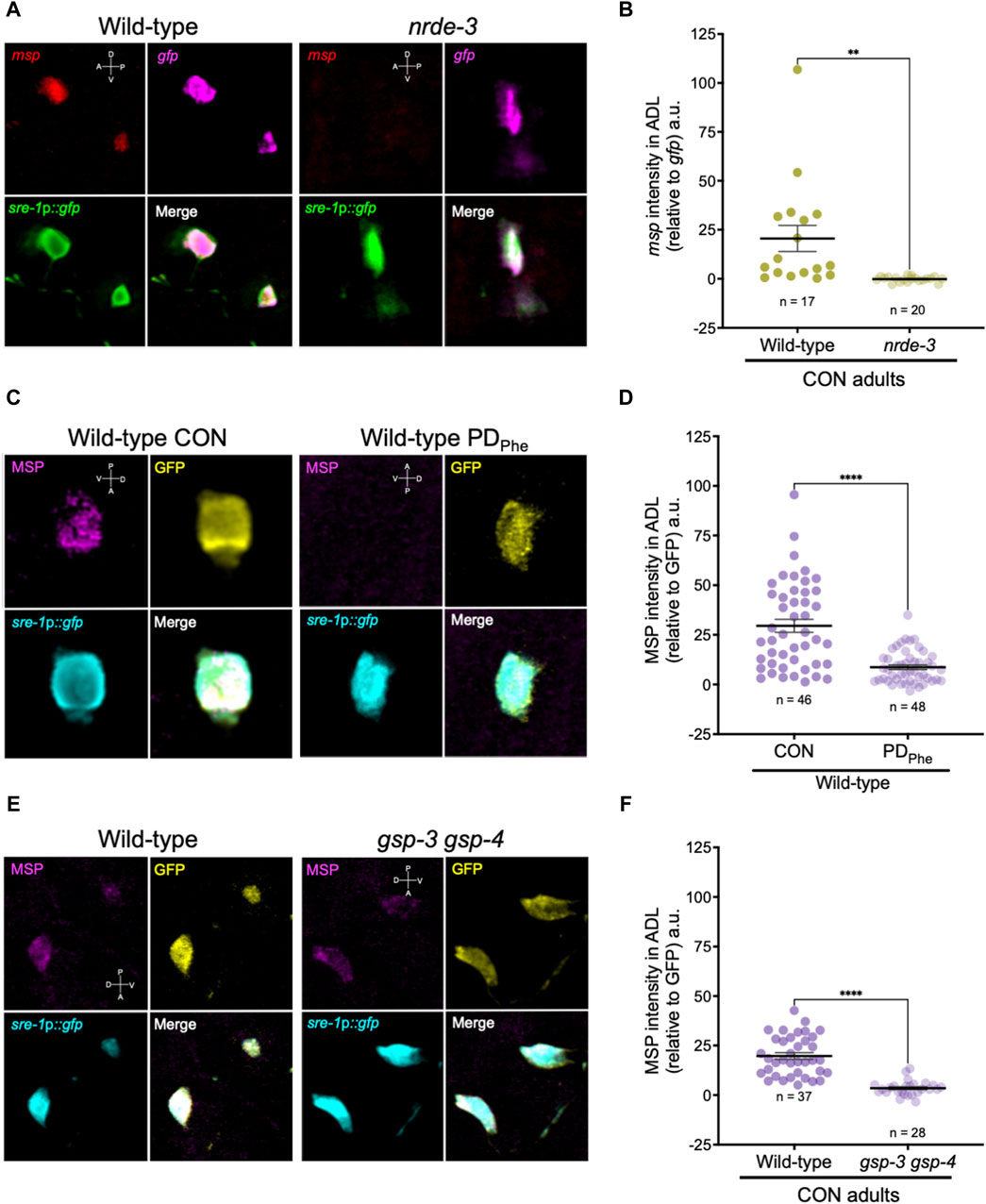
Figure 4. Major sperm proteins are expressed in ADL neurons. (A) Representative images of smFISH of ADL neurons with probes against msp (red) and gfp (magenta) mRNAs using 1-day old wild-type and nrde-3(gg66) CON adults. sre-1p::gfp transgene fluorescence shown in green. Images of animal heads shown in Supplementary Figure S2. (B) Quantification of msp relative to gfp smFISH intensity in ADL; a.u. are arbitratry units. ** p < 0.01, Student’s t-test of three independent experiments. (C) Representative images of immunostainings using antibodies against MSP (magenta) and GFP (yellow) on 1-day old wild-type CON and PDPhe adults. sre-1p::gfp transgene fluorescence shown in cyan. Images of animal heads are shown in Supplementary Figure S3. (D) Quantification of MSP immunofluorescence intensity relative to the intensity of GFP immunofluorescence. **** p < 0.0001, Student’s t-test of two independent experiments. (E) Representative images of immunostainings using antibodies against MSP (magenta) and GFP (yellow) on 1-day old wild-type and gsp-3(tm1647) gsp-4(y148) CON adults. sre-1p::gfp transgene fluorescence shown in cyan. Images of animal heads are shown in Supplementary Figure S4. (F) Quantification of MSP immunofluorescence intensity relative to the intensity of GFP immunofluorescence. **** p < 0.0001, Student’s t-test of three independent experiments. Number of animals are indicated by n. D, V, A, P represent the dorsal, ventral, anterior, and posterior orientation of the animal. Additional data included in Supplementary Table S20.
Next, we examined whether the msp mRNAs were being translated by performing indirect immunofluorescence in ADL neurons of wild-type control and PDPhe adults expressing the sre-1p::gfp transgene. Using a monoclonal antibody that recognizes MSP-19, -31, -40-, 45, -50, -51, -53, -59, -61, -65, -81, -113, and -142 (Developmental Studies Hybridoma Bank, University of Iowa, United States; 4A5), we found that control ADL neurons expressed detectable MSPs, and PDPhe ADL neurons exhibited a significant downregulation of MSP (Figures 4C,D; Supplementary Figures S3A, S3C). As a positive control, we also performed indirect immunofluorescence with anti-GFP antibody and observed similar levels of GFP fluorescence intensity between wild-type control and PDPhe ADL neurons. As an additional control, we verified that MSP staining was visible in both control and PDPhe adult gonads (Supplementary Figures S3B, S3D). Overall, these results demonstrate that MSPs are expressed in ADL neurons of continuously developed hermaphrodites in a NRDE-3 dependent manner and are downregulated specifically in ADL neurons of adults that experienced crowding-induced dauer.
3.5 Sperm-expressed phosphatase is required for attraction and avoidance behaviorsAfter verifying the expression of germline genes in ADL neurons, we next sought to determine what role they play in ADL function. The msp genes comprise a 48-membered gene family with highly conserved sequences that encode 14 kilodalton (kDa) proteins (Scott et al., 1989). MSPs are the most abundant protein in nematode sperm, comprising ∼17% of their total protein and concentrating up to 3-fold in their sole locomotive structure, the pseudopod, compared to the cell body (Klass and Hirsh, 1981; Ward and Klass, 1982; King et al., 1992; Smith, 2014). Nematode sperm lack a flagellum, and MSPs substitute for a function that would otherwise be performed by structural proteins such as actin and myosin by forming a pseudopod (Nelson and Ward, 1981; Nelson et al., 1982; Roberts and Stewart, 1995; 2000; Bottino et al., 2002). MSPs polymerize into filaments at the leading edge of pseudopod in mature spermatozoa and disassemble at its trailing edge in an assembly-disassembly mechanism results in a treadmill-like form of locomotion (Sepsenwol et al., 1989; Sepsenwol and Taft, 1990; Roberts and King, 1991; King et al., 1994; Smith, 2014). In addition to enabling sperm movement, MSPs are also extracellular signaling molecules secreted by sperm that function as hormones to trigger oocyte maturation and ovulation, despite them lacking sequence motifs associated with secretory proteins (Miller et al., 2001; 2003; Kosinski et al., 2005). Although originally discovered in nematodes, the phylogenetic scope of proteins with MSP-like domains extends from plants to humans (Burke and Ward, 1983; Skehel et al., 1995; Laurent et al., 2000; Skehel et al., 2000).
Mature sperm are considered transcriptionally and translationally dormant on account of their extremely compacted protamine-bound haploid genome and cytoplasmic shedding during the terminal stage of spermatogenesis; thus, mechanisms involving post-translational modifications are important for regulating fertility (Hecht, 1998). In mammals, the serine/threonine PP1 phosphatases play critical roles in modulating sperm development and locomotion (Fardilha et al., 2011; Ferreira et al., 2022). C. elegans has two serine/threonine PP1 phosphatases, GSP-3 and GSP-4, which are required for MSP filament disassembly, and a double mutation in gsp-3 and gsp-4 results in sterility due to chromosome segregation defects and sperm immobility (Wu et al., 2012). Because of the technical difficulty to inactivate ADL-expressed msp genes simultaneously, we first investigated the potential of MSP function in ADL by characterizing the behavioral phenotypes of gsp-3(tm1647) and gsp-4(y148) mutants. Like the msp genes, gsp-3 and gsp-4 are only expressed in wild-type control ADL neurons in a NRDE-3 dependent manner (Supplementary Table S17). First, we examined MSP levels in a gsp-3 gsp-4 double mutant strain expressing the sre-1p::gfp transgene and found that MSPs continue to be detected in ADL neurons, albeit at significantly lower levels than wild-type adults (Figures 4E,F; Supplementary Figures S4A, S4C). Previous work showed that GSP-3 and GSP-4 do not affect the transcription of msp genes in sperm (Supplementary Figures S4B, S4D) (Wu et al., 2012); thus, the observed decrease in MSP in the gsp-3 gsp-4 mutant strain may be due to their effects on stability of the protein. Next, we asked whether GSP-3 and GSP-4 play a role in regulating the expression of additional genes by examining the fraction of ADL neurons that exhibit GFP due to expression an osm-9p::gfp transcriptional reporter transgene. We showed previously that in wild-type adults carrying the osm-9p::gfp transgene, GFP was expressed in ADL neurons (ADLON) of control adults but was significantly downregulated in PDPhe adults (ADLOFF), while the expression in AWA neurons remained unaffected (Figures 5A,B) (Sims et al., 2016). While gsp-4 control and PDPhe adults exhibited similar GFP expression as wild-type ADL neurons, gsp-3 control adults had a significant reduction of animals expressing GFP, resulting in the loss of osm-9p::gfp expression difference between wild-type control and PDPhe ADL neurons (Figure 5B). Our results are consistent with GSP-3, potentially through the action of MSPs, being required to regulate gene expression in ADL neurons of continuously developed hermaphrodites.
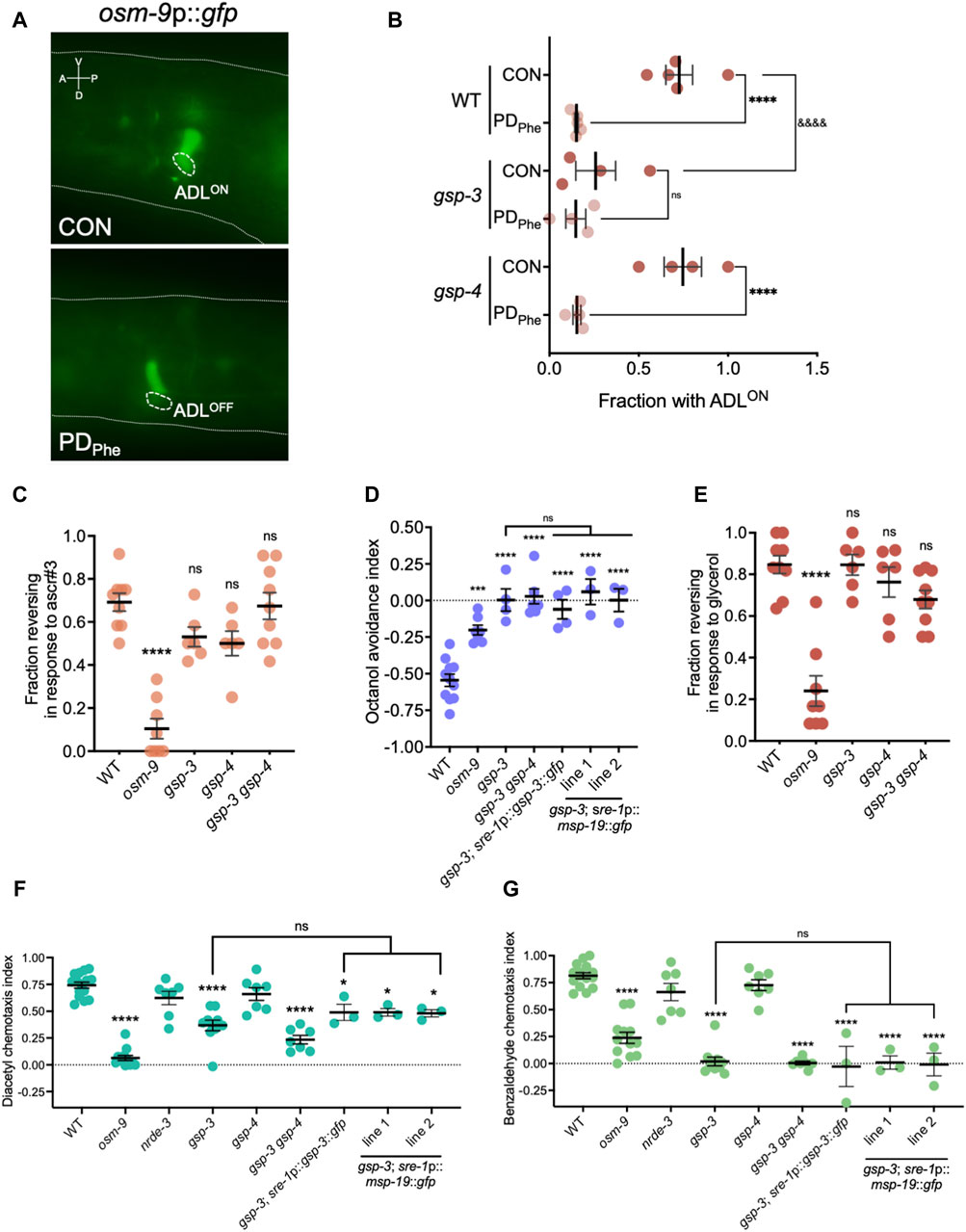
Figure 5. Sp
留言 (0)