Hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors, known as statins, have been used as first-choice drugs for the primary and secondary prevention of cardiovascular diseases (Adhyaru and Jacobson, 2018). Numerous studies and guidelines have emphasized the importance of statin use. A recent meta-analysis reported that the use of statins in patients with increased cardiovascular disease risk reduced their risk for all-cause mortality by 0.9-fold and cardiovascular events by 0.7-fold (Chou et al., 2022). The 2019 American College of Cardiology/American Heart Association guideline recommended that patients with high atherosclerotic cardiovascular disease (ASCVD) risk should be treated with statins to reduce the risk. High-intensity statin therapy should be initiated to prevent ASCVD when adult patients’ low-density lipoprotein (LDL) cholesterol levels are not less than 190 mg/dL (Arnett et al., 2019).
Statin therapy is effective and tolerable; however, problems with statin toxicity remain. Of all adverse events related to statins, statin-related myotoxicity (SRM) is the most common and occurs in approximately 10%–25% of cases (Vinci et al., 2021). SRM phenotypes vary, ranging from mild creatine kinase (CK) elevation to severe rhabdomyolysis. SRM also includes myalgia, muscle cramps, myopathy, and immune-mediated statin myopathy (Turner and Pirmohamed, 2019).
There are several hypotheses of musculoskeletal adverse events. Several studies suggested that mitochondrial dysfunction and coenzyme Q 10 (CoQ10) depletion caused by statins may be associated with SRM (De Pinieux et al., 1996). CoQ10 is produced by the mevalonate pathway and functions as an electron carrier in the mitochondrial electron transfer system, which protects against reactive oxygen species (Dohlmann et al., 2022). Because statins inhibit HMG-CoA reductase, which participates in the mevalonate pathway, CoQ10 levels might become decreased in the muscles leading to adverse events like muscle cramps, myalgia, and others (Kennedy et al., 2020).
CoQ10 is synthesized step by step via the mevalonate pathway and numerous genes are required for its biosynthesis (Doimo et al., 2014). Several studies analyzed the association between CoQ10-related genetic variants and various diseases induced by decreased CoQ10 levels (Mantle et al., 2023). Among them, COQ2 rs4693075 was mainly studied, but the previous studies were limited and controversial (Oh et al., 2007; Puccetti et al., 2010; Carr et al., 2013; Hubacek et al., 2017; Bakar et al., 2018; Ramakumari et al., 2018; Chowdhury et al., 2019). Moreover, no study has investigated the effect of other CoQ10-related genetic variants on SRM. Therefore, this study aimed to identify the association between SRM and genetic factors related to CoQ10 biosynthesis including COQ2, COQ3, COQ5, COQ6, and COQ7 in Korean stroke patients receiving statins.
Materials and methodsStudy patientsWe performed a retrospective analysis of prospectively collected samples from February to May 2021 at Ewha Womans University Seoul Hospital and Ewha Womans University Mokdong Hospital. This study was approved by the Institutional Review Board (IRB) of each hospital in agreement with the 1975 Declaration of Helsinki and its later amendments (IRB numbers: 2020-11-014 and 2021-02-026, respectively) Written informed consent was obtained from all patients before enrollment.
The inclusion criteria of patients of this study were those aged not less than 20 years old who had been treated with statins (atorvastatin, rosuvastatin, pitavastatin, pravastatin, or simvastatin) for the secondary prevention for ASCVD after stroke. Patients administered statins for at least 4 weeks were included in the control group. Cases of myotoxicity were defined by the following criteria: (Adhyaru and Jacobson, 2018): intolerable myalgia with CK < 4 × upper limit of normal (ULN) (SRM 2); (Chou et al., 2022); myopathy with a CK level between 4 × ULN and 10 × ULN (SRM 3); or (Arnett et al., 2019) severe myopathy with a CK level between 10 × ULN and 50 × ULN (SRM 4) (Alfirevic et al., 2014). The ULN for CK levels was 120 U/L (Neal et al., 2009). Patients were excluded if they met the following criteria: (Adhyaru and Jacobson, 2018): they did not have muscle symptoms with CK elevation less than 4 × ULN (SRM 0), (Chou et al., 2022), had tolerable myalgia (SRM 1), (Arnett et al., 2019), had CK not less than 4 × ULN at baseline, (Vinci et al., 2021), had elevated CK or muscle pain within 7 days of other illnesses (surgery, cancer, or heart disease like myocardial infarction), or (Turner and Pirmohamed, 2019) their DNA samples were insufficient for analysis.
We collected data from electronic medical records, including patients’ sex, age, weight, body mass index, total cholesterol, triglyceride, low-density lipoprotein, high-density lipoprotein, CK, smoking, alcohol use, comorbidity, concomitant drugs, and class of statin.
Genotyping methodsFive candidate genes (COQ2, COQ3, COQ5, COQ6, and COQ7) were selected to investigate the relationship between CoQ10-related genetic associations and SRM. A total of 37 single nucleotide polymorphisms (SNPs) were chosen based on previous findings (Acosta et al., 2016; Stefely and Pagliarini, 2017; Cunningham et al., 2022), and minor allele frequencies and linkage disequilibrium were determined in Asian populations (Barrett et al., 2005; Ward and Kellis, 2016). We excluded SNPs having the relationship of LD in Asian populations (r2 ≥ 0.8) based on HaploReg.
DNA was extracted from patient saliva or blood. We extracted the DNA from blood samples with the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) or from saliva with OraGene-600 kits (DNA Genotek, OTT, Canada) and PrepIT reagents (DNA Genotek, OTT, Canada). All SNPs were identified as dbSNP rsID and analyzed by TaqMan genotyping assay. The TaqMan allele discrimination technique was used to perform RT-PCR on ABI 7300 instrument (Applied Biosystems, Carlsbad, CA, United States of America). The PCR was performed in a 25 μL optical 8-cap strip containing 0.2 ng/μ L of DNA samples and 13.75 μL of PCR mix. The PCR reagent mixture included 12.5 μL of the TaqMan Genotyping Master Mix and 1.25 μL of the 20X TaqMan SNP Genotyping Assay Mix (Applied Biosystems in Foster City, California, United States of America). The catalog number of used assay was 4351379. Ten minutes after denaturing at 95°C, the PCR was run for 15 s at 92 °C for 40 cycles and 60 s at 60°C for 40 cycles.
Statistical analysisThe chi-squared and Fisher’s exact tests were used to compare the categorical variables between patients who underwent SRM and those who did not. The unpaired t-test was used to compare continuous variables. Multivariable logistic regression analysis with backward elimination was used to identify the independent risk factors for SRM using variables with p < 0.05 in the univariate analysis, including sex and age. Two models were constructed using demographic factors only (model I), and demographic and genetic factors (model II). The unadjusted and adjusted odds ratios (ORs) with 95% confidence intervals (CI) were calculated from univariate and multivariable analyses, respectively. Haplotype analysis was carried out on gene SNPs exhibiting significance in the multivariable analysis using Haploview software (version 4.2; Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, United States of America). The Hosmer-Lemeshow goodness-of-fit test was performed for the fit of the prediction model. The discrimination of the model was evaluated further by calculating the area under the receiver operating characteristic curve (AUROC). Sensitivity analysis was conducted to evaluate outcomes by adding patients with SRM 0 or SRM 1 into the case group. All statistical analyses were performed using R software (version 4.2.2; R Foundation for Statistical Computing, Vienna, Austria). p < 0.05 was considered statistically significant.
ResultsA total of 801 stroke patients were enrolled in the study (Figure 1). We excluded 91 patients who did not have muscle symptoms with CK elevation or had tolerable myalgia without CK elevation. Seven patients were excluded because their baseline CK levels were not less than 480 U/L. Eleven patients were excluded because their CK levels were elevated by other medical issues including myocardial infarction and surgical procedures. We also excluded four patients as their DNA samples were insufficient for analysis. As a result, 688 patients were included in the analysis. Of the included patients, 56 experienced statin-associated muscle symptoms. Twelve patients underwent myopathy (SRM 3) after statin treatment, and two patients experienced severe myopathy (SRM 4). Forty-two patients had intolerable myalgia (SRM 2). The remaining 632 patients who never experienced SRM were classified as the control group.
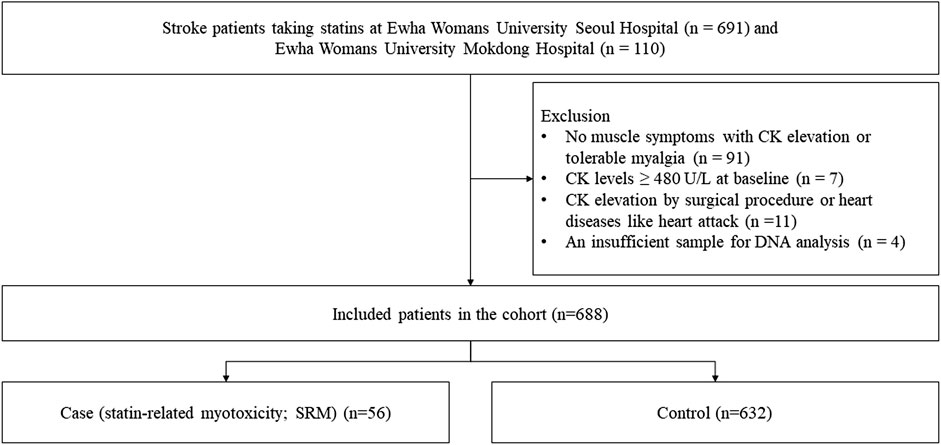
Figure 1. Flow chart of patient selection.
In the case group comprising 56 patients experiencing SRM, three patients (5.4%) discontinued statin therapy, while 27 patients (48.2%) transitioned to alternative statins. Among the nine patients initially prescribed atorvastatin, three switched to rosuvastatin, five to pitavastatin, and one to pravastatin. Within the cohort of 15 patients receiving rosuvastatin, nine shifted to atorvastatin, two to pitavastatin, and one to pravastatin. Of the three patients using pitavastatin, one transitioned to atorvastatin, one to rosuvastatin, and the remaining patient to pravastatin. All patients exhibited good tolerance to the substituted statins. Moreover, seven patients (12.5%) in total underwent dose reduction of statins, with six patients demonstrating good tolerance to the adjusted doses. In the control group, none experienced SRM, and all participants exhibited good tolerance to statin therapy.
The demographic and clinical characteristics of the included patients are presented in Table 1. The mean age of the study population was 63.1 years, and male patients comprised 69.6% of the cohort. The most administered statin was atorvastatin (53.3%), followed by rosuvastatin (39.0%). The duration of therapy in the SRM group was shorter than in the control group (520.1 days vs. 828.5 days, p = 0.004). The most co-medicated drugs were antiplatelet drugs (84.3%) followed by angiotensin-converting enzyme inhibitors or angiotensin II receptor blockers (52.6%). Statin types, ezetimibe, and diuretics were significant factors for SRM. The CK values according to genotypes are presented in the Supplementary Table S1.
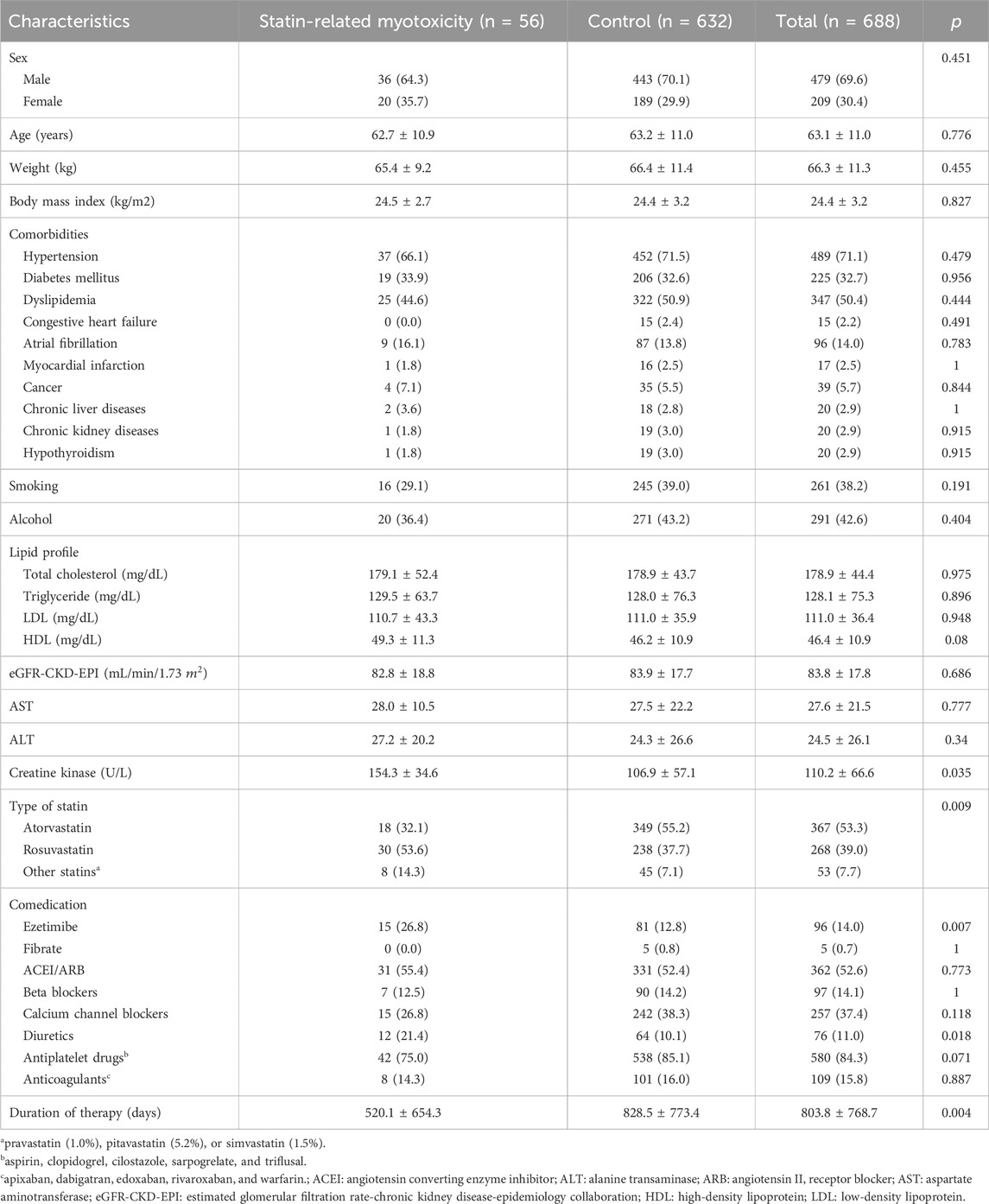
Table 1. Baseline characteristics of patients included in this study.
Table 2 shows the association between CoQ10-related gene SNPs and SRM. Wild-type allele (G) carriers of COQ2 rs4693075 had a higher risk of SRM than mutant-type homozygote carriers (CC) (13.2% vs. 6.3%, p = 0.006). Regarding COQ3 genes, rs11548336 was significantly associated with statin-associated muscle symptoms. Mutant-type allele (C) carriers of rs11548336 had a lower risk of SRM than wild-type homozygote (TT) carriers (6.5% vs. 12.0%, p = 0.023). Variant allele (A) carriers of COQ5 rs10849757 were more associated with SRM risk than wild-type homozygote carriers (GG) (9.7% vs. 3.9%, p = 0.024).
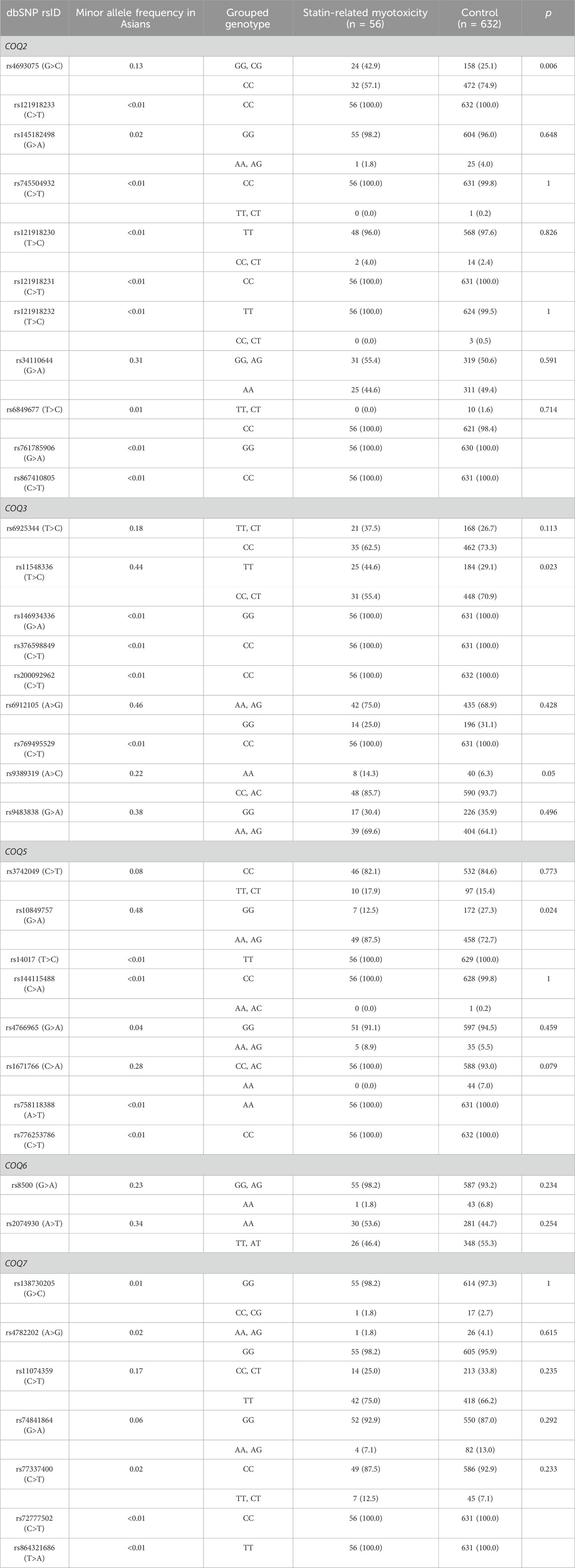
Table 2. Effects of COQ2, COQ3, COQ5, and COQ7 grouped genotypes on statin-related myotoxicity.
Two models were constructed for multivariable logistic regression analysis using factors with p < 0.05 in the univariate analysis, along with age and sex (Table 3). Model I was constructed based on clinical factors only, and model II was based on clinical factors and genetic factors. For model 1, patients treated with rosuvastatin and other statins (pravastatin, pitavastatin, or simvastatin) had a 2.2- and 3.1-fold higher risk of myotoxicity than those treated with atorvastatin after covariates were adjusted, respectively. Among concomitant drugs, ezetimibe and diuretics had more than 2-fold higher SRM risk than those who were not. As shown in model II, statin type and ezetimibe remained significant factors even after adjusting for genetic factors. Among the SNPs studied, COQ2 rs4693075, COQ3 rs11548336, and COQ5 rs10849757 were significantly associated with SRM risk. Patients carrying the G allele of rs4693075 experienced a 2.9-fold increase in SRM compared with those carrying the CC genotype (95% CI: 1.6–5.3). Patients carrying the TT genotype of rs11548336 and A allele of rs10849757 had a 1.9-fold (95% CI: 1.1–3.5) and 3.3-fold (95% CI: 1.5–8.3) higher risk of muscle-related toxicity, respectively.
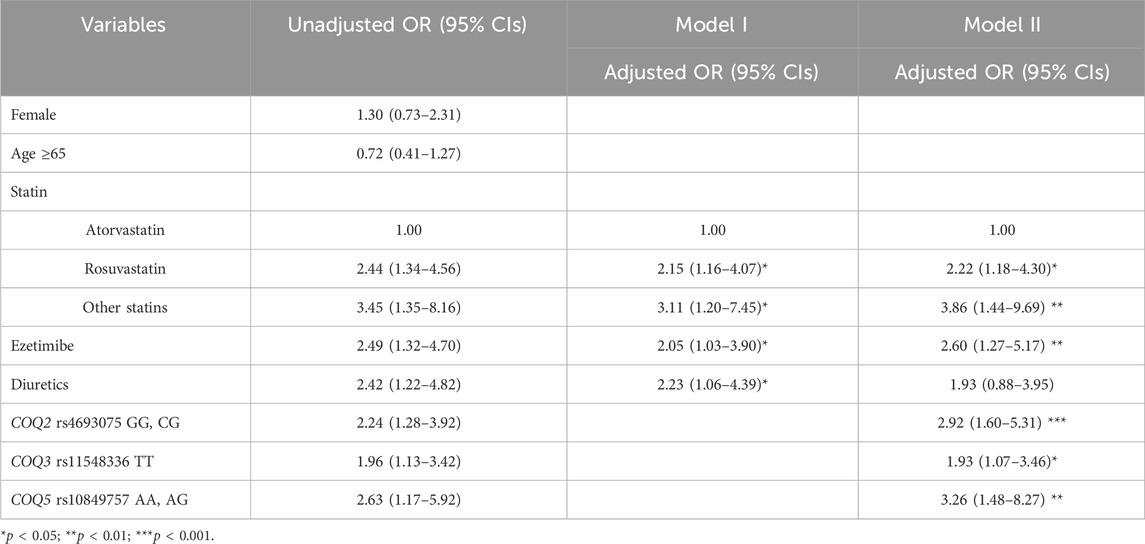
Table 3. Univariate and multivariable regression analyses to identify predictors for statin-related myopathy.
To identify genetic associations between haplotypes and SRM, further haplotype analyses were conducted on genes exhibiting significance in the multivariable analysis. As shown in Supplementary Figure S1, the following SNPs were in a high relationship of LD: rs4693075 and rs34110644 for COQ2 gene; rs9483838, rs6925344, and rs11548336 for COQ3 gene; rs1671766 and rs10849757 for COQ5 gene. There were significant differences between case and control groups in the following haplotype frequencies, which carry alleles demonstrated in the multivariable analysis: COQ2 CG, GG haplotype; COQ3 GTT haplotype; COQ5 CA, and AG haplotype (Supplementary Table S2). COQ2 GG (rs4693075 and rs34110644) haplotype, COQ3 GTT (rs9483838, rs6925344, and rs11548336) haplotype, and COQ5 CA (rs1671766 and rs10849757) haplotype were highly associated with SRM risk.
The Hosmer-Lemeshow test showed that model I and model II were a good fit (x2 = 0.600 and p = 0.741; x2 = 9.075 and p = 0.247, respectively). The AUROC values were 0.663 (95% CI: 0.590–0.736) for model I and 0.747 (95% CI: 0.684–0.809) for model II (Figure 2). We also performed a sensitivity analysis, adding 91 patients (73 with SRM 0 and 18 with SRM 1) into the case group. The final model, in which the same variables as in model II were entered, showed the same trend as the original model: adjusted ORs were 1.76 (95% CI: 1.17–2.67) for rosuvastatin, 2.37 (95% CI: 1.17–4.62) for other statins, 3.04 (95% CI: 1.91–4.82) for ezetimibe, and 1.71 (95% CI: 0.99–2.88) for diuretics. In the case of SNPs, the adjusted ORs were 2.00 (95% CI: 1.32–3.02) for the rs4693075 G allele, 1.79 (95% CI: 1.21–2.65) for the rs11548336 TT genotype, and 2.20 (95% CI: 1.36–3.69) for the rs10849757 A allele.
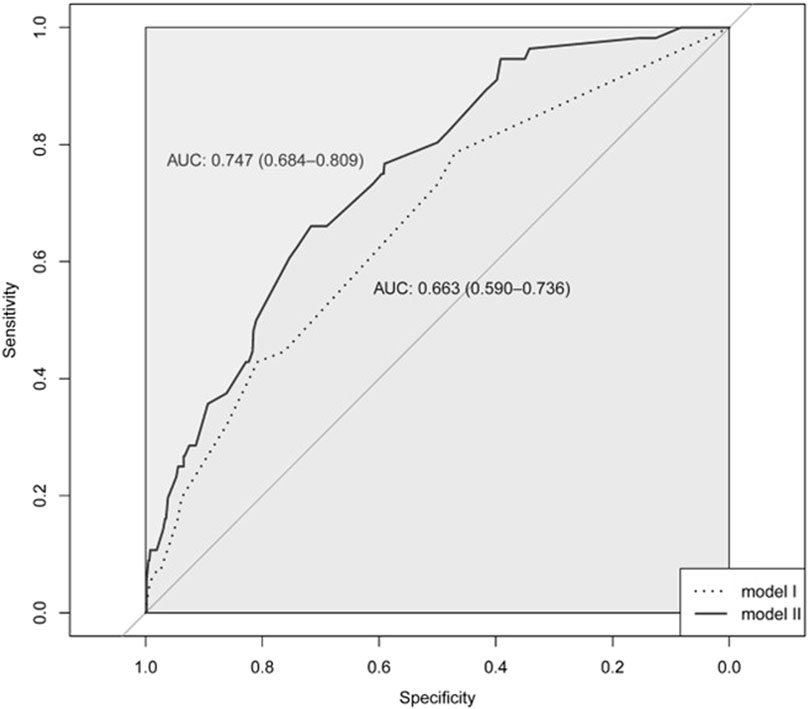
Figure 2. The receiver operating characteristic (ROC) curve for model I and model II.
DiscussionCoQ 10 is an essential cofactor protecting against oxidative stress in mitochondria and is endogenously synthesized through the mevalonate pathway (Molyneux et al., 2008). It is well known that statins reduce CoQ 10 levels, thereby inducing myotoxicity (Apostolopoulou et al., 2015). Additionally, mutation of COQ genes has been correlated with diminished CoQ10 levels. Several case reports have demonstrated that patients with COQ2, COQ5, or COQ7 mutations had decreased CoQ10 levels alongside muscle-related disorders (Jakobs et al., 2013; Malicdan et al., 2018; Wang et al., 2022). This study demonstrated that the CoQ10-related gene polymorphisms, COQ2 rs4693075, COQ3 rs11548336, and COQ5 rs10849757, were significantly related to statin-induced musculoskeletal adverse events. Patients administered rosuvastatin had a higher incidence of SRM than those administered atorvastatin. The concurrent administration of ezetimibe also increased SRM risk compared to no use.
The COQ2, COQ3, COQ5, COQ6, and COQ7 genes encode enzyme synthesizing CoQ10 in eukaryotic mitochondria, which are responsible for rate-limiting steps in the mevalonate pathway (Acosta et al., 2016). COQ2 enzymes condense isoprene to a benzoquinone followed by methylation by COQ3, decarboxylation by COQ5, hydroxylation by COQ6, and deamination by COQ7. Genes involved in the mevalonate pathway have been studied for CoQ10 deficiency, including SRM (Potgieter et al., 2013). For the COQ2 gene, Oh et al. reported that patients with the wild-type homozygote had a 2.3-fold higher risk of SRM than those with the variant-type allele (Oh et al., 2007). Our research revealed a similar result, indicating that the incidence of SRM was higher in patients with the wild-type allele than in those with the variant-type homozygote. However, several studies have reported conflicting results, in which there was no significant association between rs4693075 and statin intolerance, or that the variant allele had a higher risk (Puccetti et al., 2010; Carr et al., 2013; Hubacek et al., 2017; Bakar et al., 2018; Ramakumari et al., 2018; Chowdhury et al., 2019). Therefore, further studies are required to examine ethnic differences in the incidence of SRM.
The present study found that COQ3 rs11548336 and COQ5 rs10849757 were associated with SRM. Patients with wild-type alleles of COQ3 rs11548336 had a higher risk of SRM than mutant allele carriers, which might be due to lower gene expression by the wild-type allele in skeletal muscles, according to the GTEx portal (GTEx ConsortiumLaboratory Data Analysis &Coordinating Center LDACC—Analysis Working GroupStatistical Methods groups—Analysis Working GroupEnhancing GTEx eGTEx groupsNIH Common FundNIH/NCI et al., 2017). rs10849757 is an intron variant of the COQ5 gene (Ward and Kellis, 2016), and the variant allele increased the SRM risk in this study as opposed to the results that variant alleles showed the highest gene expression in skeletal muscle tissues according to the GTEx portal. CoQ10 deficiency caused by a mutation in the COQ5 gene was investigated recently, and the lack of COQ5 biosynthesis led to decreased CoQ10 concentrations in skeletal muscles (Malicdan et al., 2018). No studies have investigated the association between rs10849757 of COQ5 and SRM but considering the possibilities of changing the extent of gene expression by intron variants (Barrett et al., 2012), further studies should be investigated.
As we selected stroke patients for the study, most patients in the cohort were treated with atorvastatin or rosuvastatin. A high dose of atorvastatin and rosuvastatin is a strongly effective therapy for the secondary prevention of ASCVD among ischemic stroke patients (Kleindorfer et al., 2021). A recent randomized controlled trial proved that highly intensive statin treatment was effective at lowering lipid levels among patients after stroke, and this strong evidence was the basis for the statin treatment guidelines (Amarenco et al., 2020). However, the most prevalent adverse event, SRM, is a major issue for patients treated with statins (Backes et al., 2017). The incidence of SRM varies depending on statin classes. Sakaeda et al. reported that SRM after rosuvastatin treatment occurred approximately 1.9–2.7-fold higher than after atorvastatin treatment (Sakaeda et al., 2011). Similarly, Mueller et al. reported that the overall hazard ratio for rosuvastatin was 1.17 compared to atorvastatin (Mueller et al., 2021). In line with other results, our study showed that rosuvastatin had a higher incidence of SRM than atorvastatin. Why rosuvastatin has a higher incidence of SRM than atorvastatin is not clear, but it might be related to the different pharmacokinetic and pharmacodynamic properties of individual statins (Nikolic et al., 2020).
Co-administrated drugs may also affect musculoskeletal symptoms as drug-drug interactions increase the SRM risk. Interestingly, ezetimibe use with statins was correlated with SRM in the present study. Whether the concurrent use of ezetimibe with statins increases the SRM risk or not is controversial. Few studies have reported the effect of ezetimibe on the adverse events of statins, and the results did not have the same tendency. Cases of CK > 10 × ULN occurred in 0.1% of patients with combination statin and ezetimibe therapy, and in 0.4% of patients with statin monotherapy in clinical trials (Author Anonymous, 2023). In contrast, myalgia occurred more often in ezetimibe-statin groups than in statin-alone groups. The mechanism of ezetimibe-induced muscle symptoms is uncertain, but it might include myotoxicity caused by fatty acid oxidation impairment when ezetimibe and statins are administered together (Havranek et al., 2006). Unfortunately, we were unable to analyze the risk of the combination with niacin that could increase SRM risk because patients in this study did not take niacin (HPS2-THRIVE Collaborative Group, 2013). Fibrate could also increase the SRM risk (Graham et al., 2004), but there was no significant difference between the case and control groups in our study due to the small number of patients with fibrate (n = 5). However, our study showed that diuretic use increased SRM risk. According to Hopewell et al., diuretic use with statins might be associated with a higher risk of SRM (Hopewell et al., 2020). This might be due to an electrolyte imbalance such as hypokalemia or physiological alterations including volume contraction (Mosenkis and Townsend, 2005).
Although we implemented the study on the relationship between SRM and CoQ10-related gene polymorphism, there were some limitations. First, this was the retrospective design based on past data. Second, it was conducted only on Koreans, which means further studies on Asians and other ethnic groups are needed. Third, the mechanisms of the SRM difference between rosuvastatin and atorvastatin as well as the pharmacological effect of ezetimibe on SRM, remain unclear. Fourth, CK levels of the case group were unaffected by gene polymorphisms, including rs4693075, rs11548336, and rs10849757. Considering that there were some missing values in CK levels, this needs to be validated through further studies. Fifth, we could not identify how different SNPs associated with lower or higher risk of statin-induced myopathy related to the molecular function of CoQ genes. Despite these limitations, this is the first study to show the influence of COQ2, COQ3, and COQ5 genetic polymorphisms on SRM. As we showed the effects of clinical factors and genetic factors on SRM, this study might contribute to interpreting the cause of statin-induced musculoskeletal disorders and reducing the rate of statin withdrawal due to SRM.
Data availability statementThe data presented in the study are deposited in the Mendeley Data repository, https://data.mendeley.com/datasets/53454fx4hy/1.
Ethics statementThe studies involving humans were conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of EwhaWomans University Seoul Hospital and Ewha Womans University Mokdong Hospital (IRB number: 2020-11-014 and 2021-02-026, respectively). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributionsY-AP: Conceptualization, Formal Analysis, Writing–original draft. YC: Conceptualization, Resources, Writing–original draft. DL: Formal Analysis, Writing–original draft. JK: Formal Analysis, Writing–original draft. MP: Formal Analysis, Writing–original draft. S-AC: Formal Analysis, Writing–original draft. T-JS: Conceptualization, Investigation, Resources, Writing–review and editing. HG: Conceptualization, Funding acquisition, Investigation, Supervision, Writing–review and editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (grant number: NRF-2023R1A2C1007463).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1358567/full#supplementary-material
ReferencesAcosta, M. J., Vazquez Fonseca, L., Desbats, M. A., Cerqua, C., Zordan, R., Trevisson, E., et al. (2016). Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta 1857 (8), 1079–1085. doi:10.1016/j.bbabio.2016.03.036
PubMed Abstract | CrossRef Full Text | Google Scholar
Alfirevic, A., Neely, D., Armitage, J., Chinoy, H., Cooper, R. G., Laaksonen, R., et al. (2014). Phenotype standardization for statin-induced myotoxicity. Clin. Pharmacol. Ther. 96 (4), 470–476. doi:10.1038/clpt.2014.121
PubMed Abstract | CrossRef Full Text | Google Scholar
Amarenco, P., Kim, J. S., Labreuche, J., Charles, H., Abtan, J., Béjot, Y., et al. (2020). A comparison of two LDL cholesterol targets after ischemic stroke. N. Engl. J. Med. 382, 9. doi:10.1056/NEJMoa1910355
PubMed Abstract | CrossRef Full Text | Google Scholar
Arnett, D. K., Blumenthal, R. S., Albert, M. A., Buroker, A. B., Goldberger, Z. D., Hahn, E. J., et al. (2019). 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American heart association task force on clinical practice guidelines. Circulation 140 (11), e596–e646. doi:10.1161/CIR.0000000000000678
PubMed Abstract | CrossRef Full Text | Google Scholar
Backes, J. M., Ruisinger, J. F., Gibson, C. A., and Moriarty, P. M. (2017). Statin-associated muscle symptoms-Managing the highly intolerant. J. Clin. Lipidol. 11 (1), 24–33. doi:10.1016/j.jacl.2017.01.006
PubMed Abstract | CrossRef Full Text | Google Scholar
Bakar, N. S., Neely, D., Avery, P., Brown, C., Daly, A. K., and Kamali, F. (2018). Genetic and clinical factors are associated with statin-related myotoxicity of moderate severity: a case-control study. Clin. Pharmacol. Ther. 104 (1), 178–187. doi:10.1002/cpt.887
PubMed Abstract | CrossRef Full Text | Google Scholar
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi:10.1093/bioinformatics/bth457
PubMed Abstract | CrossRef Full Text | Google Scholar
Barrett, L. W., Fletcher, S., and Wilton, S. D. (2012). Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 69 (21), 3613–3634. doi:10.1007/s00018-012-0990-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Carr, D. F., O'Meara, H., Jorgensen, A. L., Campbell, J., Hobbs, M., McCann, G., et al. (2013). SLCO1B1 genetic variant associated with statin-induced myopathy: a proof-of-concept study using the clinical practice research datalink. Clin. Pharmacol. Ther. 94 (6), 695–701. doi:10.1038/clpt.2013.161
PubMed Abstract | CrossRef Full Text | Google Scholar
Chou, R., Cantor, A., Dana, T., Wagner, J., Ahmed, A. Y., Fu, R., et al. (2022). Statin use for the primary prevention of cardiovascular disease in adults: updated evidence report and systematic review for the US preventive services task force. JAMA 328 (8), 754–771. doi:10.1001/jama.2022.12138
PubMed Abstract | CrossRef Full Text | Google Scholar
Chowdhury, F. A., Baker, S. A., Islam, M. S., Nahid, N. A., Mamun, M. A. A., et al. (2019). Association between variants of COQ2 and TNF-α genes and statin-induced toxicities in Bangladeshi hyperlipidemic patients. Drugs and Ther. Perspect. 35, 621–626. doi:10.1007/s40267-019-00677-x
CrossRef Full Text | Google Scholar
Cunningham, F., Allen, J. E., Allen, J., Alvarez-Jarreta, J., Amode, M. R., Armean, I. M., et al. (2022). Ensembl 2022. Nucleic Acids Res. 50 (D1), D988–D995. doi:10.1093/nar/gkab1049
PubMed Abstract | CrossRef Full Text | Google Scholar
De Pinieux, G., Chariot, P., Ammi-Saïd, M., Louarn, F., Lejonc, J. L., Astier, A., et al. (1996). Lipid-lowering drugs and mitochondrial function: effects of HMG-CoA reductase inhibitors on serum ubiquinone and blood lactate/pyruvate ratio. Br. J. Clin. Pharmacol. 42 (3), 333–337. doi:10.1046/j.1365-2125.1996.04178.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Dohlmann, T. L., Kuhlman, A. B., Morville, T., Dahl, M., Asping, M., Orlando, P., et al. (2022). Coenzyme Q10 supplementation in statin treated patients: a double-blinded randomized placebo-controlled trial. Antioxidants (Basel) 11 (9), 1698. doi:10.3390/antiox11091698
PubMed Abstract | CrossRef Full Text | Google Scholar
Doimo, M., Desbats, M. A., Cerqua, C., Cassina, M., Trevisson, E., and Salviati, L. (2014). Genetics of coenzyme q10 deficiency. Mol. Syndromol. 5 (3-4), 156–162. doi:10.1159/000362826
PubMed Abstract | CrossRef Full Text | Google Scholar
Graham, D. J., Staffa, J. A., Shatin, D., Andrade, S. E., Schech, S. D., La Grenade, L., et al. (2004). Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA 292 (21), 2585–2590. doi:10.1001/jama.292.21.2585
PubMed Abstract | CrossRef Full Text | Google Scholar
GTEx ConsortiumLaboratory, Data Analysis &Coordinating Center LDACC—Analysis Working GroupStatistical Methods groups—Analysis Working GroupEnhancing GTEx eGTEx groupsNIH Common FundNIH/NCIet al. (2017). Genetic effects on gene expression across human tissues. Nature 550 (7675), 204–213. doi:10.1038/nature24277
PubMed Abstract | CrossRef Full Text | Google Scholar
Havranek, J. M., Wolfsen, A. R., Warnke, G. A., and Phillips, P. S. (2006). Monotherapy with ezetimibe causing myopathy. Am. J. Med. 119 (3), 285–286. PMID: 16490482. doi:10.1016/j.amjmed.2005.06.051
PubMed Abstract | CrossRef Full Text | Google Scholar
Hopewell, J. C., Offer, A., Haynes, R., Bowman, L., Li, J., Chen, F., et al. (2020). Independent risk factors for simvastatin-related myopathy and relevance to different types of muscle symptom. Eur. Heart J. 41 (35), 3336–3342. doi:10.1093/eurheartj/ehaa574
PubMed Abstract | CrossRef Full Text | Google Scholar
HPS2-THRIVE Collaborative Group (2013). HPS2-THRIVE randomized placebo-controlled trial in 25 673 high-risk patients of ER niacin/laropiprant: trial design, pre-specified muscle and liver outcomes, and reasons for stopping study treatment. Eur. Heart J. 34 (17), 1279–1291. doi:10.1093/eurheartj/eht055
PubMed Abstract | CrossRef Full Text | Google Scholar
Hubacek, J. A., Adamkova, V., Zlatohlavek, L., Steiner-Mrazova, L., and Vrablik, M. (2017). COQ2 polymorphisms are not associated with increased risk of statin-induced myalgia/myopathy in the Czech population. Drug Metab. Pers. Ther. 32 (4), 177–182. doi:10.1515/dmpt-2017-0027
PubMed Abstract | CrossRef Full Text | Google Scholar
Jakobs, B. S., van den Heuvel, L. P., Smeets, R. J., de Vries, M. C., Hien, S., Schaible, T., et al. (2013). A novel mutation in COQ2 leading to fatal infantile multisystem disease. J. Neurol. Sci. 326 (1-2), 24–28. doi:10.1016/j.jns.2013.01.004
PubMed Abstract | CrossRef Full Text | Google Scholar
Kennedy, C., Köller, Y., and Surkova, E. (2020). Effect of Coenzyme Q10 on statin-associated myalgia and adherence to statin therapy: a systematic review and meta-analysis. Atherosclerosis 299, 1–8. doi:10.1016/j.atherosclerosis.2020.03.006
PubMed Abstract | CrossRef Full Text | Google Scholar
Kleindorfer, D. O., Towfighi, A., Chaturvedi, S., Cockroft, K. M., Gutierrez, J., Lombardi-Hill, D., et al. (2021). 2021 guideline for the prevention of stroke in patients with stroke and transient ischemic attack: a guideline from the American heart association/American stroke association. Stroke 52 (7), e364–e467. doi:10.1161/STR.0000000000000375
PubMed Abstract | CrossRef Full Text | Google Scholar
Malicdan, M. C. V., Vilboux, T., Ben-Zeev, B., Guo, J., Eliyahu, A., Pode-Shakked, B., et al. (2018). A novel inborn error of the coenzyme Q10 biosynthesis pathway: cerebellar ataxia and static encephalomyopathy due to COQ5 C-methyltransferase deficiency. Hum. Mutat. 39 (1), 69–79. doi:10.1002/humu.23345
PubMed Abstract | CrossRef Full Text | Google Scholar
Mantle, D., Millichap, L., Castro-Marrero, J., and Hargreaves, I. P. (2023). Primary coenzyme Q10 deficiency: an update. Antioxidants (Basel) 12 (8), 1652. doi:10.3390/antiox12081652
PubMed Abstract | CrossRef Full Text | Google Scholar
Molyneux, S. L., Young, J. M., Florkowski, C. M., Lever, M., and George, P. M. (2008). Coenzyme Q10: is there a clinical role and a case for measurement? Clin. Biochem. Rev. 29 (2), 71–82.
PubMed Abstract | Google Scholar
Mueller, A. M., Liakoni, E., Schneider, C., Burkard, T., Jick, S. S., Krähenbühl, S., et al. (2021). The risk of muscular events among new users of hydrophilic and lipophilic statins: an observational cohort study. J. Gen. Intern Med. 36 (9), 2639–2647. doi:10.1007/s11606-021-06651-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Neal, R. C., Ferdinand, K. C., Ycas, J., and Miller, E. (2009). Relationship of ethnic origin, gender, and age to blood creatine kinase levels. Am. J. Med. 122 (1), 73–78. doi:10.1016/j.amjmed.2008.08.033
PubMed Abstract | CrossRef Full Text | Google Scholar
Nikolic, D., Banach, M., Chianetta, R., Luzzu, L. M., Pantea Stoian, A., Diaconu, C. C., et al. (2020). An overview of statin-induced myopathy and perspectives for the future. Expert Opin. Drug Saf. 19 (5), 601–615. doi:10.1080/14740338.2020.1747431
PubMed Abstract | CrossRef Full Text | Google Scholar
Oh, J., Ban, M. R., Miskie, B. A., Pollex, R. L., and Hegele, R. A. (2007). Genetic determinants of statin intolerance. Lipids Health Dis. 6, 7. doi:10.1186/1476-511X-6-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Potgieter, M., Pretorius, E., and Pepper, M. S. (2013). Primary and secondary coenzyme Q10 deficiency: the role of therapeutic supplementation. Nutr. Rev. 71 (3), 180–188. doi:10.1111/nure.12011
PubMed Abstract | CrossRef Full Text | Google Scholar
Puccetti, L., Ciani, F., and Auteri, A. (2010). Genetic involvement in statins induced myopathy. Preliminary data from an observational case-control study. Atherosclerosis 211 (1), 28–29. doi:10.1016/j.atherosclerosis.2010.02.026
PubMed Abstract | CrossRef Full Text | Google Scholar
Ramakumari, N., Indumathi, B., Katkam, S. K., and Kutala, V. K. (2018). Impact of pharmacogenetics on statin-induced myopathy in South-Indian subjects. Indian Heart J. 70 (Suppl. 3), S120–S125. doi:10.1016/j.ihj.2018.07.009
PubMed Abstract | CrossRef Full Text | Google Scholar
Sakaeda, T., Kadoyama, K., and Okuno, Y. (2011). Statin-associated muscular and renal adverse events: data mining of the public version of the FDA adverse event reporting system. PLoS One 6 (12), e28124. doi:10.1371/journal.pone.0028124
PubMed Abstract | CrossRef Full Text | Google Scholar
Turner, R. M., and Pirmohamed, M. (2019). Statin-related myotoxicity: a comprehensive review of pharmacokinetic, pharmacogenomic and muscle components. J. Clin. Med. 9 (1), 22. doi:10.3390/jcm9010022
PubMed Abstract | CrossRef Full Text | Google Scholar
Vinci, P., Panizon, E., Tosoni, L. M., Cerrato, C., Pellicori, F., Mearelli, F., et al. (2021). Statin-associated myopathy: emphasis on mechanisms and targeted therapy. Int. J. Mol. Sci. 22 (21), 11687. doi:10.3390/ijms222111687
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, Y., Gumus, E., and Hekimi, S. (2022). A novel COQ7 mutation causing primarily neuromuscular pathology and its treatment options. Mol. Genet. Metab. Rep. 31, 100877. doi:10.1016/j.ymgmr.2022.100877
PubMed Abstract | CrossRef Full Text | Google Scholar
Ward, L. D., and Kellis, M. (2016). HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881. doi:10.1093/nar/gkv1340
留言 (0)