Pathogen avoidance is a pivotal behavioural adaptation observed in diverse animal species within natural ecosystems. It serves as a mechanism to distinguish safe food sources from potential threats posed by harmful bacteria and lethal pathogens. This adaptive response, an integral component of the behavioural immune system, plays a fundamental role in individual survival, thereby influencing the persistence of populations within their ecological niches (1). Various species, including mice (2), bonobos (pygmy chimpanzees) (3), chimpanzees (4), and humans (5), demonstrate the capacity for pathogen avoidance.
Caenorhabditis elegans (C. elegans), a nematode that lives in microbe-rich soils (6, 7), is also capable of avoiding pathogens. Given its habitat, C. elegans inevitably encounters pathogenic bacteria, prompting the development of sophisticated avoidance mechanisms via natural selection. One such pathogen is the opportunistic human pathogen Pseudomonas aeruginosa (PA14) (7, 8). C. elegans is initially attracted to PA14, but over time, C. elegans avoids PA14 by aversive learning (9). Such behavioural avoidance has emerged as a valuable model for unravelling the intricacies of pathogen avoidance mechanisms. In this review, we will first discuss various stimuli that promote pathogen avoidance. Next, we will describe the neural circuitry and the underlying neural and molecular mechanisms that regulate pathogen avoidance. We will also cover the recently discovered role of small non-coding RNAs in transgenerational inheritance of pathogen avoidance and the role of small non-coding RNAs. Finally, we will conclude this review by discussing the crosstalk between innate immunity and pathogen avoidance, as well as the C. elegans avoidance of pathogens other than PA14. This review aims to comprehensively summarise these scientific research achievements and provide a systematic overview of pathogen avoidance behaviours in C. elegans.
2 Stimuli that trigger pathogen avoidance and the underlying molecular mechanisms2.1 AerotaxisAerotaxis refers to the behavioural movement of animals in response to oxygen concentrations in the environment. C. elegans migrates towards regions with 5–12% oxygen levels but avoids regions with higher (>12%) and lower (<2%) oxygen concentrations (10). The haem domain of GCY-35, a specific soluble guanylate cyclase homologue, binds to molecular oxygen. GCY-35 activity is regulated by molecular oxygen, which subsequently produces 3’,5’-cyclic guanosine monophosphate (cGMP) that acts as a second messenger and is implicated in oxygen sensation (10). TAX-4, a cyclin nucleotide-gated channel, is activated by GCY-35 through cGMP in URX, AQR, and PQR sensory neurons to promote the aggregation of animals on a bacterial lawn and their accumulation on the thickest part of the bacterial lawn (known as bordering behaviour). The processes of aggregation and boarding mediated by GCY-35 and TAX-4 are antagonised by the activity of the neuropeptide receptor NPR-1 (11, 12). Hyperoxia avoidance regulated by NPR activity requires the neurotransmitter serotonin in ADF sensory neurons. The neuronal TGF-beta homologue DAF-7 inhibits serotonin synthesis in ADF neurons, thereby regulating hyperoxia avoidance (10). Interestingly, it has been determined that alginate biosynthesis in mucoid P. aeruginosa suppresses NPR-1-mediated pathogen avoidance behaviour (12). GCY-35 and GCY-36 expressed in the head and tail neurons modifies C. elegans movement, promoting reversal and turning when oxygen levels increase. Hyperoxia avoidance is also controlled by the TRP-related channel subunits OCR-2 and OSM-9 and the transmembrane protein ODR-4, which act on the nociceptive neurons ASH and ADL (13, 14). Interactions between GLB-5 and the H-NOX domains of GCY-35 and GCY-36 are essential for rapid adaptation to low or high oxygen levels (15).
PA14, consumes oxygen, results in a decrease in the surrounding oxygen level (11, 12), which may attract and subsequently harm C. elegans. However, C. elegans have developed the capacity to adapt to their preferences through the alteration of aerotaxis to counter the challenge of lower oxygen surroundings, enabling them to avoid PA14 (Figure 1).
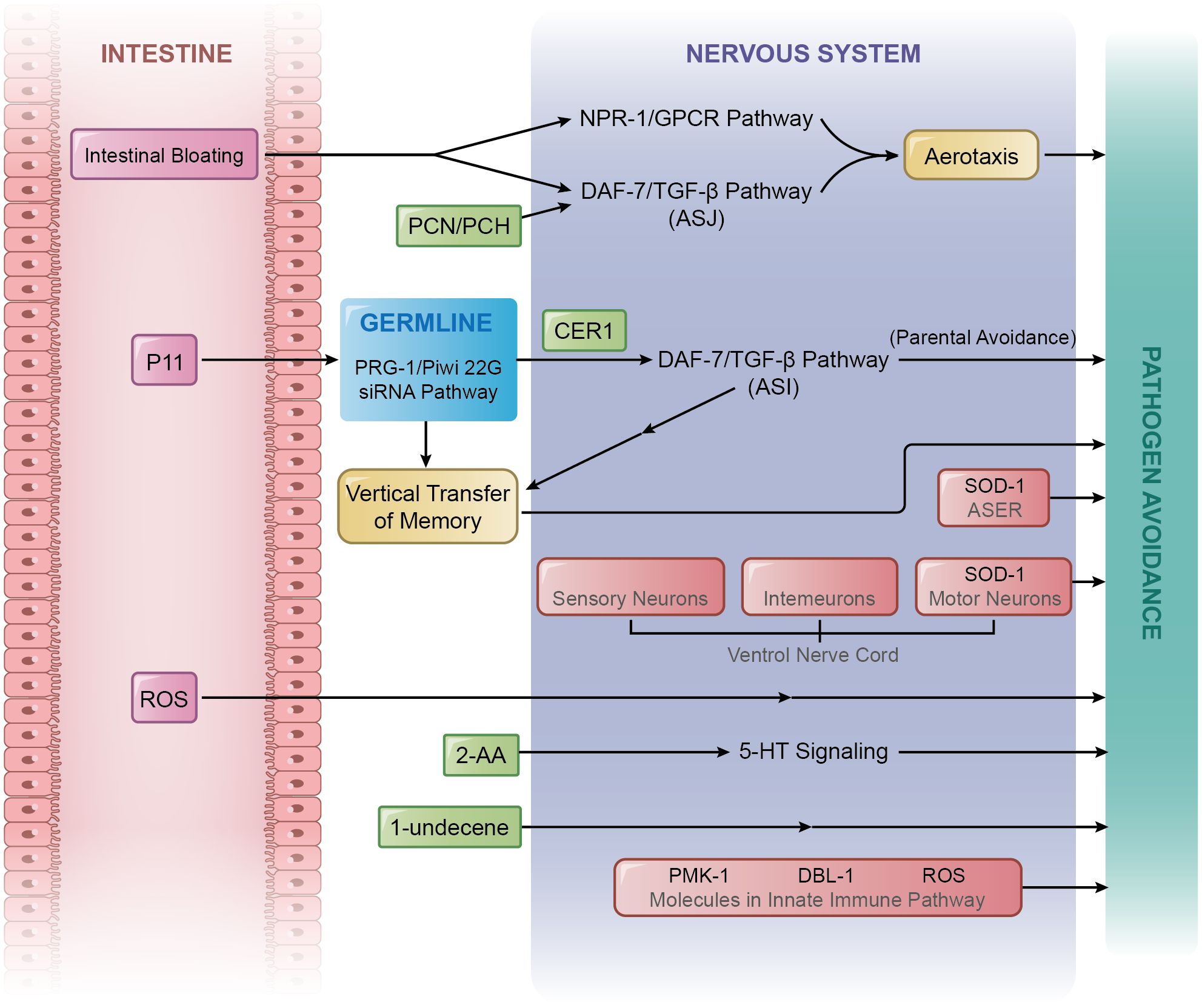
Figure 1 Mechanisms of pathogen avoidance. A variety of comprehensive mechanisms are in place to regulate pathogens avoidance, which include those caused by the alteration of aerotaxis, intestinal bloating, metabolites, transgenerational epigenetic inheritance, and crosstalk with innate immunity.
A critical component of the adaptive response is the DAF-7/TGF-β signalling pathway in ASJ neurons of C. elegans (16). Detailed chemical analysis of secondary metabolites of PA14 has identified two chemical components, phenazine-1-carboxamide (PCN) and pyochelin (PCH), as potent stimulators of daf-7 gene expression as well as the activation of the TGF-β signalling pathway in ASJ neurons. Consequently, the sensation of PCN and PCH is believed to be a crucial step in the alteration of aerotactic behaviour and promotes PA14 avoidance (16).
2.2 Intestinal bloatingIntestinal bloating is identified as the expansion of the intestinal lumen (17). Researchers have discovered a correlation between the degree of intestinal bloating and the degree of pathogen avoidance; reduced intestinal bloating led to delayed avoidance, whereas increased bloating enhanced avoidance (17). Further research confirmed that the DAF-7/TGF-β signalling pathway could be stimulated during intestinal bloating and that intestinal bloating does, indeed, contribute to pathogen avoidance triggered by aerotaxis alteration (17, 18). Moreover, intestinal bloating and ASJ neuron detection of PCN and PCH trigger aerotactic changes, initiating pathogen avoidance. A recent study reported that intestinal bloating can stimulate histone H4 Lys8 acetylation in the C. elegans germline, which requires the participation of PAR-5, a protein belonging to the 14-3-3 chaperone protein family. This process is pivotal in pathogen avoidance, potentially acting as an intermediary in the signalling pathway between the intestine and neurons (19). The ASJ neurons contribute significantly to aerotactic responses resulting in pathogen avoidance; however, the roles of AWB, AWC, or ADF neurons remain unclear. Future investigations are warranted because of the critical role these neurons play in forming the neural circuitry for pathogen avoidance. In addition to the DAF-7/TGF-β pathway, another neuroendocrine pathway, NPR-1-mediated signalling, is also essential for pathogen avoidance triggered by the alteration of aerotaxis (18, 20, 21). A neuropeptide Y receptor homologue, NPR-1 (22), is expressed in AQR, PQR, and URX sensory neurons. Its ligands are FLP-18 and FLP-21 (23). To function in these neurons, NPR-1 requires TAX-2, TAX-4, and soluble guanylyl cyclase GCY-35 to bind to molecular oxygen (10, 20, 23). Notably, expression of the npr-1 gene could also be activated by intestinal bloating (17), suggesting a parallel operational mode with the DAF-7/TGF-β pathway after intestinal bloating. The functions of both DAF-7 and NPR-1 in pathogen avoidance necessitate the expression of the transient receptor potential channel vanilloid genes, osm-9 and ocr-2 (10, 16). Furthermore, NPR-1 can be inhibited by HECW-1, an E3 ubiquitin ligase, which functions in outer labial lateral (OLL) sensory neurons, leading to the inhibition of avoidance behaviour (24) (Figure 1). These intricate networks of sensory inputs and signalling pathways underscore the complexity and adaptability of pathogen avoidance in C. elegans in response to environmental stimuli.
2.3 Secondary metabolitesSeveral types of secondary metabolites secreted by PA14 can be regarded as microbial-associated molecular patterns (25, 26). Some of these serve as stimuli for C. elegans to avoid pathogens (Figure 1). In addition to PCN and PCH, 2-aminoacetophenone (2-AA) initiates pathogen avoidance. 2-AA is a volatile chemical synthesised by PA14. When exposed to 2-AA, 5-HT signalling in neurons is stimulated. Since 5-HT receptors exist in many types of tissues, the information carried by 5-HT spreads throughout multiple parts of C. elegans, including the intestine, germline, and head. Such a reaction results in the enhanced expression of the heat shock factor protein HSF-1 (27) that is required for pathogen avoidance (28). After contact with PA14, HSF-1 assists the rapid transcription of the hsp-70 gene by association with RNA polymerase II (28). The hsp-70 gene can encode the chaperone heat shock protein HSP-70 (29). Another effective secondary metabolite of PA14 is 1-undecene, an 11‐carbon olefin, which is a Pseudomonas-specific volatile (30). It can be sensed by AWB neurons and elicit pathogen avoidance behavioural responses (31).
3 Neural circuit of pathogen avoidanceNeurons serve as information perception and integration centres in C. elegans. They manipulate all behaviours in response to varying environmental stimuli. When a threat from PA14 arises, they play an indispensable role in controlling pathogen avoidance behaviour through a series of intricate molecular events (Figure 2).
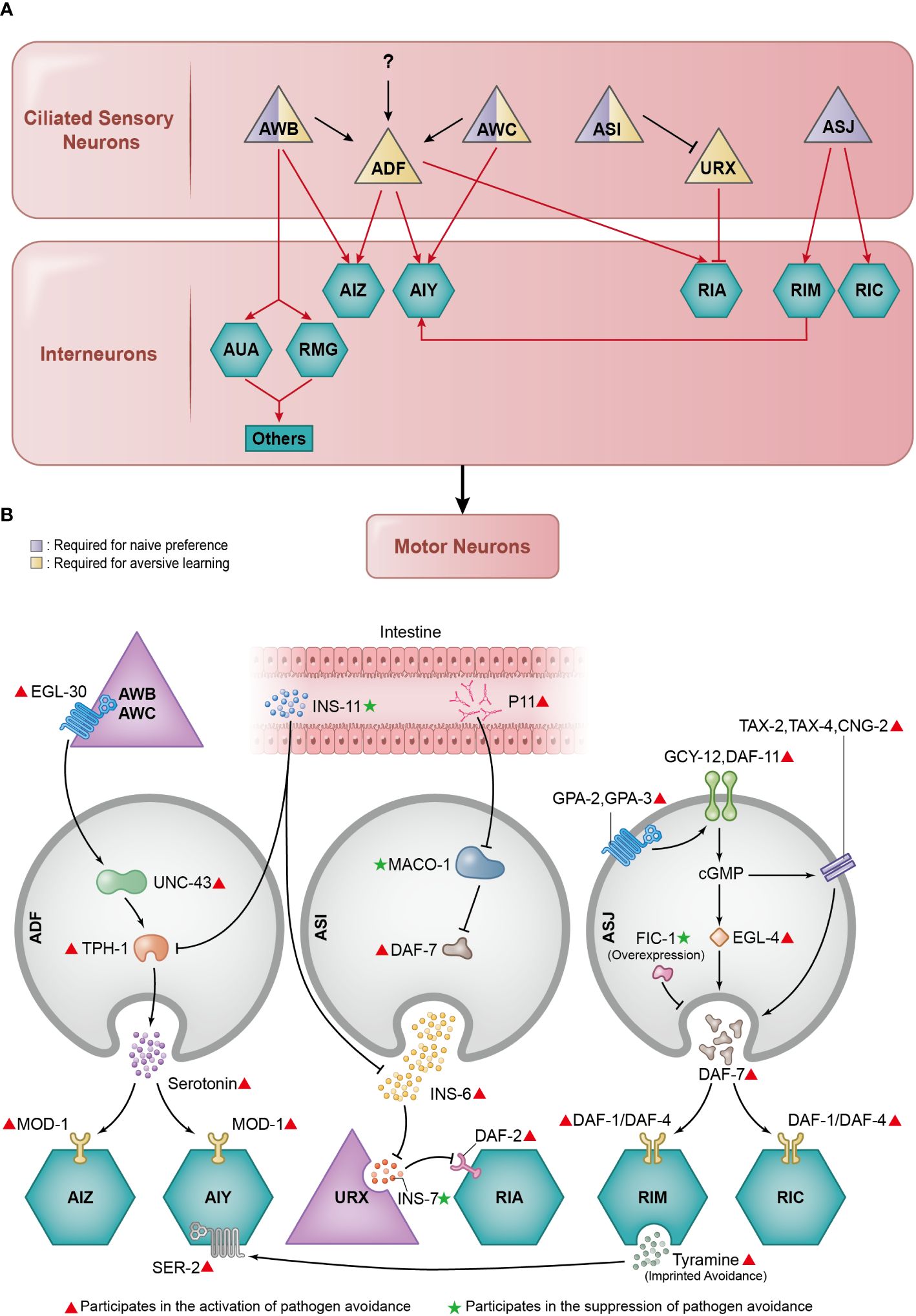
Figure 2 Essential neuronal basis of pathogen avoidance. (A) Several neural circuits contribute to the intricate process of pathogen avoidance. In essence, these circuits form a three-layer structure composed of ciliated sensory neurons, interneurons, and motor neurons. The sensory neurons play distinct roles: some are tasked with naïve preference, others with learned avoidance, and a select few handle both. (B) The regulation of pathogen avoidance is a collaborative effort between neurotransmitters and neuromodulators. Working in unison, they meticulously calibrate the process of pathogen avoidance.
Upon PA14 exposure, C. elegans undergoes two phases of pathogen avoidance behaviour: initial attraction and subsequent repulsion. Initially, C. elegans is attracted to PA14 via the process of naïve preference, but over time, C. elegans avoids PA14 through the mechanism of learned avoidance (9). At least three different interconnected neural circuits participate in this process.
The first is the AWB-AWC sensorimotor circuit, which is responsible for both initial naïve preference and learned avoidance (32). The AWB and AWC neurons are located in the anterior region of C. elegans and possess cilia that are partially exposed to the external environment (33). When the surrounding bacteria change from the standard laboratory non-pathogenic bacterium Escherichia coli OP50 to the pathogenic bacterium PA14, the intracellular calcium dynamics in AWC are inhibited, whereas those in AWB neurons are stimulated (32).
The second neural circuit is the AWB neuron-mediated learned reflexive aversion circuit, devoted to learned avoidance. After aversive learning, C. elegans exhibit aversive reflexes on exposure to PA14. Backward locomotion is necessary during aversive reflection. This is controlled by the AWB neuron-mediated learned reflexive aversion circuit, which goes from sensory neurons AWB to interneurons AUA/RMG, progresses to lower-layer interneurons AVA/AVD/AVE, and ultimately reaches motor neurons VA/DA/AS/VA/DD (34).
The third circuit is the ADF modulatory neural circuit dedicated to learned avoidance, although ADF neurons themselves have a slight effect on naïve preference (32). ADF neurons are located in the anterior region of C. elegans and also possess cilia exposed to the environment (33). Upon exposure to PA14, increased serotonin secretion is triggered. The serotonin produced by ADF further signals the serotonin-gated chloride channel MOD-1 in the downstream AIY and AIZ interneurons, inducing learned avoidance through aversive learning (9, 35). Serotonin production in the ADF neurons relies on the rate-limiting enzyme tryptophan hydroxylase-1 (TPH-1), which is upregulated upon pathogen exposure. During this process, Gqα protein EGL-30 in upstream AWB and AWC neurons induces the expression of CaMKII/UNC-43 in ADF neurons, leading to the increased expression of tph-1 (36). Notably, there is also a down-regulatory mechanism for tph-1; exposure to pathogens triggers ins-11 expression in the intestine via the p38 MAPK signalling pathway and the transcription factor EB-coding gene hlh-30. The increased INS-11 levels subsequently reduce the basal expression levels of tph-1 (37). These three preliminarily identified circuits share multiple common neurons, but how they interact with each other remains to be further investigated.
Serotonin is not only pivotal in aversive learning in C. elegans but may also play a similar role in humans. Disgust is hypothesised to function as an adaptive pathogen avoidance mechanism in humans, preventing exposure to parasites, including bacteria (38). This behaviour manifests in two phases: prior disgust, which occurs without contact with the pathogen, and posterior disgust, which occurs after contact (39). The posterior disgust phase often leads to vomiting, a process in which serotonin serves as a critical signalling molecule (39, 40).
While the aforementioned C. elegans neural circuits predominantly involve the AWB, AWC, and ADF neurons in pathogen avoidance behaviour, it is crucial to consider the roles of other ciliated sensory neurons located in the anterior region of C. elegans, such as the ASI and ASJ neurons (33). Although they do not form clearly specified circuits, as outlined before, ASI and ASJ neurons contribute significantly to pathogen avoidance. The ASI neurons are required for naïve preference (41) and learned avoidance (33). The ASJ neurons are required for naïve preference (16), but their role in learned avoidance has not been definitively established.
The ASI neurons secrete insulin-like neuropeptide INS-6, which suppresses the expression of insulin-like neuropeptide INS-7 in URX sensory neurons. INS-7, in turn, inhibits the insulin receptor DAF-2 in RIA interneurons. Since DAF-2 in the RIA interneurons positively regulates learned pathogen avoidance, the neuronal cascade initiated from the secretion of INS-6 from ASI neurons also positively regulates learned pathogen avoidance (42). The pathway responsible for the activation of ins-11 described earlier could suppress the expression of ins-6 in the ASI neurons, contributing to the reduction of aversive learning behaviours (37). On the other hand, a recent study reported that the small RNA P11 (sRNA P11) of PA14 could down-regulate the expression of the maco-1 gene, an endoplasmic reticulum (ER) membrane protein coding gene, causing activation of the DAF-7/TGF-β pathway in ASI neurons, thereby promoting pathogen avoidance (41).
In ASJ neurons, the G protein-coupled receptors (GPCR) GPA-2 and GPA-3 are activated during pathogen exposure. GPCRs then stimulate the guanylate cyclases DAF-11 and GCY-12 to produce cGMP, which activates the cGMP-dependent kinase EGL-4 and the cyclic nucleotide–gated (CNG) ion channels TAX-2, TAX-4, and CNG-2. Together, these proteins initiate the rapid transcription of the daf-7 gene (43). The daf-7 gene encodes neuromodulator DAF-7 that can bind with TGF-β receptor DAF-1/DAF-4 on downstream interneurons RIM and RIC, activating R-SMADs DAF-8/DAF-14 and then suppressing co-SMAD DAF-3, resulting in enhanced pathogen avoidance by modulation of aerotaxis (16). The expression of daf-7 in the ASJ neurons and its downstream TGF-β pathway could be suppressed by the overexpression of FIC-1 (44), which belongs to the family of AMPylase containing the Fic domain (45, 46). Additionally, ASJ neurons can sense nitric oxide (NO) to initiate pathogen avoidance, a process that requires the CNG channels TAX-2 and TAX-4 and the receptor guanylate cyclase DAF-11. When NO is present or removed, the calcium levels increase in ASJ neurons. This phenomenon is influenced by TRX-1/thioredoxin, a redox-sensing protein (47). The presence of cilia in all the previously described neurons underlines their significance in the sensation of external stimuli (48–50), illustrating the crucial role of ciliary structures in the behavioural pathogen avoidance.
Another noteworthy phase of avoidance is known as imprinted aversive learning. When exposed to PA14 shortly after hatching, C. elegans undergo long-term imprinted aversive learning. The formation of this long-term memory involves the sensory neurons AIB and interneurons RIM, whereas the retrieval of memory relies on the sensory neurons AIY and interneurons RIA. Specifically, the signals are transmitted by the neuromodulator tyramine, which is secreted by the RIM and binds to the tyramine receptor SER-2 in AIY. Additionally, imprinted aversive learning requires the participation of serotonin secretion-related genes, including tph-1 and mod-1, glutamine secretion -related genes eat-4, glr-1, and glr-3, as well as the cAMP response element-binding protein-coding homolog crh-1 (35).
4 Epigenetic inheritance of pathogen avoidanceThe phenomenon of epigenetic inheritance, in which certain behaviours are transmitted across generations, has been observed and studied in various species. In the case of C. elegans, this intriguing phenomenon is evident in the context of pathogen avoidance. When maternal C. elegans were exposed to PA14 for 24 h, enhanced naïve preference and learned avoidance response to PA14 were observed in at least four subsequent generations (51).
In many animals, the epigenetic inheritance of behaviour across generations requires multiple small noncoding RNA pathways (52, 53). Similarly, in C. elegans, the epigenetic inheritance of pathogen avoidance behaviour critically relies on small noncoding RNA pathways, specifically the PRG-1/Piwi 22G siRNA pathway. Notably, the role of DAF-7/TGF-β signalling within the ASI neurons is crucial to this process (51). Researchers revealed that PA14 exposure leads to the generation of abundant PIWI-interacting RNAs in maternal C. elegans. Subsequently, a sequence involving the Piwi argonaute protein PRG-1 (54), RNA-dependent-RNA-polymerase RRF-1 (55), and RNaseD homologue MUT-7 (56) generates secondary endo-siRNAs (22G RNAs) using PIWI-interacting RNAs as sources (51). These siRNAs then translocate to the nucleus and guide the expression of histone methyltransferase SET-25 (57) and histone receptor HPL-2 (58), leading to chromatin modifications and the epigenetic inheritance of pathogen avoidance in subsequent generations (51). Intriguingly, parental PA14 exposure duration influences progeny behaviour; a short four-hour exposure results in progeny preference for PA14, whereas an eight-hour exposure switches the response to avoidance. The Piwi argonaute protein PRG-2 (59) is involved in this modulation (60).
Further research revealed a more intricate mechanism underlying epigenetic inheritance via the PRG-1/Piwi 22G siRNA pathway (41). This process is triggered by a specific noncoding RNA, sRNA P11, which is exclusive to pathogenic PA14. It induces learned avoidance in the maternal C. elegans and their progeny. Elements in the RNA interference pathway are key to maternal learned avoidance: Double-stranded RNA (dsRNA) transporter SID-2 (61) and dsRNA endoribonuclease Dicer DCR-1 (62) are required for response to PA14 sRNA; AGO3 homolog RDE-1 (63), RNA interference-defective protein RDE-2 (64), RDE-4 (65), and MUT-7 (56) are required for response to sRNAs of both PA14 and OP50 (41); dsRNA transporter SID-1 (66) is required for response to sRNAs of both PA14 and OP50, as well as naïve preference. After the ingestion of sRNA P11, the PRG-1/Piwi 22G siRNA pathway in the germline is activated. It is essential for the transgenerational epigenetic inheritance of pathogen avoidance in progenies induced by P11. Several elements in the PRG-1/Piwi 22G siRNA pathway are essential: Piwi argonaute protein PRG-1, RNA-dependent RNA polymerases RRF-1 (67) and, RRF-3 (68), and heterochromatin regulator HPL-2 (41). In germlines, the virus-like particle transposon Cer1 loads RNAs and carries signals from germlines to the ASI neurons (69–71). This further downregulates the expression of the ER membrane protein MACO-1, which in turn up-regulates DAF-7 in the ASI neurons, resulting in maternal learned avoidance and epigenetic inheritance of pathogen avoidance in progenies (41). Besides sRNA, intestinal bloating could also induce the transgenerational inheritance of pathogen avoidance, which is accompanied by chromatin modification involving H4 Lys8 acetylation in germlines (19) (Figure 2).
5 Signalling crosstalk between pathogen avoidance and innate immunityThe pathogen avoidance behaviour of C. elegans coordinates their escape from pathogens to limit infection. In addition, C. elegans also initiate certain mechanisms of the innate immune system to eliminate bacteria to prevent pathogen invasion. Although the modes of activation and mechanisms of pathogen avoidance and innate immunity are quite different, several studies identified evidence for signalling crosstalk between these defence strategies.
Pathogen avoidance can be induced by several classical factors in the innate immune system. One key factor are reactive oxygen species (ROS). Bacteria ingested by C. elegans are usually transferred to the intestine by movement of the pharynx. Ingested PA14 accumulates in the intestine, resulting in ROS production (72, 73). ROS, which include hydroxyl radicals, superoxide anions, and hydrogen peroxide, are generated from oxygen reduction. While excessive ROS can be toxic to cells (74–77), they also serve a protective function by killing invading bacteria and fungi (72, 78), suggesting dual roles in innate immunity. Recent studies demonstrated that ROS also affect pathogen avoidance (Figure 1).
Exposure to PA14 triggers ROS generation in the C. elegans intestine, promoting the expression of the antioxidant enzyme superoxide dismutase-1 (SOD-1) in intestinal ASER neurons (79). SOD-1 protects cells by converting superoxide into less toxic oxygen or hydrogen peroxide (80, 81). The induction of SOD-1 in ASER neurons results in the inhibition of the pathogen avoidance at first. Subsequently, SOD-1 levels return to normal when C. elegans show a tendency toward pathogen avoidance. Although SOD-1 is expressed in the cell body, dendrites, and cilia of ASER, those expressed in the cilia are essential for pathogen avoidance (79). Furthermore, guanylyl cyclases GCY-22 and GCY-5 mediate SOD-1 induction in ASER neurons (82), while NPR-1 suppresses SOD-1 expression (83). Interestingly, another antioxidant enzyme, SOD-5 (81), also suppresses pathogen avoidance (83).
In addition to the ASER neurons, SOD-1 is also expressed in the ventral nerve cords, specifically in the cholinergic motor neurons. The ventral nerve cords are composed of sensory neurons, interneurons, and motor neurons. The AMPA-type ionotropic glutamate receptor GLR-1 in interneurons (84) can suppress SOD-1 expression upon pathogen exposure (84). Lack of GLR-1 results in enhanced SOD-1 expression and enhanced pathogen avoidance, indicating that GLR-1 negatively regulates pathogen avoidance in a non-cell-autonomous manner. As GLR-1 is a glutamate receptor, glutamatergic sensory neurons upstream of the interneurons where GLR-1 is expressed may also be involved in pathogen avoidance. Indeed, lack of the vesicular glutamate transporter EAT-4 (85) in glutamatergic sensory neurons could regulate pathogen avoidance. Specifically, rather than promoting or suppressing avoidance, the loss of EAT-4 causes C. elegans to lose the ability to distinguish between the pathogenic bacterium PA14 and the non-pathogenic bacterium E. coli OP50 (86).
In the intestine, PA14 induces ROS production in intestinal cells. ROS increases the formation of oxidised glutathione or GSSG. GSSG can be transported into the pseudocoelomic cavity by the gut efflux pump MRP-1, an ATP-binding cassette transporter located in the basolateral membrane of intestinal cells. Subsequently, GSSG triggers pathogen avoidance. This process requires the NMDA class glutamate receptor-1 NMR-1 in downstream neurons. Researchers verified this by demonstrating that extracellular GSSG supplementation activated pathogen avoidance, while loss of either MRP-1 or NMR-1 resulted in the inhibition of aversive learning (87). Another study found out that NMR-1 in the RIM neurons decreases INX-4 abundance via UNC-43 to diminish the strength of the gap junction in the RIM-circuit, which contributes to the pathogen avoidance (88). Given that ROS can impair intestinal cell functions, potentially leading to intestinal damage and subsequent bloating, it would be worthwhile to determine whether there are connections between the avoidance caused by intestinal bloating and ROS production.
In addition to ROS, several critical proteins within the classical immune pathway play a role in activating pathogen avoidance behaviours. One key protein is DBL-1, a ligand that initiates the DBL-1/TGF-β innate immune signalling pathway (89, 90). Research has demonstrated that DBL-1 is essential for aversive learning (91). Researchers found that the exposure to PA14 decreases the calcium response in AVA interneurons. This diminished AVA neuronal activity triggers the secretion of DBL-1, which then binds to the type I TGF-β receptor SMA-6 in the hypodermis. Subsequent TGF-β signalling facilitates aversive learning, enabling the organism to actively avoid PA14 (91). Another crucial factor is PMK-1, a core molecule in the p38 MAPK pathway (92), which acts as a negative regulator of pathogen avoidance (93). A deficiency of innate immunity caused by the loss of PMK-1 can stimulate avoidance behaviour in C. elegans. Within the OLL neurons, this loss suppresses the expression of the hecw-1 gene, which encodes an E3 ubiquitin ligase. Suppression of this ligase exerts a non-cell-autonomous effect, increasing NPR-1 expression in the RMG neurons and ultimately activating the pathogen avoidance response (93).
Multiple factors involved in pathogen avoidance can induce innate immunity. NPR-1 can positively control innate immunity by inhibiting the activity of neurons AQR, PQR, and URX, which transfer neuroendocrine signals through pseudocoelomic body fluid to non-neural tissues and negatively regulate the innate immunity (94). Likewise, intestinal bloating upregulates multiple innate immune genes, such as clec-60, lys-3, lys-4, ilys-3, cpr-2, and F53A9.8 (17). Furthermore, the secondary metabolite PCN can bind to the nuclear hormone receptor NHR-86/HNF4 in intestinal epithelial cells. This binding triggers the expression of multiple immune genes, initiating a transcriptional program that enhances innate immunity (95).
6 Pathogen avoidance of non-Pseudomonas bacteriaWhile PA14 is the most studied pathogen for avoidance behaviours, there are also extensive studies on the mechanisms of pathogen avoidance of non-Pseudomonas bacteria species, such as Bacillus thuringiensis (BT) and Enterococcus faecalis (EF).
BT, a gram-positive, rod-shaped aerobic bacterium, produces Cry toxin, a δ-endotoxin that can undergo oligomerisation (96, 97). Cry toxins are widely used as a bioinsecticide (97). C. elegans tends to avoid BT with an observed capacity to differentiate between strains of varying pathogenicity, preferentially evading more virulent strains. C. elegans with npr-1(ur89) mutations exhibit stronger avoidance behaviour. The RNA-seq profile of npr-1(ur89) mutants suggests the potential involvement of Ebox transcription factors, oxidative stress genes, p38 MAPK signalling, C-type lectins, and insulin-like signalling in the avoidance of BT (98). Two studies confirmed the participation of C-type lectins and insulin-like signalling in pathogen avoidance. One study revealed that the mutation of the C-type lectin gene C54G4.4 exhibited increased BT avoidance (99). Another study demonstrated that two main components of insulin-like signalling, the receptor DAF-2 and the transcription factor DAF-16, are critical for BT avoidance (100). Notably, one of the Cry toxins secreted by BT, Cry6Aa2, induces avoidance in the absence of BT (101).
EF is a gram-positive, spherical or ovoid-shaped and facultative anaerobic bacterium (102, 103). C. elegans initially exhibit avoidance of the EF lawn, followed by periodic returns, forming a cyclic behavioural pattern with peak avoidance observed approximately four hours post-exposure (104). Exposure to EF causes intestinal bloating, which results in pathogen avoidance mediated by AWB and AWC neurons. Avoidance is also contingent on the presence of the NPR-1, TAX-2, and TAX-4 proteins in conjunction with ASE neurons. Additionally, GON-2 and GTL-2, two transient receptor potential melastatin channels, have been identified as mediators of this avoidance response (104). The sensorimotor circuit for learned avoidance of EF is initiated by the AWB sensory neurons, proceeding through the interneurons AUA/RMG, extending to the lower layer interneurons AVA/AVD/AVE, and ultimately reaching the motor neurons VA/DA/AS/VA/DD (34).
7 Concluding remarksFrom the phenotypic to the molecular level, studies on pathogen avoidance in C. elegans have come a long way. The interpretation of pathogen avoidance has enriched our knowledge of these sophisticated behaviours. In this paper, we reviewed the stimuli that trigger pathogen avoidance, including alterations in aerotaxis, intestinal bloating, and metabolites. Furthermore, we summarised the neural circuits in pathogen avoidance, transgenerational epigenetic inheritance of pathogen avoidance, signalling crosstalk between pathogen avoidance and innate immunity, and C. elegans avoidance of non-pathogenic bacteria. Diverse pathogen avoidance mechanisms provide comprehensive and adaptive protection, enabling multiple reactions to occur in complex environments. Collectively, these mechanisms contribute to the evolutionary advantages of C. elegans. Future studies could investigate how each mechanism contributes to this complex behaviour. It would also be valuable to investigate the interactions that may occur among pathogen avoidance mechanisms. Given the evolutionary conservation between C. elegans and humans, these studies may shed light on our understanding of gut neural signalling in humans, thus providing insight into potential therapies for treating bacterial infections.
Author contributionsML: Writing – original draft, Writing – review & editing. YT: Writing – original draft, Writing – review & editing. HT: Writing – original draft, Writing – review & editing. WT: Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (21991084), National Key Research and Development Program of China (2019YFA090580), and Science, Technology and Innovation Commission of Shenzhen Municipality (JCYJ20210324121000001).
AcknowledgmentsWe would like to thank R. Pocock and V. Anggono for critical reading and corrections of the manuscript.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
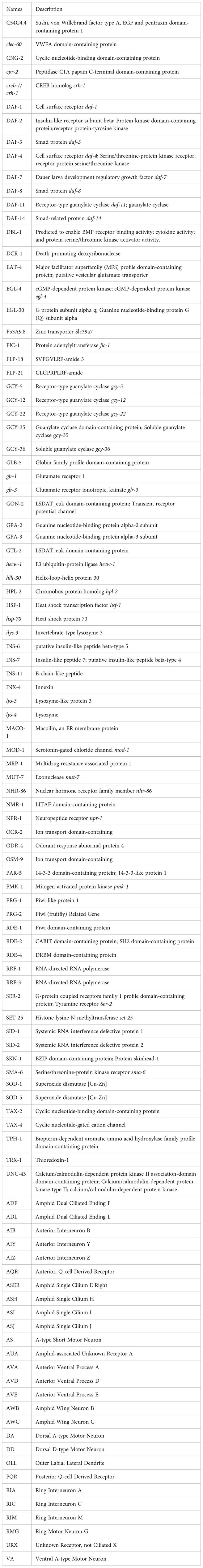
Glossary
1. Schaller M. The behavioural immune system and the psychology of human sociality. Philos Trans R Soc London Ser B Biol Sci. (2011) 366:3418–26. doi: 10.1098/rstb.2011.0029
CrossRef Full Text | Google Scholar
2. Boillat M, Challet L, Rossier D, Kan C, Carleton A, Rodriguez I. The vomeronasal system mediates sick conspecific avoidance. Curr Biol CB. (2015) 25:251–5. doi: 10.1016/j.cub.2014.11.061
CrossRef Full Text | Google Scholar
3. Sarabian C, Belais R, MacIntosh AJJ. Feeding decisions under contamination risk in bonobos. Philos Trans R Soc London Ser B Biol Sci. (2018) 373. doi: 10.1098/rstb.2017.0195
CrossRef Full Text | Google Scholar
4. Goodall J. Social rejection, exclusion, and shunning among the gombe chimpanzees. Ethology Sociobiology. (1986) 7:227–36. doi: 10.1016/0162-3095(86)90050-6
CrossRef Full Text | Google Scholar
5. Pacheco-López G, Bermúdez-Rattoni F. Brain-immune interactions and the neural basis of disease-avoidant ingestive behaviour. Philos Trans R Soc London Ser B Biol Sci. (2011) 366:3389–405. doi: 10.1098/rstb.2011.0061
CrossRef Full Text | Google Scholar
7. Samuel BS, Rowedder H, Braendle C, Félix MA, Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci United States America. (2016) 113:E3941–9. doi: 10.1073/pnas.1607183113
CrossRef Full Text | Google Scholar
8. Tan MW, Ausubel FM. Caenorhabditis elegans: A model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr Opin Microbiol. (2000) 3:29–34. doi: 10.1016/s1369-5274(99)00047-8
PubMed Abstract | CrossRef Full Text | Google Scholar
9. Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in. Caenorhabditis elegans Nat. (2005) 438:179–84. doi: 10.1038/nature04216
CrossRef Full Text | Google Scholar
10. Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PloS Biol. (2006) 4:e274. doi: 10.1371/journal.pbio.0040274
PubMed Abstract | CrossRef Full Text | Google Scholar
11. Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. (2004) 430:317–22. doi: 10.1038/nature02714
PubMed Abstract | CrossRef Full Text | Google Scholar
12. Reddy KC, Hunter RC, Bhatla N, Newman DK, Kim DH. Caenorhabditis elegans npr-1-mediated behaviors are suppressed in the presence of mucoid bacteria. Proc Natl Acad Sci United States America. (2011) 108:12887–92. doi: 10.1073/pnas.1108265108
CrossRef Full Text | Google Scholar
13. de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. (2002) 419:899–903. doi: 10.1038/nature01169
PubMed Abstract | CrossRef Full Text | Google Scholar
14. Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol CB. (2006) 16:649–59. doi: 10.1016/j.cub.2006.03.023
CrossRef Full Text | Google Scholar
15. Abergel Z, Chatterjee AK, Zuckerman B, Gross E. Regulation of neuronal oxygen responses in C. elegans is mediated through interactions between globin 5 and the H-nox domains of soluble guanylate cyclases. J Neurosci. (2016) 36:963–78. doi: 10.1523/jneurosci.3170-15.2016
PubMed Abstract | CrossRef Full Text | Google Scholar
16. Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. (2014) 159:267–80. doi: 10.1016/j.cell.2014.09.011
PubMed Abstract | CrossRef Full Text | Google Scholar
17. Singh J, Aballay A. Microbial colonization activates an immune fight-and-flight response via neuroendocrine signaling. Dev Cell. (2019) 49:89–99.e4. doi: 10.1016/j.devcel.2019.02.001
PubMed Abstract | CrossRef Full Text | Google Scholar
19. Hong C, Lalsiamthara J, Ren J, Sang Y, Aballay A. Microbial colonization induces histone acetylation critical for inherited gut-germline-neural signaling. PloS Biol. (2021) 19:e3001169. doi: 10.1371/journal.pbio.3001169
PubMed Abstract | CrossRef Full Text | Google Scholar
20. Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. (2009) 323:382–4. doi: 10.1126/science.1166527
PubMed Abstract | CrossRef Full Text | Google Scholar
22. de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. (1998) 94:679–89. doi: 10.1016/s0092-8674(00)81609-8
PubMed Abstract | CrossRef Full Text | Google Scholar
23. Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol CB. (2005) 15:905–17. doi: 10.1016/j.cub.2005.04.017
CrossRef Full Text | Google Scholar
28. Ooi FK, Prahlad V. Olfactory experience primes the heat shock transcription factor hsf-1 to enhance the expression of molecular chaperones in C. elegans. Sci Signaling. (2017) 10. doi: 10.1126/scisignal.aan4893
CrossRef Full Text | Google Scholar
29. Zatsepina OG, Evgen'ev MB, Garbuz DG. Role of a heat shock transcription factor and the major heat shock protein hsp70 in memory formation and neuroprotection. Cells. (2021) 10. doi: 10.3390/cells10071638
PubMed Abstract | CrossRef Full Text | Google Scholar
30. Rui Z, Li X, Zhu X, Liu J, Domigan B, Barr I, et al. Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc Natl Acad Sci United States America. (2014) 111:18237–42. doi: 10.1073/pnas.1419701112
CrossRef Full Text | Google Scholar
31. Prakash D, Ms A, Radhika B, Venkatesan R, Chalasani SH, Singh V. 1-undecene from Pseudomonas aeruginosa is an olfactory signal for flight-or-fight response in Caenorhabditis elegans. EMBO J. (2021) 40:e106938. doi: 10.15252/embj.2020106938
PubMed Abstract | CrossRef Full Text | Google Scholar
32. Ha HI, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, Qin Y, et al. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron. (2010) 68:1173–86. doi: 10.1016/j.neuron.2010.11.025
PubMed Abstract | CrossRef Full Text | Google Scholar
33. Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. eLife. (2014) 3:e01948. doi: 10.7554/eLife.01948
PubMed Abstract | CrossRef Full Text | Google Scholar
34. Filipowicz A, Lalsiamthara J, Aballay A. Dissection of a sensorimotor circuit underlying pathogen aversion in C. elegans. BMC Biol. (2022) 20:229. doi: 10.1186/s12915-022-01424-x
PubMed Abstract | CrossRef Full Text | Google Scholar
36. Qin Y, Zhang X, Zhang Y. A neuronal signaling pathway of camkii and gqα Regulates experience-dependent transcription of tph-1. J Neurosci. (2013) 33:925–35. doi: 10.1523/jneurosci.2355-12.2013
留言 (0)