BCG is the primary treatment for high-risk non-muscle invasive bladder cancer (NMIBC).1 Yet, at least one-third of NMIBCs treated with BCG will not respond and progress to more advanced stages of bladder cancer. In 2020, following the results of KEYNOTE-057,2 3 in which 40.6% of patients had a complete response at 3 months, pembrolizumab monotherapy was approved for use in patients with BCG unresponsive high-risk NMIBC. Unfortunately, response to pembrolizumab is not durable, and overall, 80% of treated patients will have recurred or progressed by 12 months.
Since FDA approval, pembrolizumab has become a potential treatment for BCG unresponsive high-risk NMIBC. Yet, there is limited information on how to best identify patients who will benefit from this course of treatment. Identifying biomarkers that predict response to pembrolizumab could facilitate the selection of NMIBCs and spare unresponsive patients the unnecessary toxicity associated with immune checkpoint treatment.
We have previously performed a multi-omics evaluation of a small phase I trial of BCG unresponsive NMIBCs treated with BCG and intravesical pembrolizumab.4 While this study primarily focused on tumor response mechanisms in the unique setting of intravesical administration of pembrolizumab and BCG, we were intrigued to compare this to intravenous pembrolizumab. We previously identified an increase in T cells and decreased expression of exhaustion markers associated with response to intravesical pembrolizumab.4 To evaluate the response mechanisms of intravenous pembrolizumab, we performed digital spatial profiling of tumors from responders and non-responders before and after treatment. Our results describe the spatial transcriptomic differences in pretreatment NMIBCs and offer initial insights to identify individuals who may respond to intravenous pembrolizumab.
MethodsSample identification and collectionAfter obtaining institutional review board approval, the Northwestern Medicine Enterprise Data Warehouse was queried to identify patients with BCG unresponsive NMIBC who were treated with at least three cycles of intravenous pembrolizumab. Patients were selected for inclusion in the study if adequate pretreatment and post-treatment formalin-fixed paraffin-embedded (FFPE) bladder biopsies were available. We identified five patients who met the inclusion criteria (online supplemental table 1). FFPE blocks were sectioned at a thickness of 5 µm and mounted on slides for DSP analysis. Clinical response was defined by patients with negative blue-light cystoscopy, cytology, and mapping biopsy after 3 months of treatment with pembrolizumab.
Digital spatial profilingUsing methods previously described,4 5 slides were deparaffinized, and target retrieval was performed, followed by RNA probe library in situ hybridization using the NanoString GeoMX Whole Transcriptome Atlas (WTA) (NanoString, Seattle, WA). Slides were stained with immunofluorescent antibodies from the NanoString solid tumor tumor microenvironment (TME) morphology kit (Pan-CK for epithelial cells, CD45 for immune cells, and SYTO 83 for nuclear staining) (NanoString) and loaded onto the GeoMx digital spatial profiler (NanoString). Regions of interest were manually selected for transcriptomic profiling, and photocleaved DNA oligonucleotides were collected in individual wells of a 96-well plate. Illumina i5, and i7 dual indexing primers were added to the oligonucleotide tags during library preparation. Sequencing was performed on an Illumina NovaSeqX (Illumina Inc, San Diego, CA). Reads were trimmed, stitched, aligned, and de-duplicated. Fastq files were converted to DCC files using the GeoMx NGS Pipeline V.2.3.3.10, which were then loaded onto the GeoMx instrument and converted into target counts for each segment. Raw counts were filtered based on two criteria: to remove targets detected below the limit of quantitation and to remove segments with fewer than 50 nuclei. Filtered raw counts were Q3 normalized for comparison across all segments and were used for further analysis (online supplemental table 2).
Bioinformatics and data visualizationAll analyses were performed in R V.4.2.3. Principal component analysis was performed using PCAtools V.2.10.0. Heatmaps were generated using ComplexHeatmap V.2.14.0. Differential expression analysis was performed using limma V.3.54.2. Volcano plots were generated using ggplot2 V.3.4.4. Pathway analysis was conducted using msigdb6 gene sets downloaded using msigdbR V.7.5.1 package and analyzed using fgsea V.1.24.0. Gene signatures for the intravesical cohort were generated by comparing PanCK+ and stroma-specific expression profiles between responders and non-responders. Enrichment of these gene sets within the current intravenous pembrolizumab cohort was tested using the GSEA function, and GSEA plots were generated using the gseaplot function in clusterProfiler v4.6.2. Exhaustion score was calculated as a mean expression of PDCD1, HAVCR2, LAG3, CTLA4, and TIGIT genes. Inflammation score was generated as a mean expression of genes within the msigdb hallmark interferon-alpha (IFN-α) and IFN-γ response gene sets.6 Immune deconvolution of the stromal segments was performed using SpatialDecon V.1.8.0. Immune Infiltration score was generated as a sum of the different immune cell types (T/NK cells, B cells, myeloid cells, neutrophils, and mast cells) identified by immune deconvolution.
ResultsCohortWe have previously performed bulk RNA sequencing to identify expression signatures of CPI-treated tumors across multiple cohorts.7 Yet, due to the limited size of the tumor epithelium in NMIBCs with carcinoma in situ (CIS), bulk RNA-seq lacks the resolution required to dissect the granular details of the tumor/TME in this setting. Therefore, here, we performed spatial profiling of tumor sections before and after intravenous pembrolizumab to identify (1) pretreatment features associated with response or resistance and (2) describe how intravenous pembrolizumab alters the tumor/TME interaction in BCG unresponsive NMIBCs. Five patients treated with intravenous pembrolizumab were evaluated: three non-responders and two responders. The demographics and clinical history of the cohort are listed in online supplemental table 1. We evaluated pre-pembrolizumab clinical features using multiple described nomograms and could not predict response.8–10 Yet, non-responders were more likely to be multi-focal and were older than 70 (online supplemental table 3).
Spatial transcriptomic analysisA total of 119 areas of interest (AOIs) were profiled across five patients at two treatment time points (pre and post), capturing 60 tumor and 59 stromal segments. As depicted in online supplemental figure 1A, we observed a distinct segregation of PanCK+ and stromal segments. Comparing gene expression, we find elevated levels of keratins within the PanCK+ segments and increased expression of stromal (ACTA2, COL1A1, COL4A1, COL6A1) and immune markers (IGHG4, IGHG2) in the stromal segments (online supplemental figure 1B).
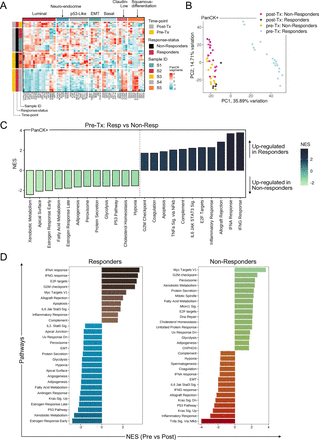
 Figure 1
Figure 1 (A) Heatmap illustrating gene expression patterns across bladder cancer subtypes within PanCK+ segments. (B) Principal component analysis plot showing the distribution and relationships among PanCK+ segments in the cohort. (C) Bar plot showing significantly enriched pathways in pretreatment PanCK+ segments, distinguishing responders from non-responders. (D) Bar plot highlighting pathways changing in responders and non-responders pretreatment to post-treatment.
Characteristics of PanCK+ tumor segments that are predictive of response and Impact of therapy on gene expression in PanCK+ tumor segmentsThe reported response to pembrolizumab in BCG unresponsive bladder NMIBCs is 40% at 3 months. To identify expression signatures of the tumor segments that may affect the treatment response, we first characterized the 60 PanCK+ AOIs. Comparing the expression of canonical bladder cancer subtype markers across the cohort (figure 1A), we identified elevated levels of claudin-low and squamous differentiation markers in pretreatment PanCK+ segments from responders. In contrast, pretreatment PanCK+ AOIs from non-responders were enriched for luminal markers. Claudin-low tumors have been previously identified to be immune infiltrated with an expression profile predicted to respond to immune checkpoint blockade.11
We identified minimal heterogeneity within AOIs from each patient (online supplemental figure 1C). Pretreatment responders formed a distinct cluster independent of the rest of the cohort (figure 1B) and demonstrated upregulation of genes related to IFN-α and IFN-γ response, TNF-alpha signaling, and the IL6-JAK-STAT signaling pathways (figure 1C, online supplemental table 4). These findings suggest a heightened inflammatory tumor epithelium within the pretreatment responder segments. In contrast, PanCK+ segments from non-responders had elevated markers of p53 pathway genes, as well as estrogen response (figure 1C, online supplemental table 4).
We recently described a transcriptome-based evaluation of the response to immune checkpoint inhibitors in muscle-invasive bladder cancer.7 We hypothesized that the underlying mechanisms of pembrolizumab activity may remain consistent across disease stages. To test this hypothesis, we applied the previously identified five gene signatures to tumor AOIs within this cohort.7 We find Cluster1-MIBC-CPI signatures, which were associated with resistance to pembrolizumab (15% complete response) and enriched in luminal immune cold-MIBC tumors with FGFR3 mutations, to be upregulated in pretreatment PanCK+ segments from non-responders in this cohort. Robertson et al found Clusters 2 and 3-MIBC-CPI were associated with immune infiltration and a favorable response to immunotherapy (63% complete response).7 We find Cluster3 and Cluster2-MIBC-CPI signatures upregulated in pretreatment PanCK+ segments from responders in the intravenous pembrolizumab cohort (online supplemental figure 1D). This suggests that the CPI clusters may be conserved in the tumor epithelium across stage.
We were interested in the dynamic changes caused by pembrolizumab. Spatial profiling of longitudinally collected specimens pretherapy and post-therapy allowed us to isolate the impact of therapy on the tumor epithelium and the TME separately. Specifically, 807 genes were differentially regulated in responsive tumors, compared with 22 in non-responsive tumors (online supplemental figure 1E). To identify the gene sets that change in response to pembrolizumab, we compared PanCK+ segments pretreatment to those collected post-treatment. As seen in figure 1D, post-treatment PanCK+ segments from responders showed a net increase in inflammation-related pathways. In contrast, post-treatment PanCK+ segments from non-responders upregulated cell cycle gene sets such as G2M checkpoint and E2F targets (figure 1D). Our analysis indicates that PanCK+ segments exhibiting signs of inflammation before intravenous pembrolizumab administration may display enhanced responsiveness to this therapy.
Characteristics of PanCK− stromal segments that are predictive of responseNext, we profiled the features of the TME that could contribute to pembrolizumab’s responsiveness. Pretreatment TME AOIs are separated by response (figure 2A). Inflammatory gene sets such as IFN-α and IFN-γ response genes were strongly upregulated in responders, whereas non-responders upregulated markers of EMT, myogenesis, and angiogenesis (figure 2B). We used immune deconvolution to identify individual immune cell populations within each stromal AOI. Prior to treatment, TMEs from responders had elevated levels of neutrophils, T cells, and NK cells in the TME relative to non-responders (figure 2C).

 Figure 2
Figure 2 (A) Principal component analysis visualizing the distribution of stromal areas of interest in the cohort. (B) Bar plot highlighting pathways significantly enriched in pretreatment stromal segments from responders and non-responders. (C) Violin boxplots comparing the cellular abundance of specified immune populations within the indicated conditions. (D) Violin boxplots compare exhaustion scores between responders and non-responders in both pretreatment and post-treatment samples. (E) GSEA plot showing the enrichment of stromal gene signatures generated in the intravesical cohort in differentially expressed genes from stromal segments from the intravenous pembrolizumab cohort comparing responders to non-responders. (F) Volcano plot depicting differential expression of protein markers in responder’s versus non-responders to pembrolizumab therapy in the PURE-01 cohort.
Overexpression of PD-1 on the cell surface is a well-established marker of T-cell exhaustion.12 Comparing the expression of exhaustion markers across the different conditions, we find elevated levels of exhaustion markers in the pretreatment stromal compartment from responders relative to non-responders (figure 2D). Next, we find that stromal gene signatures identified in a study of intravesical BCG+pembrolizumab therapy were similarly enriched in this cohort of pembrolizumab monotherapy (figure 2E).
We further validated these findings in a proteomics digital spatial profiling dataset of muscle invasive tumors treated with neo-adjuvant pembrolizumab (PURE-01 study7). Tumors with complete response had elevated levels of cytotoxic T cell marker CD8, immune checkpoint markers LAG3, Tim-3, and PD-L1 relative to the tumors that did not respond to this therapy (figure 2F).
Comparison of response strategies for high-risk NMIBCOur group has previously described the spatial comparisons of the first-in-human administration of BCG and intravesical pembrolizumab.4 Given this cohort’s unique administration of pembrolizumab, we wanted to evaluate the differences in urothelial gene expression profile between intravesical pembrolizumab and BCG compared with intravenous pembrolizumab.
As seen in figure 3A, responders to the combination intravesical therapy exhibited low levels of inflammation in the PanCK+ segments, whereas responders to the intravenous pembrolizumab monotherapy showed elevated levels of inflammation in the pretreatment PanCK+ segments. Responders in both cohorts exhibited elevated levels of immune infiltration in the stroma.

 Figure 3
Figure 3 (A) Correlation plot comparing inflammation score for PanCK+ segments and infiltration score in the neighboring stromal segments between responders and non-responders for the intravesical BCG+pembrolizumab and intravenous pembrolizumab cohort. (B) GSEA plot showing the enrichment of stromal gene signatures generated in the intravesical cohort in differentially expressed genes from stromal segments from the intravenous pembrolizumab cohort comparing responders to non-responders.
Testing gene signatures, we find that the PanCK+ signature that predicts response in the intravesical pembrolizumab and BCG cohort identified non-responders in the intravenous pembrolizumab cohort, and the PanCK+ signature that predicts lack of response in the intravesical combination therapy cohort are enriched in responders in the pembrolizumab monotherapy cohort (figure 3B).
Overall, our findings suggest tumor segments that are inflamed before therapy might benefit more from intravenous pembrolizumab, whereas pretreatment non-inflamed BCG unresponsive tumors might be a better candidate for treatment with a combination therapy of BCG and pembrolizumab.
DiscussionImmunotherapy is being used to manage bladder cancers at all stages.13 Overall, the response rate to CPIs in metastatic urothelial cancer is ~20%, with patients expressing PDL1/PD1 more likely to have a more durable response.14 15 In early-stage BCG unresponsive NMIBC, a similar response was described for patients with CIS, with or without papillary tumors in KEYNOTE0572 3 and SWOG S1605.16 These results suggest similar drug activity and provide a rationale for identifying the specific patients who might benefit long-term from CPI.
Here, we attempt to profile early-stage NMIBCs to identify response mechanisms to CPI. Despite the frequency of BCG-responsive BCa, few patients are cured (NED for >24 months). Our goal with this investigation was to identify pre-treatment features associated with response.
We evaluated the tumor and TME by response status and identified pretreatment signatures associated with response to intravenous pembrolizumab. We validated signatures generated from the PURE01-CPI trial and the intravesical BCG and pembrolizumab trial. Our data confirms that resistance to pembrolizumab is a consequence of limited immune infiltration into the TME.4 7 17
It is notable that the pretreatment epithelial signature predictive of response to intravenous pembrolizumab in BCG unresponsive disease seen here differs from our previous work with a combination of intravesical BCG and pembrolizumab. Given the sequences of therapy in these two studies (BCG failure followed by intravenous pembrolizumab vs BCG failure followed by simultaneous administration of intravesical BCG and pembrolizumab), most of the benefit from BCG may involve inducing an inflammatory anti-tumor response.
In contrast, pembrolizumab “releases the breaks” on an already present yet ineffective inflammatory response from BCG. This was confirmed by a decrease in exhaustion markers in responsive tumors. Finally, our results suggest that the immune response to pembrolizumab is conserved across bladder cancer stages.
A limitation of our study was the limited number of patients in our cohort evaluated. Yet, to our knowledge, this is the largest cohort of patients with NMIBC evaluated by spatial profiling. Further studies, with larger numbers of patients and with other checkpoint therapies, may provide justification for pretreatment evaluation of tumor and TME to predict response to therapy. As further liquid and genomic biomarkers are identified, we anticipate a greater role in decision-making.18 19
Our findings highlight the need to assess the transcriptomic state of the BCG unresponsive tumors prior to deciding the course of treatment. We anticipate that the application of an expression-based biomarker like the one described here can identify tumors that are likely to respond to pembrolizumab. Further evaluation of more patients treated with different CPIs is needed to refine our results.
ConclusionWe performed a spatial-based evaluation of tumors treated with pembrolizumab. We identified distinct expression signatures associated with the response and resistance of the tumor and TME. Future studies evaluating the accuracy of these signatures will help validate our findings and facilitate biomarker application in patients with NMIBC.
Data availability statementData are available upon reasonable request. Data are included in the supplemental file.
Ethics statementsPatient consent for publicationNot applicable.
Ethics approvalAll research was approved by Northwestern IRB. The study was approved by the Northwestern IRB. The study was considered exempt by Northwestern because of the low risk to patients, using retrospective samples obtained for clinical care (category 4). The IRB study is STU00215723.
留言 (0)