With the overuse of antibiotics, the problem of bacterial resistance has become increasingly serious, followed by the emergence and prevalence of various superbugs, including carbapenem-resistant Enterobacteriaceae, carbapenem-resistant Acinetobacter baumannii, which pose a huge threat to human health annually and places a huge burden on healthcare systems (Hsu et al., 2017; Tacconelli et al., 2018). Tigecycline, as third generation tetracycline drug, exerts antibacterial effects by binding to the 30S subunit of the ribosome and inhibiting peptide elongation (Olson et al., 2006). It is recognized as a crucial option against multi-drug resistant bacteria, often referred to as the “last resort.”
However, recently, a large number of tetracycline resistance genes have emerged and become popular (Fang et al., 2020). The plasmid-mediated tet(X4) gene, which mediates the high level of tigecycline resistance, was discovered in 2019 (He et al., 2019). In addition, tet(X4) is prevalent worldwide, primarily in China (Zhang et al., 2022), and has been found in a variety of sample sources, including patients, farms, slaughterhouses, sewage, and migratory birds (Chen et al., 2019; Cui et al., 2022; Dong, 2022). Thus, the spread of the tet(X4) is a public health concern that cannot be ignored.
An important reason for the pandemic of resistance genes is that they tend to transfer in individual samples, which is associated with a variety of transfer elements (Partridge et al., 2018). Previous studies have shown that the transfer of tet(X4) is related to several transfer elements, including ISCR2, IS26, and IS1R (Liu et al., 2022). The transmission mode of tet(X4) requires further investigation. The type IV secretion system (T4SS), which mediates the horizontal transfer of antibiotic resistance and virulence genes (Christie, 2016), has been found in the tet(X4)-positive plasmid (Du et al., 2020). However, the insertion of large fragments containing T4SS associated genes into tet(X4) plasmids has rarely reported.
In this study, we described the genomic characteristics of tet(X4)-positive E. coli from animals and humans in China and the potential element insertions related to T4SS which may associated with high-frequency transfer of tet(X4). Therefore, it emphasizes that the sustainability surveillance of tet(X4) is imperative.
Materials and methods Epidemiological studyWe undertook a retrospective observational cross-sectional study to assess the prevalence of tet(X4)-positive E. coli in animals, farmers, inpatients and healthy volunteers. A total of 775 nonduplicate samples were collected from two farms and one hospital in Guangdong province, China from 2019 to 2020, including 475 nasal swab samples of pigs in two farms, with farm 1 contributing 419 samples and farm 2 contributing 56 samples, 67 fecal swab samples of pigs, 48 skin swab samples of pig farm workers in farm 1, 172 fecal swab samples of inpatients and 13 fecal swab samples of healthy volunteers.
Samples were cultured on brain heart infusion (BHI) plates containing tigecycline (2 mg/L) and incubated overnight at 37°C. Subsequently, the tigecycline-resistant colonies with the typical colonial morphology of Enterobacteriaceae – namely moist-looking colonies which were smooth, translucent with a regular edge were screened for the presence of tet(X4) by PCR using the primers tet(X4)-F (5’-AGGAACAGGACACGAATTGC-3′) and tet(X4)-R (5’-TTACTGGCGGAGCCGTCTA-3′) (Ding et al., 2020). Species identification of tet(X4)-positive isolates was conducted by MALDI-TOF MS (BrukerDaltonik GmbH, Bremen, Germany) and 16S rDNA sequencing.
Antimicrobial susceptibility testingThe minimum inhibitory concentrations (MICs) of tetracycline (TET), gentamicin (GEN), amikacin (AMK), ampicillin (AMP), imipenem (IMP), ceftazidime (CAZ), cefotaxime (CTX), chloramphenicol (CHL), ciprofloxacin (CIP), fosfomycin (FOS), and trimethoprim-sulfamethoxazole (SXT) were determined using the agar dilution method, excepted tigecycline (TGC) and colistin (CT) were determined using the broth dilution method. The results were interpreted in accordance with the Clinical and Laboratory Standards Institute guidelines (CLSI, 2022), as well as the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2021).
Whole-genome sequencing and bioinformatics analysisGenomes of tet(X4)-positive E. coli were sequenced using Illumina NovaSeq 6000 platform. After completing quality control of raw sequencing reads using fastp v0.20 (Chen et al., 2018), the contigs were generated using SPAdes v3.13.1 (Bankevich et al., 2012). Contigs <500 bp in length were filtered out. Coding sequences were annotated using Prokka v1.12. Specific genes were detected by comparing contigs to various corresponding database using Abricate v0.8.7, including Resfinder database for resistance genes, Virulence Factor Database for virulence genes, and Plasmidfinder database for replicon types. MLST (Multilocus Sequence Typing) was identified using mlst v2.23.0.
Long-reads sequencing and plasmid analysisTo analyze the tet(X4)-carrying plasmids, long-reads sequencing was performed using the PacBio platform. Raw data were assembled using Unicycler v0.4.8. The method of gene annotation and identification was the same as that used for the whole genome analysis mentioned above. Insertion sequences were identified using Isfinder. T4SS genes were identified using oriTfinder. The comparison of the genetic environments of tet(X4) were performed using Easyfig v2.1 and R package “gggenes.”
Phylogenetic analysisPan-genome was constructed using Roary v3.12.0 with the “- e -- mafft” parameter (Page et al., 2015). Core-genome single nucleotide polymorphism were extracted and filtered using snp-sites (Page et al., 2016) and VCFtools (Danecek et al., 2011), respectively. The phylogenetic tree based cgSNP was constructed using RAxML v8.2.10 (Stamatakis, 2014) with GTR + G model and was visualized using iTOL (Letunic and Bork, 2021). The Minimum spanning tree was constructed using PHYLOViZ 2.0 (Nascimento et al., 2017).
Results Molecular epidemiology of tet(X4)-positive Escherichia coliAmong the 775 multi-source samples, 146 (18.84%) tet(X4)-positive E. coli were isolated. Out of these, 144 isolates were obtained from pigs originating from farm 1 (106/486, 21.81%) and farm 2 (38/56, 67.86%). Additionally, 2 isolates were derived from skin swabs of farm workers and inpatients, respectively. Specifically, 26.95% (128/475) of tet(X4)-positive E. coli were obtained from nasal swabs of pigs, followed by 23.88% (16/67) from fecal swabs of pigs. In contrast, tet(X4)-positive E. coli appeared sporadically in the skin swabs of pig farm workers (1/48, 2.08%) and anal swabs of inpatients (1/172, 0.58%). It was tet(X4) negative in fecal swabs of healthy volunteers.
We tested the antimicrobial susceptibility of 146 tet(X4)-positive E. coli strains to 13 commonly used antibiotics. The MIC results showed that 145 tet(X4)-producing isolates were resistant to tigecycline and tetracycline. The MIC values for tigecycline range from 8 to 128 mg/L. Although carrying the tet(X4) gene, the strain 20PN649-3 was sensitive to tigecycline with a MIC of 0.5 mg/L. In addition, most of tet(X4)-positive E. coli were resistant to chloramphenicol (144/146, 98.63%), ampicillin (142/146, 97.26%) and trimethoprim/sulfamethoxazole (122/146, 83.56%). Resistance rates were lower for cefotaxime (45/146, 30.82%), ceftazidime (40/146, 27.40%), gentamicin (40/146, 27.40%), ciprofloxacin (28/146, 19.18%), fosfomycin (1/146, 0.68%) and imipenem (1/146, 0.68%). All tet(X4)-positive E. coli were susceptible to amikacin and colistin (Figure 1; Supplementary Table S1). These results indicated that these tet(X4)-positive E. coli were resistant not only to tetracycline antibiotics but also to a range of other antibiotics.
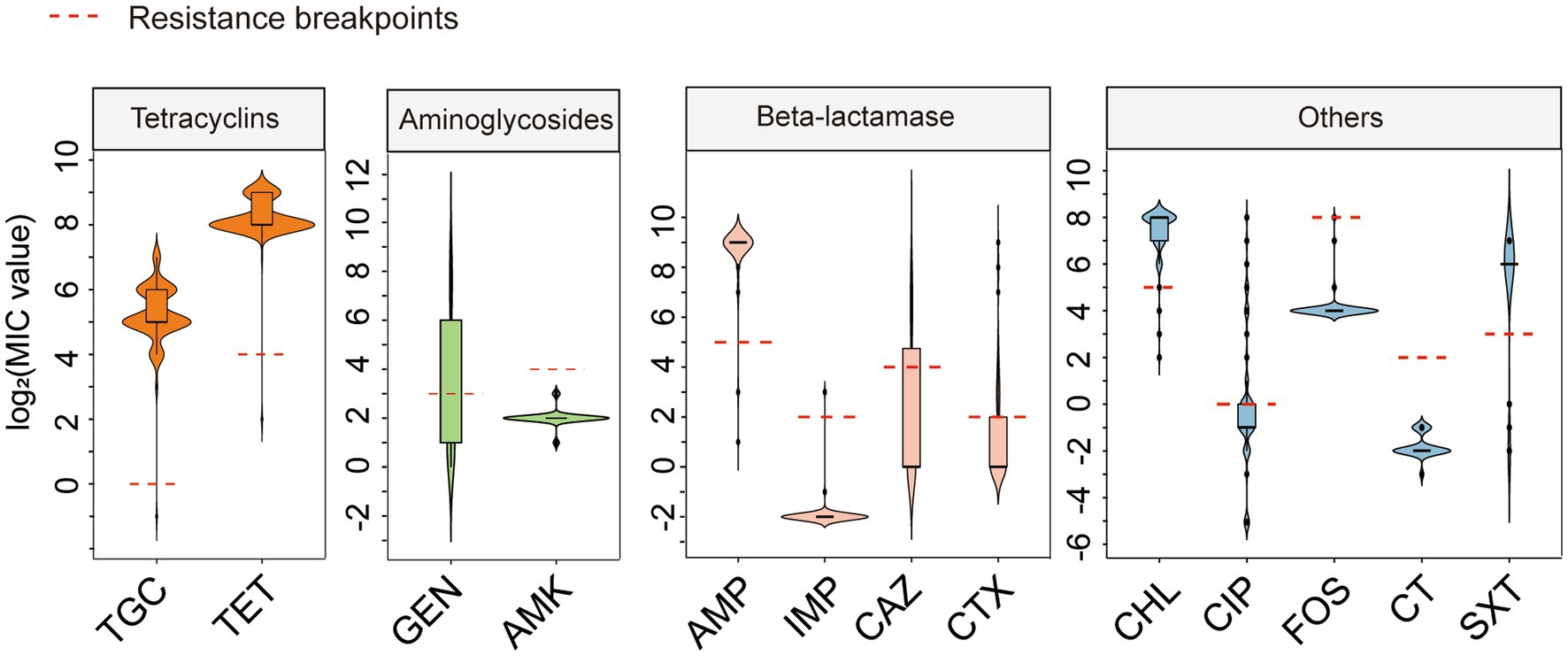
Figure 1. Antimicrobial resistance profiles of 56 tet(X4)-positive E. coli. TGC, Tigecycline; TET, Tetracycline; GEN, Gentamicin; AMK, Amikacin; AMP, Ampicillin; IMP, Imipenem; CAZ, Ceftazidime; CTX, cefotaxime; CHL, Chloramphenicol; CIP, Ciprofloxacin; FOS, Fosfomycin; CT, Colistin; SXT, Sulfamethoxazole/Trimethoprim. Red dashed line: MIC breakpoint based on CLSI and EUCAST guidelines: TGC > 0.5, TET ≥ 16, GEN ≥ 8, AMK ≥ 16, AMP ≥ 32, IMP≥4, CAZ ≥ 16, CTX ≥ 4, CHL ≥ 32, CIP ≥ 1, FOS ≥ 256, CT ≥ 4, STX > 4.
Phylogenetic analysis of tet(X4)-positive Escherichia coli strains from various sourcesFifty-six tet(X4)-positive E. coli including 2 isolates from humans and 54 isolates randomly chosen from 144 tet(X4)-positive E. coli from pigs were further sequenced by whole genome sequencing. Among these, 54 strains were derived from two different farms (farm 1, n = 53 and farm 2, n = 1). To analyze the genomic differences between tet(X4)-positive E. coli strains from different sources, 8 tet(X4) positive E. coli strains of different sources obtained from GenBank were included in the dataset (Supplementary Table S2A). As mentioned above, 64 tet(X4)-positive E. coli strains used for phylogenetic analysis were sourced from healthy individual’s stool, inpatient fecal samples, pig swabs, chicken, pork, and wastewater (Figure 2A). The origins of these samples varied and clustered into 30 distinct STs (Figures 2B,C). In the strains collected in this study, the STs with the highest prevalence were ST877 (8/56, 14.29%), ST871 (6/56, 10.71%), ST195 (7/56, 12.50%), ST10 (4/56, 7.14%), and ST398 (4/56, 7.14%). The sample collected from inpatient belongs to ST9867. The ST871 strain S221-6 from inpatients in the public dataset showed close proximity in the phylogenetic tree to the corresponding ST871 strains collected in our study. Similarly, the ST10 strain YPE10 from pork in the public dataset exhibited a close relationship in the phylogenetic tree with the ST10 strains collected in our study. This finding indicated that tet(X4) had undergone extensive dissemination among multiple lineages of E. coli.
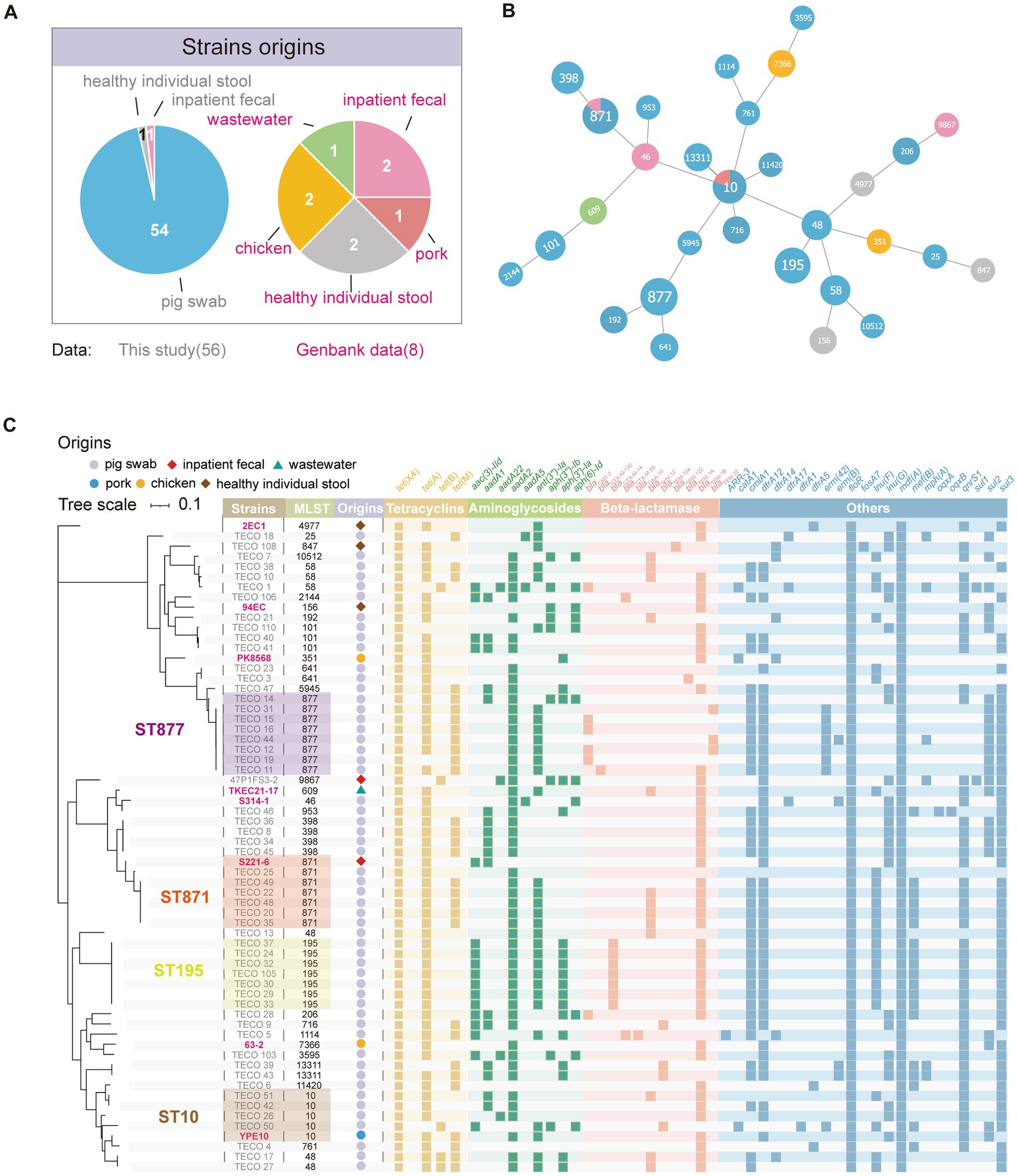
Figure 2. Population structure and resistance gene characteristics of 65 tet(X4)-positive E. coli. (A) Pie chart illustrating the distribution of sample sources. GenBank data accession numbers mentioned in this figure are listed in Supplementary Table S2A. (B) Minimum spanning tree based on MLST (Multilocus Sequence Typing). Each node in the tree represents a specific MLST type. The colors assigned to the nodes correspond to the strain’s origins, following the same color scheme as presented in (A). (C) Phylogenetic tree and heatmap of resistance genes among the strains. The reference strains obtained from GenBank are highlighted in the tree.
Antimicrobial resistance gene (ARG) profiles of tet(X4)-positive Escherichia coliA total of 60 resistance genes have been identified via whole genome sequencing. In the strains collected in this study, except for the tet(X4) gene found in all samples, most strains also carried tet(A) (49/56, 87.5%) (Figure 2C). Several resistance genes associated with aminoglycoside antibiotic resistance, including the widely carried aad2 and ant(3″)-la genes, were identified. For beta-lactam antibiotics, a variety of resistance genes were detected, including blaCMY-2 (6/56, 10.71%), blaCTX-M-14 (7/56, 12.5%), blaSHV-12 (12/56, 21.43%), blaTEM-1B (36/56, 64.29%). Furthermore, all strains harbored the florfenicol resistance gene floR and the macrolides resistance gene mdf(A). Additionally, high carriage rates were observed for the chloramphenicol resistance gene cmlA1 (40/56, 71.43%), trimethoprim resistance gene dfrA12 (41/56, 73.21%), fluoroquinolone resistance gene qnrS1 (40/56, 71.43%), and sulphonamide resistance gene sul3 (46/56, 82.14%) (Supplementary Table S3).
Various genetic elements of tet(X4) in Escherichia coliThe transfer of tet(X4) is frequently associated by surrounding mobile elements. To investigate the genomic region housing tet(X4) and to observe the associated mobile elements, we conducted a comparative analysis of the genetic environment surrounding tet(X4) in the 3 isolates investigated in this study. Notably, we identified distinct structures in the 3 tet(X4)-haboring plasmids including IS26-catD-tet(X4)-ISCR2 (pTECO25-2), IS1R-catD-tet(X4)-ISCR2 (pTECO1-1), and IS1R-catD-tet(X4)-IS26 (p47P1FS3-2-1) (Figure 3). This finding revealed that ISCR2, IS1R and IS26 combined into three different genetic contexts, indicating a diverse surrounding environment worthy of further exploration. Subsequently, following the published manuscripts from 2019 to 2022 related to tet(X4)-positive E. coli, we downloaded 398 tet(X4)-positive E. coli assembly data from GenBank, among which there were 41 samples with a single copy of tet(X4) and a complete surrounding genetic environment (Supplementary Tables S2B,C). Thirteen tet(X4)-harboring genetic contexts were identified (Figure 4). The major genetic contexts identified were IS1R-catD-tet(X4)-ISCR2 (Type1) and ISCR2-catD-tet(X4)-ISCR2 (Type2), which are consistent with the findings of other researchers (Liu et al., 2022). Mobile elements such as IS1R, ISCR2, IS26 and their corresponding truncated elements are distributed around tet(X4), facilitating the propagation of tet(X4).
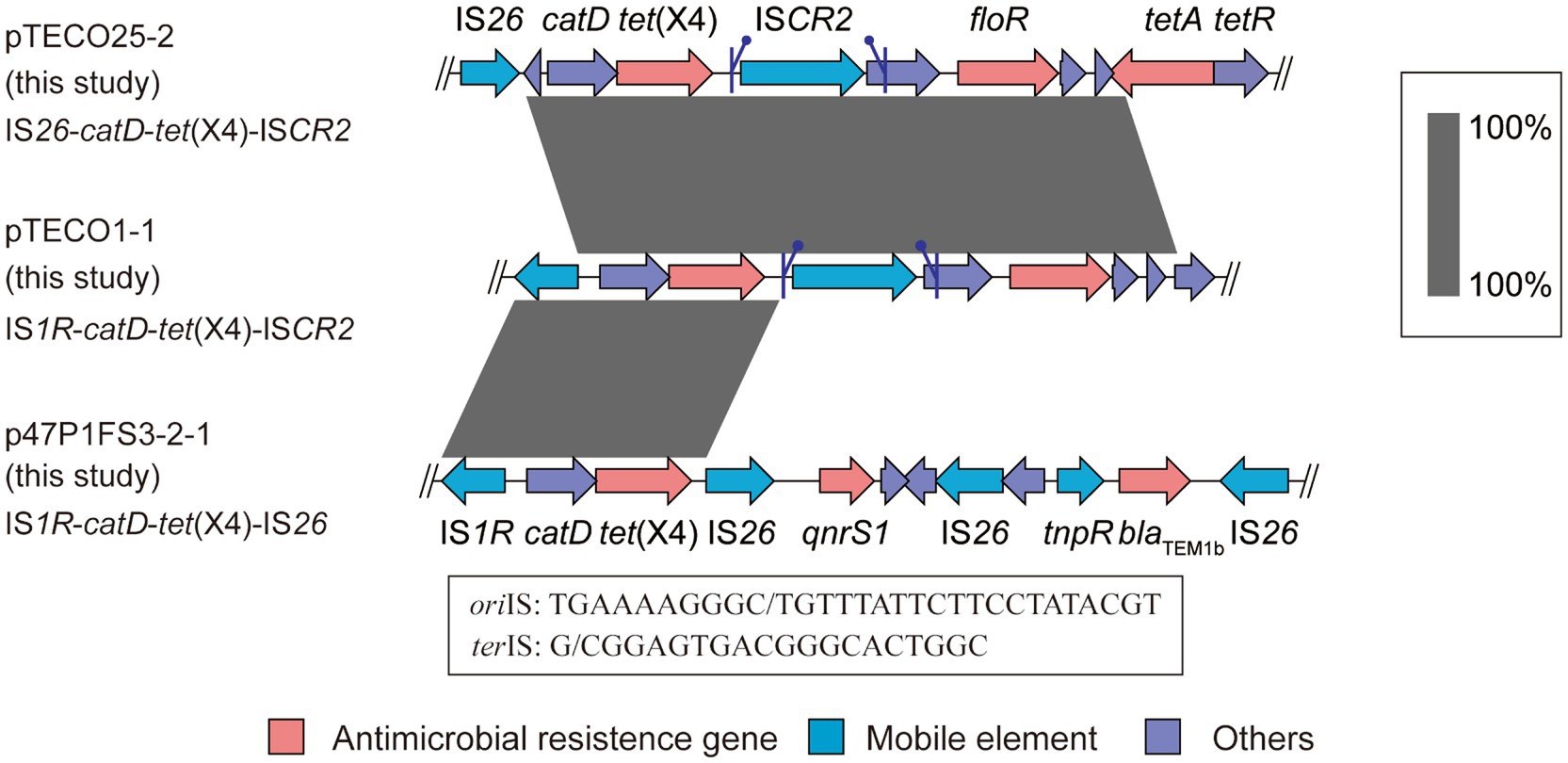
Figure 3. Comparison of genetic contexts of tet(X4). The schematic diagram provides a comparative analysis of genetic contexts surrounding tet(X4) in three strains subjected to short-read sequencing (TECO25, TECO1, and 47P1FS3-2). The symbols flanking the ISCR2 represent the putative oriIS and terIS of the ISCR2, respectively.
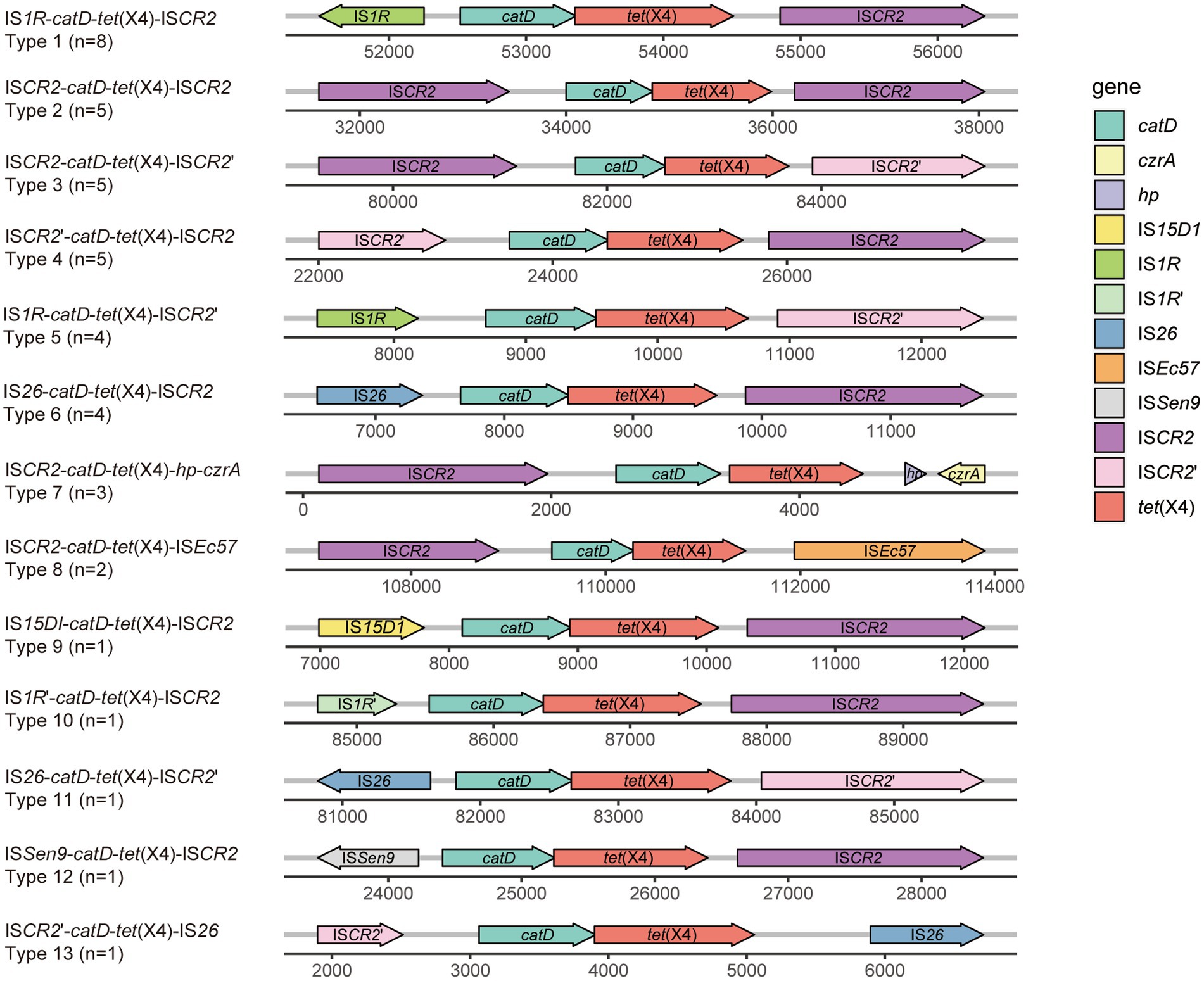
Figure 4. Different types of the genetic environments of tet(X4). The figure represents a comparison of the genetic environments of tet(X4) among 41 tet(X4)-positive E. coli obtained from GenBank. GenBank data accession numbers mentioned in this figure are listed in Supplementary Table S2C. The IS1R’ in this figure indicates that the IS1R gene fragment is incomplete, potentially due to truncation caused by certain insertion sequences. The representation method mentioned above is equally applicable to the incomplete genes present in this figure and Figure 5, including ISCR2’, ISPst2’, TnAs1’, and TnAs3’.
Comparison of plasmids carrying tet(X4)To provide a comprehensive view of the plasmids carrying tet(X4), we presented the complete plasmid structures containing tet(X4) from 3 strains (TECO25, TECO1, and 47P1FS3-2). These plasmids are pTECO1-1 (279,481 bp), p47P1FS3-2-1 (178,895 bp), and pTECO25-1 (29,625 bp). p47P1FS3-2-1, which had IncFIA(HI1)-IncHI1B(R27)-IncHI1A replicon type, carried the resistance genes tet(X4), qnrS, blaTEM-1B, ant(3″)-la, lnu(G). pTECO1-1 (178,895 bp), had an IncFIA(HI1)-IncFII(pCoo)-IncHI1B(R27)-IncHI1A replicon type. Additionally, pTECO1-1 carried floR, aac(3)-IId, lnu(F), aadA2 genes, in addition to all the resistance genes on p47P1FS3-2-1. pTECO25-1, with replicon type IncX1, carried the resistance genes aadA2, Inu(F), tet(X4), floR, tet(A) (Figure 5). The blaCMY-2 and blaCTX-M-14, mentioned earlier, were not located on these tet(X4)-positive plasmids.
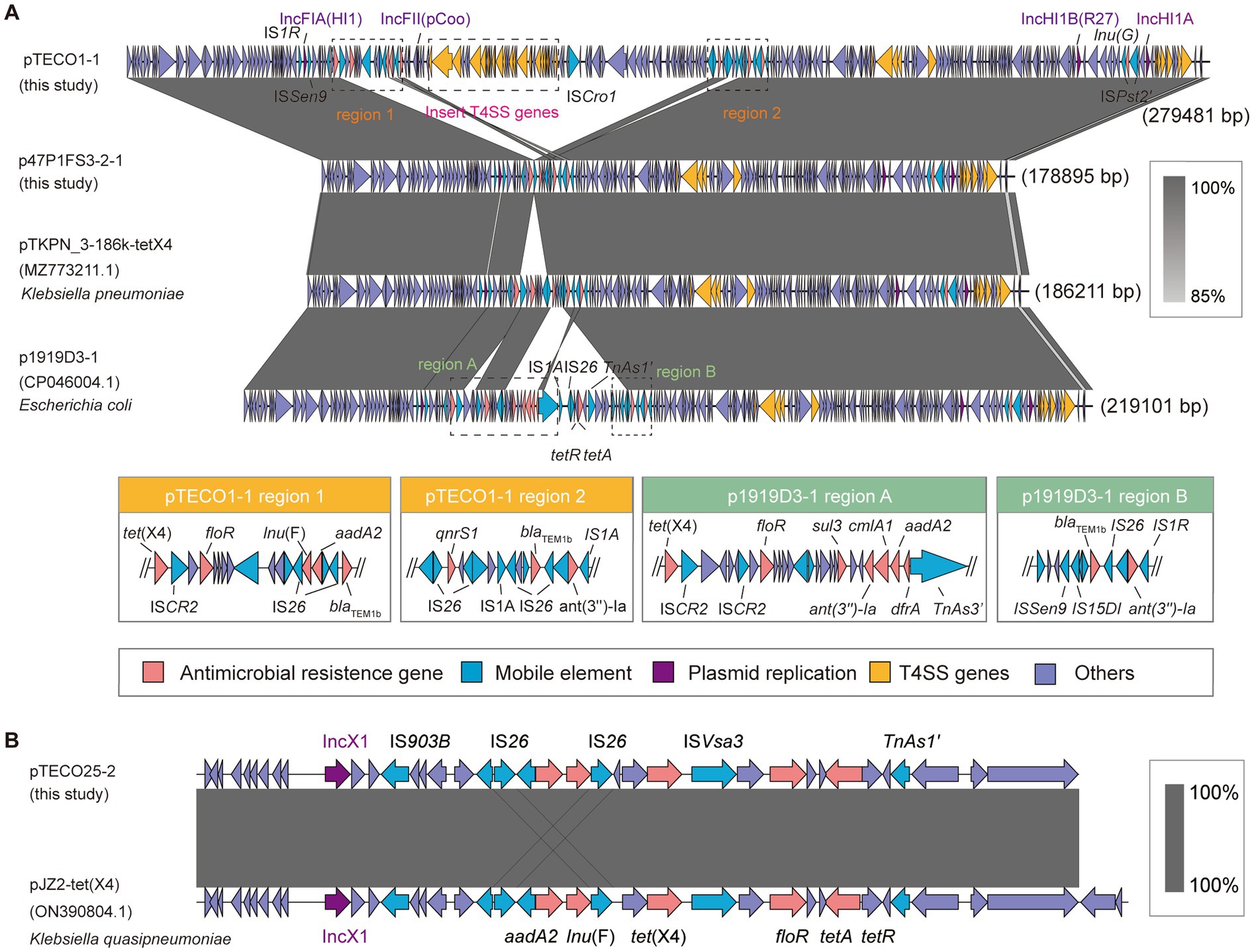
Figure 5. Structures of tet(X4)-positive plasmid. (A) The schematic diagram provides a comparative analysis of pTECO1-1, p47P1FS3-2-1, pTKPN_3-186k-tetX4 (MZ773211.1), and p1919D3-1 (CP046004.1). Different colored arrows representing different gene categories. Some regions where resistance gene insertions occur in pTECO1-1 and p1919D3-1 are indicated separately. (B) The schematic diagram provides a comparative analysis of pTECO25-1, pJZ2-tet(X4) (ON390804.1).
BLAST analysis of p47P1FS3-2-1 against the NCBI database revealed that the plasmid p1919D3-1 (CP046004.1) had 98% coverage and 100% identity, whereas the plasmid pTKPN_3-186k-tet(X4) (MZ773211.1) had 100% coverage and identity. Their replicon types were all IncFIA(HI1)-IncHI1B(R27)-IncHI1A, suggesting that these strains shared a plasmid backbone with our strains. BLAST alignment results showed that these 4 plasmids were highly conserved at both ends, and had different fragment insertions downstream of tet(X4) mediated by variable insertion sequences, such as IS26, IS1R, ISCR2, and IS15DI (Figure 5A). This region was diverse, including a multidrug resistance region in p1919D3-1 and a region containing a plasmid replicon in pTECO1-1. Comparison of tet(X4)-positive plasmids showed a 100-kbp large fragment insertion at the back end of tet(X4), containing a replicon and a 40-kbp length of the T4SS transfer-related gene cluster (Supplementary Figure S1). Klebsiella quasipneumoniae pJZ2-tet(X4) (ON390804.1) was identified by performing BLAST of pTECO25-1 against the NCBI database, with 100% coverage, 99.8% identity (Figure 5B), indicating that cross-species transmission of this plasmid has occurred.
DiscussionThe environment serves as a vast reservoir for drug resistance genes and plays a crucial role in their transmission (Lin et al., 2021). Since its discovery, tet(X4) has been extensively detected in environment samples (Zhang et al., 2022). Regarding the transmission of tet(X4) from the environment to human, potential routes have been proposed in previous studies. These include: contamination of agricultural land or water sources such as reservoirs and wetlands through animal manure, subsequently leading to human exposure via crops or aquatic organisms, such as freshwater fish and shrimp (Li et al., 2019; Dao et al., 2022). Additionally, the presence of contaminated retail meat into the market had been identified as a potential transmission pathway (Sun et al., 2021). Direct contact or fecal-oral transmission among individuals involved in the food processing chain is also considered a possible route (Wang et al., 2020). Our study highlights the possibility of transmission from pork processing plants to workers, emphasizing the importance of implementing prevention and control measures among individuals working in relevant industries. The plasmids p1919D3-1 and pTKPN_3-186k-tet(X4) were derived from strains isolated from swab samples of pig feces from Henan province and pig nasal swabs from Guangdong province, China, respectively. The similarity of these plasmid backbones to the plasmid p47P1FS3-2-1 originating from the patient in our study suggests the potential transmission and evolution of tet(X4)-positive plasmids across different regions in China and between animals and humans.
tet(X4) is highly prevalent in China, with the detection of strains carrying tet(X4) including E. coli, Klebsiella pneumoniae, Citrobacter braakii, Enterobacter cloacae (Li et al., 2021; Wu et al., 2022). The prevalence of tet(X4)-positive E. coli in China exhibited significant diversity in STs with regional variations. In two provinces in northwestern China, Shanxi and Ningxia, the dominant STs of tet(X4)-positive E. coli were ST6704, ST2035, ST48, ST1602, and ST877 (Sun et al., 2020). In western China, ST10, ST34, ST48, and ST195 have been identified as predominant STs (Feng et al., 2022), while in the southern region, ST10, ST48, ST877, and ST2144 were relatively more prevalent (Cui et al., 2022). In our study, we isolated tet(X4)-positive E. coli strains from Guangdong Province in southern China, where we observed high proportions of ST877, ST871, ST195, and ST10. The diverse range of STs indicates that the horizontal transfer of plasmids carrying tet(X4) has likely occurred, contributing to the genetic variation and dissemination of this resistance gene. Moreover, the two tet(X4)-positive E. coli strains isolated from the patients in this study exhibited a unique ST (ST9867), which has not been previously reported, indicating the transfer of tet(X4) to a novel ST and its subsequent spread to human populations.
A transposable unit is formed when tet(X4) is transferred, and is generally composed of mobile elements, tet(X4) and partner gene catD, in which mobile elements are the key mediators of the transfer. Previous studies have demonstrated that ISCR2, through its upstream and downstream oriIS and terIS elements, plays a crucial role in mediating the transfer of tet(X4) by forming a transposon unit that facilitates rolling-circle transposition (Toleman et al., 2006; Sun et al., 2020). Additionally, under the influence of other IS elements and homologous recombination events, ISCR2 generates a highly diverse genomic environment. This includes the formation of tandem multicopies due to transposase misreading of terIS, capture of other resistance genes, and truncation by other IS elements (Liu et al., 2022; Zhai et al., 2022). In this study, all the isolated strains carrying a single copy of tet(X4). Using the long-reads sequencing, we identified three transposon units comprising the different mobile elements. Combining our data with available online resources, we found that, in addition to ISCR2, IS1R and IS26 elements were also frequently involved in mediating the transfer of tet(X4). Furthermore, we discovered that the presence of the florfenicol resistance floR gene frequently accompanied tet(X4) downstream, indicating that the dissemination of tet(X4) plasmids may be influenced by the selective pressure exerted by florfenicol.
One way in which antibiotic resistance plasmids acquire additional resistance genes is through the frequent insertion of genetic fragments. During horizontal gene transfer, antibiotic resistance plasmids can acquire additional resistance genes by inserting new fragments, such as transposons or other mobile elements. In this study, four plasmids shared the same plasmid backbone, IncFIA(HI1)-IncHI1B(R27)-IncHI1A. However, compared to plasmid p47P1FS3-2-1, the other three plasmids underwent insertion of resistance gene fragments, including aadA2, qnrS1, and Inu(F). The insertion of diverse resistance genes provided the tet(X4)-positive plasmid with increased adaptability. In addition, the acquisition of an exogenous sequence containing a substantial number of T4SS-related genes by pTECO1-1 was a notable event. However, the significance of these findings requires further investigation.
This study has several limitations. First, the sample size was relatively small, as it included samples collected in Guangdong, China. Additionally, this study focused primarily on pigs, which lacked diversity in sample sources. This study also revealed the insertion of T4SS genes into a tet(X4)-positive plasmid, however, their functional significance requires further experimental verification.
ConclusionIn this study, we described the high colonization rate of tet(X4)-positive E. coli in animals, along with its presence among the farm workers. Comparative analysis revealed that the successful genetic elements and the insertion of T4SS-associated genes in tet(X4)-positive plasmids, which may contribute to the rapid dissemination of tet(X4). Overall, this study serves as a timely warning regarding the potential acceleration of tigecycline resistance spread and underscores the critical importance of comprehensive surveillance spanning from animals to humans for effective global control of antibiotic resistance.
Data availability statementThe sequences obtained in this study have been deposited in the GenBank database under BioProject number PRJNA1022166. Other public sequencing data used in this study are presented in Supplementary Table S2.
Ethics statementEthical approval for this study was sought and given by both pig farm workers, inpatients and healthy volunteers. Individual consent forms were translated into Mandarin Chinese and the study vocally explained to each participant. All participants had the right to withdraw from the study at any stage.
Author contributionsLS: Writing – review & editing, Methodology, Investigation, Writing – original draft, Conceptualization. CW: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Conceptualization. CL: Writing – original draft, Methodology, Writing – review & editing, Validation, Formal analysis. RH: Writing – original draft, Investigation, Writing – review & editing, Validation, Formal analysis. GC: Writing – review & editing, Software, Methodology. DS: Writing – review & editing, Validation, Formal analysis. YY: Writing – review & editing, Software, Methodology. YF: Writing – review & editing, Validation, Formal analysis. GZ: Writing – review & editing, Software, Methodology. BY: Writing – review & editing, Validation, Formal analysis. MD: Resources, Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization. G-BT: Resources, Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization. L-LZ: Resources, Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant numbers 82325033, 8183010 to G-BT; 82102441 to L-LZ), National Key Research and Development Program (grant number 2023YFC2307102 to G-BT), Project of High-level Health Teams of Zhuhai in 2018 (The Innovation Team for Antimicrobial Resistance and Clinical Infection to G-BT), the Innovation Team of Chinese Medicine Administration from Sichuan Province (grant number 2023ZD02 to G-BT and MD) and Guangzhou basic and applied research foundation (grant number 202201020614 to BY).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1423352/full#supplementary-material
Footnotes ReferencesBankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
PubMed Abstract | Crossref Full Text | Google Scholar
Chen, C., Cui, C.-Y., Zhang, Y., He, Q., Wu, X.-T., Li, G., et al. (2019). Emergence of mobile tigecycline resistance mechanism in Escherichia coli strains from migratory birds in China. Emerg. Microbes Infect. 8, 1219–1222. doi: 10.1080/22221751.2019.1653795
PubMed Abstract | Crossref Full Text | Google Scholar
CLSI . (2022). Performance standards for antimicrobial susceptibility testing, 32nd. Annapolis Junction, MD: Clinical and Laboratory Standards Institute.
Cui, C.-Y., Li, X.-J., Chen, C., Wu, X.-T., He, Q., Jia, Q.-L., et al. (2022). Comprehensive analysis of plasmid-mediated tet(X4)-positive Escherichia coli isolates from clinical settings revealed a high correlation with animals and environments-derived strains. Sci. Total Environ. 806:150687. doi: 10.1016/j.scitotenv.2021.150687
PubMed Abstract | Crossref Full Text | Google Scholar
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
PubMed Abstract | Crossref Full Text | Google Scholar
Dao, T. D., Kasuga, I., Hirabayashi, A., Nguyen, D. T., Tran, H. T., Vu, H., et al. (2022). Emergence of mobile tigecycline resistance gene tet(X4)-harbouring Shewanella xiamenensis in a water environment. J. Glob. Antimicrob. Resist. 28, 140–142. doi: 10.1016/j.jgar.2021.12.022
PubMed Abstract | Crossref Full Text | Google Scholar
Ding, Y., Saw, W.-Y., Tan, L. W. L., Moong, D. K. N., Nagarajan, N., Teo, Y. Y., et al. (2020). Emergence of tigecycline-and eravacycline-resistant tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75, 3480–3484. doi: 10.1093/jac/dkaa372
PubMed Abstract | Crossref Full Text | Google Scholar
Dong, N. (2022). Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study. Sci. Total Environ. 818:151767. doi: 10.1016/j.scitotenv.2021.151767
Crossref Full Text | Google Scholar
Du, P., Liu, D., Song, H., Zhang, P., Li, R., Fu, Y., et al. (2020). Novel IS26-mediated hybrid plasmid harbouring tet(X4) in Escherichia coli. J. Glob. Antimicrob. Resist. 21, 162–168. doi: 10.1016/j.jgar.2020.03.018
PubMed Abstract | Crossref Full Text | Google Scholar
EUCAST . (2021). European committee on antimicrobial susceptibility testing breakpoint tables for interpretation of MICs and zone diameters, version 11.0. Sweden: EUCAST.
Fang, L., Chen, C., Cui, C., Li, X., Zhang, Y., Liao, X., et al. (2020). Emerging high-level Tigecycline resistance: novel tetracycline Destructases spread via the Mobile tet(X). Bio Essays 42:e2000014. doi: 10.1002/bies.202000014
PubMed Abstract | Crossref Full Text | Google Scholar
Feng, J., Su, M., Li, K., Ma, J., Li, R., Bai, L., et al. (2022). Extensive spread of tet(X4) in multidrug-resistant Escherichia coli of animal origin in western China. Vet. Microbiol. 269:109420. doi: 10.1016/j.vetmic.2022.109420
PubMed Abstract | Crossref Full Text | Google Scholar
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
PubMed Abstract | Crossref Full Text | Google Scholar
Hsu, L.-Y., Apisarnthanarak, A., Khan, E., Suwantarat, N., Ghafur, A., and Tambyah, P. A. (2017). Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in south and Southeast Asia. Clin. Microbiol. Rev. 30, 1–22. doi: 10.1128/CMR.00042-16
PubMed Abstract | Crossref Full Text | Google Scholar
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
PubMed Abstract | Crossref Full Text | Google Scholar
Li, R., Li, Y., Peng, K., Yin, Y., Liu, Y., He, T., et al. (2021). Comprehensive genomic investigation of Tigecycline resistance gene tet(X4)-bearing strains expanding among different settings. Microbiol. Spectr. 9, e01633–e01621. doi: 10.1128/spectrum.01633-21
PubMed Abstract | Crossref Full Text | Google Scholar
Li, R., Liu, Z., Peng, K., Liu, Y., Xiao, X., and Wang, Z. (2019). Cooccurrence of two tet(X) variants in an Empedobacter brevis strain of shrimp origin. Antimicrob. Agents Chemother. 63, e01636–e01619. doi: 10.1128/AAC.01636-19
PubMed Abstract | Crossref Full Text | Google Scholar
Lin, Z., Yuan, T., Zhou, L., Cheng, S., Qu, X., Lu, P., et al. (2021). Impact factors of the accumulation, migration and spread of antibiotic resistance in the environment. Environ. Geochem. Health 43, 1741–1758. doi: 10.1007/s10653-020-00759-0
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, D., Wang, T., Shao, D., Song, H., Zhai, W., Sun, C., et al. (2022). Structural diversity of the ISCR2-mediated rolling-cycle transferable unit carrying tet(X4). Sci. Total Environ. 826:154010. doi: 10.1016/j.scitotenv.2022.154010
PubMed Abstract | Crossref Full Text | Google Scholar
Nascimento, M., Sousa, A., Ramirez, M., Francisco, A. P., Carriço, J. A., and Vaz, C. (2017). PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33, 128–129. doi: 10.1093/bioinformatics/btw582
PubMed Abstract | Crossref Full Text | Google Scholar
Olson, M. W., Ruzin, A., Feyfant, E., Rush, T. S., O’Connell, J., and Bradford, P. A. (2006). Functional, biophysical, and structural bases for antibacterial activity of Tigecycline. Antimicrob. Agents Chemother. 50, 2156–2166. doi: 10.1128/AAC.01499-05
PubMed Abstract | Crossref Full Text | Google Scholar
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
PubMed Abstract | Crossref Full Text | Google Scholar
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genomics 2:e000056. doi: 10.1099/mgen.0.000056
PubMed Abstract | Crossref Full Text | Google Scholar
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
PubMed Abstract | Crossref Full Text | Google Scholar
Sun, C., Cui, M., Zhang, S., Liu, D., Fu, B., Li, Z., et al. (2020). Genomic epidemiology of animal-derived tigecycline-resistant Escherichia coli across China reveals recent endemic plasmid-encoded tet(X4) gene. Commun. Biol. 3:412. doi: 10.1038/s42003-020-01148-0
PubMed Abstract | Crossref Full Text | Google Scholar
Sun, H., Wan, Y., Du, P., Liu, D., Li, R., Zhang, P., et al. (2021). Investigation of tigecycline resistant Escherichia coli from raw meat reveals potential transmission among food-producing animals. Food Control 121:107633. doi: 10.1016/j.foodcont.2020.107633
Crossref Full Text | Google Scholar
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
PubMed Abstract | Crossref Full Text | Google Scholar
Toleman, M. A., Bennett, P. M., and Walsh, T. R. (2006). IS CR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70, 296–316. doi: 10.1128/MMBR.00048-05
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, Y., Liu, F., Zhu, B., and Gao, G. F. (2020). Discovery of tigecycline resistance genes tet(X3) and tet(X4) in live poultry market worker gut microbiomes and the surrounded environment. Sci. Bull. 65, 340–3
留言 (0)