Florfenicol (Ff) is a synthetic broad-spectrum antimicrobial agent widely employed in veterinary medicine (Wu et al., 2021). Classified as an amphenicol, it irreversibly binds to the peptidyl transferase center of the 50S ribosomal subunit of prokaryotes. This binding inhibits the elongation of the peptide chain, ultimately hindering protein synthesis. Ff is primarily bacteriostatic in Enterobacteriaceae and Staphylococcus aureus (Wei et al., 2016; Somogyi et al., 2023; Trif et al., 2023; Guo et al., 2024) and bactericidal at clinical concentrations against Haemophilus influnzae, Streptococcus suis, Mannheimia haemolytica, and Pasteurella multocida (Graham et al., 1988; Illambas et al., 2013; Lei et al., 2018; Somogyi et al., 2023). Ff was granted approval by the Food and Drug Administration (FDA) for the treatment of bovine respiratory infections caused by pathogens such as Pasteurella multocida (White et al., 2000) and by the European Medicines Agency for the control of respiratory tract infections of bacterial origin in cattle and pigs (Kehrenberg and Schwarz, 2006). Unfortunately, the extensive usage of florfenicol in veterinary practice has led to a notable increase in resistance among zoonotic pathogens to this antimicrobial (White et al., 2000; Wasyl et al., 2013; Zhan et al., 2019; Kerek et al., 2023).
Acinetobacter is an opportunistic pathogen in humans and is often overlooked as a veterinary pathogen (Wareth et al., 2019). Its natural habitat is the environment, particularly soil and water. The bacterium is implicated in both community- and healthcare-acquired infections (Villalón et al., 2019; Wareth et al., 2019). Among the Acinetobacter species, Acinetobacter baumannii is the most clinically relevant species, causing most of the infections. Recently, other members of ACB complex members including Acinetobacter pittii, Acinetobacter nosocomialis, Acinetobacter seifertii, and Acinetobacter lactucae have also been isolated from patients (Migliaccio et al., 2023). These species cause infection in patients with comorbidities such as chronic lung disease, impaired immunity, malignancy, advanced age, diabetes mellitus, or renal diseases (Mancilla-Rojano et al., 2020).
The widespread and increased use of florfenicol in livestock, aquaculture, and poultry (Zeng et al., 2019; Guo et al., 2024) has accelerated the rate at which pathogens develop resistance to it (Zhan et al., 2019; Yang et al., 2022; Trif et al., 2023). The consumption of florfenicol in Europe falls under the category of class “C” (Caution) (Categorisation of Antibiotics Used in Animals Promotes Responsible Use to protect Public and Animal Health and European Medicines Agency, 2024). Florfenicol is widely used and, in some circumstances, reported to account for as much as 24.0% and 24.2% of all class C antimicrobials used during the weaning and fattening phases in pig rearing (Trevisi et al., 2022). Unsurprisingly, determinants of florfenicol resistance have been found in the environment associated with pig farms or in freshwater aquaculture (Fernández-Alarcón et al., 2010; Zhao et al., 2016; Zeng et al., 2019; Li et al., 2020; Fu et al., 2022; Wang et al., 2022; Lin et al., 2023). Several Ff resistance mechanisms have been identified, with the floR gene playing a significant role in conferring resistance. The floR gene encodes the FloR protein (with 12 hydrophobic transmembrane regions), which forms a proton motive force (PMF)-driven efflux pump that removes both Ff and chloramphenicol from bacterial cells using active transport (Adesoji and Call, 2020; Li et al., 2020). The presence of the floR gene has been reported in various genomes. For instance, Ff resistance has been associated with the presence of the floR gene on a transferable plasmid in Klebsiella pneumoniae in clinical isolates from China (Lu et al., 2018). The presence of floR variants (floR-T1 and floR-T2) has been reported in the multidrug resistance (MDR) region as an integrative and conjugative element (ICE) in Pseudomonas aeruginosa (Qian et al., 2021). Escherichia coli isolates from clinical samples revealed the presence of the floR gene in transposon-like fragments with recombination-related genes along with tetA and tetR genes, which regulate tetracycline resistance (Møller et al., 2016; Lu et al., 2018). The occurrence of floR in drug-resistant regions on chromosomes with IS91 family transposase in Proteus vulgaris has been reported (Li et al., 2020). The presence of the floR gene flanked by insertion sequences and other resistance genes in clinical isolates of A. baumannii has also been reported (Wareth et al., 2021; Zafer et al., 2021). These observations highlight a possible horizontal gene transfer-mediated zoonotic transmission of resistance genes associated with Ff resistance to human pathogens. High Ff resistance has furthermore been observed in environmental samples, as exemplified by Ff resistance isolated from four Acinetobacter spp. reported from water samples in Nigeria (Adesoji and Call, 2020).
In this study, we used multiple genetic and bioinformatic approaches to investigate the presence of the floR gene in Acinetobacter spp. isolates of human origin and the environment to identify the genes associated with the floR resistance cassette and to further demonstrate the potential transmission of resistance determinants to human pathogens and environmental microbes. The diversity of the strains carrying the floR gene suggests the occurrence of multiple transmission events leading to amphenicol resistance.
2 Materials and methods 2.1 Bacterial isolate collectionDuring a study conducted over 10 years (from 2009 to 2019), a total of 39 patients from three separate rural clinics in Thailand (Supplementary material) had isolated events of Acinetobacter spp. infections. The majority (34/39, 87.2%) of the bacterial isolates were isolated from blood samples of the infected patients. Furthermore, 5.1% (2/39) were isolated from cerebrospinal fluid samples (CSF), 5.1% (2/39) from urine samples, and 2.5% (1/39) from a urinary catheter. In addition to the clinical samples, 10 environmental samples were also collected from the same geographic area for environmental testing for infection prevention control (IPC), i.e., to contribute to the prevention of healthcare-associated infections (HAIs) through the detection of environmental contamination with key drug-resistant pathogens. Testing was carried out at one of the clinics to detect and monitor environmental contamination with target drug-resistant pathogens. Routine diagnostic media and culture conditions were used to isolate Acinetobacter spp. from clinical specimens, which included the use of Biomerieux BacT/Alert blood culture bottles for blood samples, Oxoid Brilliance™ UTI Clarity™ agar for urine samples, and CHROMagar ESBL for environmental samples.
2.2 Whole-genome sequencing and identificationDNA extraction was performed using the BioBasic One-4-All Genomic DNA Miniprep kit (cat: BS88503). The extracted DNA was sent to NovogeneAIT Genomics Singapore Pte Ltd. for whole genome sequencing (WGS). The isolates were sequenced using 2 × 151 bp paired-end reads on an Illumina HiSeq 4000 platform. The quality of the raw reads was analyzed using FastQC. The paired reads were assembled using SPAdes (v3.6.0) (Bankevich et al., 2012). Automatic annotation was carried out using RAST (Aziz et al., 2008).
2.3 Bioinformatic analysisSpeciation of the bacterial isolates was performed using average nucleotide identity (ANI). The genomes of the isolates were compared to the annotated Acinetobacter sequences available in GenBank. Species allocation was done with a limit of 95% identity to the annotated sequence (Jain et al., 2018). Abricate ResFinder was used to search for acquired resistance genes in the genome (Zankari et al., 2012). The web version of SimpleSynteny (Veltri et al., 2016) was used to construct the synteny plots. Circle plots were constructed using CIRCOS software using Command Line (Krzywinski et al., 2009).
2.4 Antibiotic susceptibilityThe minimum inhibitory concentration (MIC99) values for Ff and chloramphenicol were determined according to the Clinical and Laboratory Standards Institute (CLSI) 2023 broth dilution guidelines, as the floR gene is also known to confer resistance against chloramphenicol (Bolton et al., 1999). According to the CLSI guidelines, a growth of ≥32 μg/mL of chloramphenicol is considered to be a chloramphenicol-resistant strain for non-Enterobacteriaceae. A growth of ≥ 8 μg/mL of Ff is considered as the Ff-resistant strain (Verner-Jeffreys et al., 2017). ASP 6 (Acinetobacter variabilis) served as a negative control for the MIC.
3 Results and discussionWhole-genome sequencing (WGS) confirmed that all the isolates belonged to the genus Acinetobacter. The acquired resistance gene annotation of the isolates revealed that 10 out of the 39 clinical isolates (25.6%) and 2 out of the 10 (20%) environmental isolates harbored the floR gene in the bacterial genome (Table 1). Among the clinical isolates of Acinetobacter spp., 10% (1/10) of the isolated A. baumannii, 25% (1/4) of the isolated A. pittii, 60% (3/5) of the isolated A. nosocomialis, 40% (4/10) of the isolated A. variabilis, 25% (1/4) of the isolated Acinetobacter junii, and 100% (1/1) of the isolated Acinetobacter johnsonii were found to be harboring the floR gene. Among the environmental isolates of Acinetobacter spp., 100% (1/1) of the isolated Acinetobacter schindleri and 50% (1/2) of the isolated A. variabilis isolated were found to be harboring the floR gene. Through antibiotic susceptibility testing, we confirmed that all the clinical strains were resistant to florfenicol and chloramphenicol. The two environmental samples were borderline resistant to Ff and were either susceptible (ASP3) or intermediate (ASP18) to chloramphenicol (Table 1). Interestingly, ASP3 also encodes cmlA1, an additional chloramphenicol-resistant genetic marker (Li et al., 2014), while remaining susceptible to chloramphenicol. The susceptibility profiles were confirmed by independent biological replicates. An analysis to understand this conflicting observation for ASP3 is underway and is not part of this study. The Ff resistance due to the presence of the floR gene is responsible for cross-resistance to chloramphenicol (Zafer et al., 2021). The use of chloramphenicol is restricted in high-income countries, but it is still a drug of choice in many low- and middle-income countries for ophthalmic use and other infections (WHO Model List of Essential Medicines−22nd list, 2021, 2021). Thus, the spread of the floR gene will alter the use of chloramphenicol in many low- and middle-income countries.
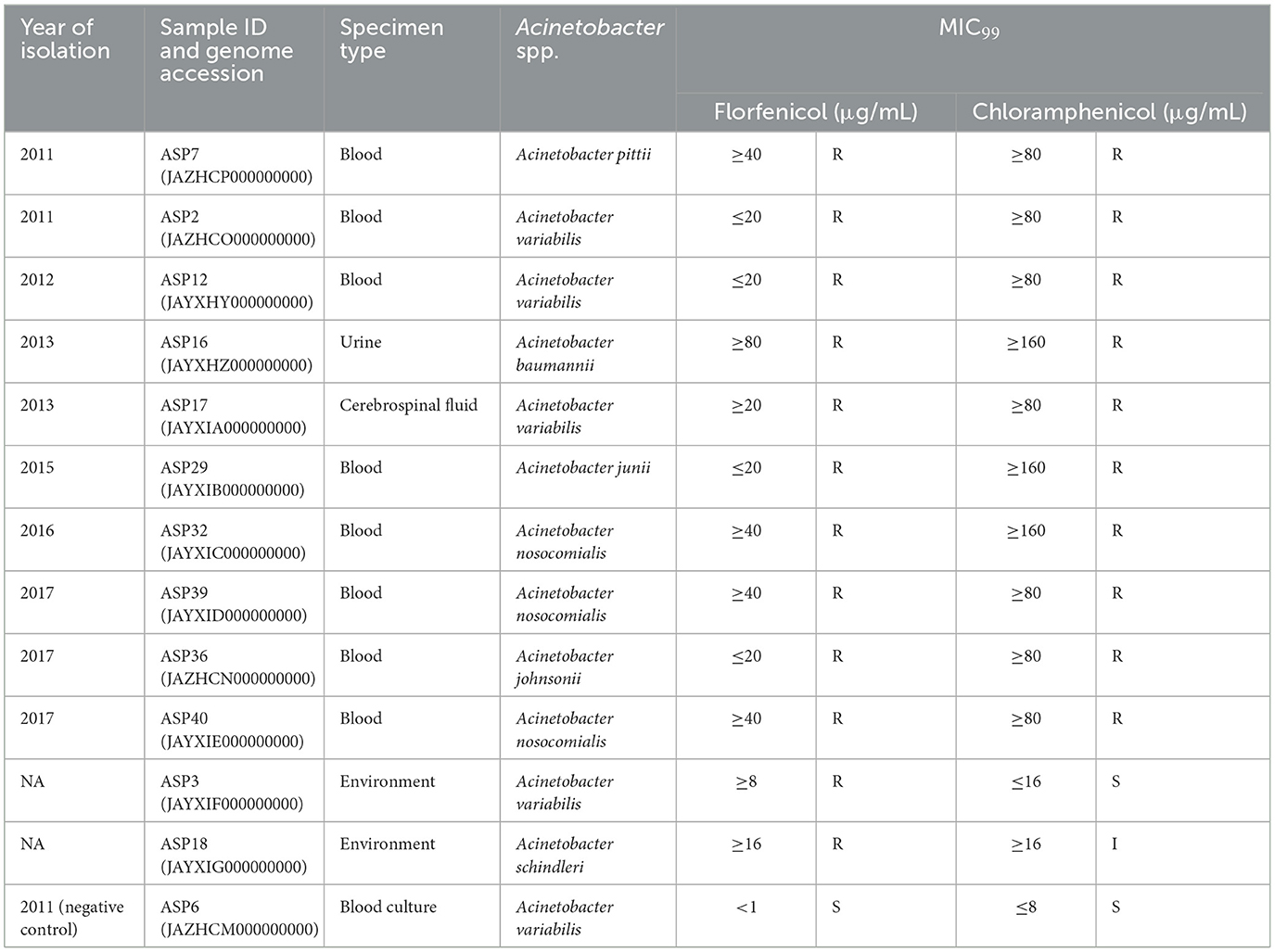
Table 1. floR-positive isolates from 2009 to 2019 and their MIC99 values for florfenicol and chloramphenicol.
The identified floR gene from all 12 isolates was found to be 100% identical to the floR gene of Vibrio cholerae (NCBI Reference Sequence: NG_047869.1) through a basic local alignment search tool (BLAST). Among the floR-positive isolates, three samples from clinical isolates (ASP2, 12, and 17) and one sample from an environmental strain (ASP3) belonged to A. variabilis. Five isolates belonged to the Acinetobacter–calcoaceticus–baumannii (ACB) complex and one isolate each to A. junii, A. johnsonii, and A. schindleri. Through chromosomal analysis, we observed the same floR gene in all isolates, but the flanking regions differed from each other (Figure 1). In all isolates, the floR gene appeared to be clustered within the variable region rich in mobile genetic elements (MGEs), as depicted in the circle plots (Figure 2). The phylogenetic analysis (Figure 3) of the isolates showed that the strains harboring the floR genes were highly diverse, with the exception of the A. nosocomialis isolates, which formed one cluster. WGS and chromosome analysis, phylogeny, and speciation suggest that with the exception of A. nosocomialis, the acquisition of floR is an independent event as they are found in different Acinetobacter species.

Figure 1. Synteny plot of the floR gene and associated acquired antibiotic resistance gene across the chromosome. The floR gene is highlighted with a red box. The values at the bottom of the beige ribbons represent the chromosomal location of the genes. Each floR-positive sample has been compared to the corresponding floR-negative strain from the study cohort, with the exception of ASP29, for which no corresponding counterpart was available within the cohort. (env): environmental isolates.
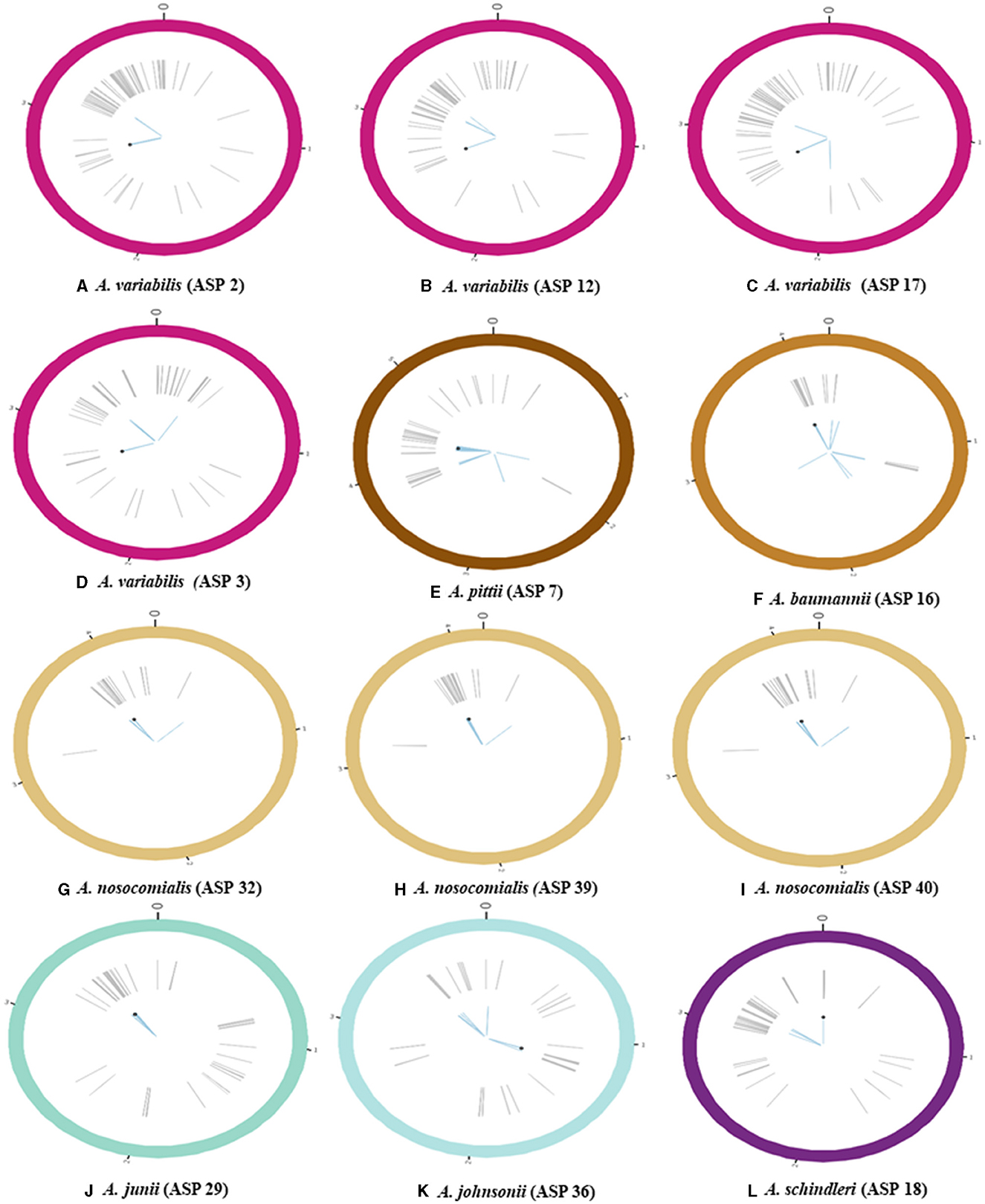
Figure 2. Schematic representation of bacterial chromosome harboring the gene conferring florfenicol (floR) resistance (all the values on the chromosomes are in mbp); (A–D) circle plots represent the bacterial chromosome of A. variabilis; (E–I) circle plots represent species belonging to the ACB complex [(E) A. pittii, (F) A. baumannii, and (G–I) A. nosocomialis]; (J, K) circle plots correspond to other Acinetobacter spp. [(J) A. junii and (K) A. johnsonii]; (L) the circle plot represents A. schindleri. Gray lines indicate the position of various mobile genetic elements (MGEs), blue-colored lines highlight annotated acquired antibiotic resistance genes, and the black point represents the location of floR on the chromosome.
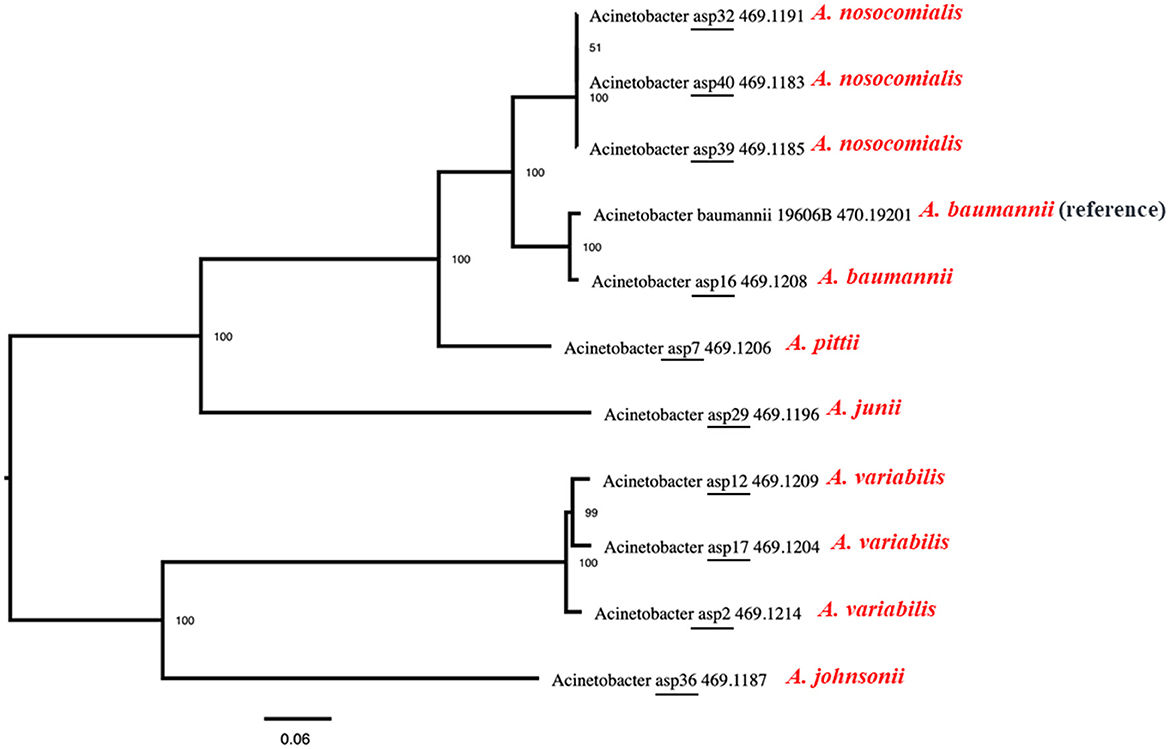
Figure 3. Maximum-likelihood phylogeny of the Acinetobacter species isolated in this study with the reference genome of ATCC Acinetobacter baumannii 19606. Average nucleotide identity was used to generate the phylogenetic tree, and the data show that the strains harboring the floR genes are highly diverse, with the exception of the A. nosocomialis isolates, which form one cluster.
Figure 1 represents the chromosomal location of the floR gene along with all the acquired resistance genes in comparison to the floR negative strains as reference strains. The insertion of the floR gene in A. baumannii is in a variable region rich in MGEs (Figure 2). The insertion is near the bla gene when compared to the floR-negative strain. This association of the insertion near the bla gene is also observed across the ACB complex isolates. Through circle and synteny plots, we can observe that the floR gene is not accompanied by conserved flanks; however, the floR gene was always present in the region rich in MGEs. Similarly, for A. variabilis isolates, the floR gene was inserted near the sul2 gene when compared to the floR-negative isolate. Notably, for both the ACB complex, A. variabilis, and other isolates, the floR gene insertion was found within the highly variable region.
The genomic annotation of WGS revealed that floR was present in recombination hotspots in all the isolates, accompanied by various MGEs, including insertion sequences (IS), transposons (Tn), and others (Figure 2). These recombination hotspots, also present in the floR-negative reference strains, harbor various other antibiotic resistance markers and prophage regions, indicating multiple events of gene uptake and a high frequency of recombination.
It is hard to determine a clonal link as multiple recombinations, insertions, and deletions might have occurred prior to and/or after the acquisition of floR. The isolates are phylogenetically far apart, the transfer of the floR gene is highly unlikely to have been occurred during speciation. Notably, all clinical samples have the macrolide resistance cassette (msrE and mphE) and the sulfamethoxazole resistance gene (sul2) in the proximity of floR in highly variable regions of the genome. Clinical isolates of A. variabilis harbor additional erythromycin and streptomycin B resistance genes (msrE) and erythromycin and macrolide resistance genes (ery and mphE). Furthermore, the floR-positive A. variabilis also harbors tetracycline (TET) and doxycycline (DOX) resistance genes. The environmental samples have the sulfamethoxazole resistance gene (sul2) located near the floR gene in both strains. Overall, WGS analysis and speciation alone suggest that, in the sample collection of this study, the acquisition of the floR gene is an independent event. It is highly possible that its selection and transmission is associated with the HGT of other drug-resistant markers. Other studies have also suggested similar possibilities (Kehrenberg et al., 2006; Verner-Jeffreys et al., 2017), but, as in this study, those inferences lack solid evidence.
4 ConclusionAcinetobacter spp. isolates from humans and the environment encode the floR gene, which confers resistance to both florfenicol and chloramphenicol. Chloramphenicol-resistant A. baumannii (XDRAB), encoding the cmlA1 gene, has been previously reported from Thailand. Chloramphenicol resistance was found to be widely prevalent in the non-target organisms in aquaculture environments in Southeast Asia in one study, but the mechanism of resistance was not reported (Huys et al., 2007; Chopjitt et al., 2020). In this study, speciation of the bacterial isolates using average nucleotide identity (ANI), combined with phylogenetic analysis, indicates that the strains reported are not epidemiologically related and, yet, they all carry the floR gene, suggesting the occurrence of independent events leading to the acquisition of the resistant markers. In all instances, the floR gene was found within variable regions, as depicted in the synteny plots. We elucidate that the wide use of florfenicol in livestock, poultry, and aquaculture production may be a driver for florfenicol resistance in veterinary and human pathogens. The consumption of florfenicol in animal farming has rapidly increased since its introduction (Wang et al., 2019). The presence of floR in both clinical and environmental samples indicates that the floR gene is persistent in the environment and can be incorporated into the genomes of different species. In all isolates, floR was located in the variable region of the bacterial genome, flanked by multiple mobile elements that host many other resistance genes. Therefore, by imposing the selection of the floR gene through the use of florfenicol, along with the selection for chloramphenicol resistance, we may be facilitating the HGT of other flanking-resistant markers, virulence factors, or other genetic elements. Florfenicol resistance may also facilitate reverse zoonosis and the reversible transfer of drug-resistant markers to veterinary pathogens and commensals. Overall, the presence of the floR gene in human and environmental isolates indicates that there is an ongoing genetic exchange between zoonotic pathogens, human pathogens, and environmental microbiomes.
Data availability statementThe data presented in the study are deposited in the NCBI GenBank repository, accession numbers JAYXHY000000000, JAYXHZ000000000, JAYXIA000000000, JAYXIB000000000, JAYXIC000000000, JAYXID000000000, JAYXIE000000000, JAYXIF000000000, JAYXIG000000000, JAZHCM000000000, JAZHCN000000000, JAZHCO000000000, JAZHCP000000000.
Ethics statementThe study did not require approval by an Ethics Committee as individual patient data, nor clinical specimens, were used. Only anonymized stored isolates obtained from routine hospital laboratory procedures were used in the study.
Author contributionsBT: Writing—original draft, Visualization, Project administration, Methodology, Investigation, Formal analysis, Data curation. LM: Writing—original draft, Visualization, Project administration, Methodology, Investigation, Formal analysis, Data curation. WW: Writing—review & editing, Data curation. TN: Writing—review & editing, Data curation. FN: Writing—review & editing, Data curation. CL: Writing—review & editing, Data curation. PB: Writing—review & editing, Validation, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Start-up grant (R-571-000-040-733) awarded to PB, National University of Singapore, and Core Funding from IDLabs, A*STAR by TF IPC Ltd under the grant titled “Temasek Foundation Infectious Diseases Programme for Surveillance and Diseases X Resilience”; and was funded in whole, or in part, by the Wellcome Trust (220211).
AcknowledgmentsLM is a recipient of the Singapore International Graduate Award (SINGA). We would like to thank the SMRU Clinic and laboratory staff for their role in obtaining the isolates. We thank Dr. Amanda Bifani for proofreading this manuscript.
Conflict of interestLM and PB are employed, and BT was employed by A*STAR Infectious Diseases Labs (A*IDL).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1368813/full#supplementary-material
ReferencesAdesoji, A. T., and Call, D. R. (2020). Molecular analysis of florfenicol-resistant bacteria isolated from drinking water distribution systems in Southwestern Nigeria. J. Glob. Antimicrob. Resist. 23, 340–344. doi: 10.1016/j.jgar.2020.10.005
PubMed Abstract | Crossref Full Text | Google Scholar
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
PubMed Abstract | Crossref Full Text | Google Scholar
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
PubMed Abstract | Crossref Full Text | Google Scholar
Bolton, L. F., Kelley, L. C., Lee, M. D., Fedorka-Cray, P. J., and Maurer, J. J. (1999). Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37, 1348–1351. doi: 10.1128/JCM.37.5.1348-1351.1999
PubMed Abstract | Crossref Full Text | Google Scholar
Chopjitt, P., Wongsurawat, T., Jenjaroenpun, P., Boueroy, P., Hatrongjit, R., Kerdsin, A., et al. (2020). Complete genome sequences of four extensively drug-resistant acinetobacter baumannii isolates from Thailand. Microbiol. Resour. Announc. 9:20. doi: 10.1128/MRA.00949-20
PubMed Abstract | Crossref Full Text | Google Scholar
Fernández-Alarcón, C., Miranda, C. D., Singer, R. S., López, Y., Rojas, R., Bello, H., et al. (2010). Detection of the floR gene in a diversity of florfenicol resistant gram-negative bacilli from freshwater salmon farms in Chile. Zoonoses Public Health 57, 181–188. doi: 10.1111/j.1863-2378.2009.01243.x
PubMed Abstract | Crossref Full Text | Google Scholar
Fu, C., Ding, H., Zhang, Q., Song, Y., Wei, Y., Wang, Y., et al. (2022). Comparative analysis of antibiotic resistance genes on a pig farm and its neighboring fish ponds in a lakeside district. Environ. Pollut. 303:119180. doi: 10.1016/j.envpol.2022.119180
PubMed Abstract | Crossref Full Text | Google Scholar
Guo, X., Chen, H., Tong, Y., Wu, X., Tang, C., Qin, X., et al. (2024). A review on the antibiotic florfenicol: occurrence, environmental fate, effects, and health risks. Environ. Res. 244, 117934. doi: 10.1016/j.envres.2023.117934
PubMed Abstract | Crossref Full Text | Google Scholar
Huys, G., Bartie, K., Cnockaert, M., Hoang Oanh, D. T., Phuong, N. T., Somsiri, T., et al. (2007). Biodiversity of chloramphenicol-resistant mesophilic heterotrophs from Southeast Asian aquaculture environments. Res. Microbiol. 158, 228–235. doi: 10.1016/j.resmic.2006.12.011
PubMed Abstract | Crossref Full Text | Google Scholar
Illambas, J., Potter, T., Sidhu, P., Rycroft, A. N., Cheng, Z., Lees, P., et al. (2013). Pharmacodynamics of florfenicol for calf pneumonia pathogens. Vet. Rec. 172, 340–340. doi: 10.1136/vr.101155
PubMed Abstract | Crossref Full Text | Google Scholar
Jain, C., Rodriguez, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 1–8. doi: 10.1038/s41467-018-07641-9
PubMed Abstract | Crossref Full Text | Google Scholar
Kehrenberg, C., Meunier, D., Targant, H., Cloeckaert, A., Schwarz, S., Madec, J. Y., et al. (2006). Plasmid-mediated florfenicol resistance in Pasteurella trehalosi. J. Antimicrob. Chemother. 58, 13–17. doi: 10.1093/jac/dkl174
PubMed Abstract | Crossref Full Text | Google Scholar
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
PubMed Abstract | Crossref Full Text | Google Scholar
Kerek, Á., Török, B., Laczkó, L., Kardos, G., Bányai, K., Somogyi, Z., et al. (2023). In Vitro Microevolution and co-selection assessment of florfenicol impact on escherichia coli resistance development. Antibiotics 12:1728. doi: 10.3390/antibiotics12121728
PubMed Abstract | Crossref Full Text | Google Scholar
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
PubMed Abstract | Crossref Full Text | Google Scholar
Lei, Z., Liu, Q., Yang, S., Yang, B., Khaliq, H., Li, K., et al. (2018). PK-PD integration modeling and cutoffvalue of florfenicol against Streptococcus suis in pigs. Front. Pharmacol. 9:325571. doi: 10.3389/fphar.2018.00002
PubMed Abstract | Crossref Full Text | Google Scholar
Li, B., Zhao, Z. C., Wang, M. H., Huang, X. H., Pan, Y. H., Cao, Y. P., et al. (2014). Antimicrobial resistance and integrons of commensal Escherichia coli strains from healthy humans in China. J. Chemother. 26, 190–192. doi: 10.1179/1973947813Y.0000000113
PubMed Abstract | Crossref Full Text | Google Scholar
Li, P., Zhu, T., Zhou, D., Lu, W., Liu, H., Sun, Z., et al. (2020). Analysis of resistance to florfenicol and the related mechanism of dissemination in different animal-derived bacteria. Front. Cell Infect Microbiol. 10:369. doi: 10.3389/fcimb.2020.00369
PubMed Abstract | Crossref Full Text | Google Scholar
Lin, X., Tan, A., Deng, Y., Liu, W., Zhao, F., Huang, Z., et al. (2023). High occurrence of antibiotic resistance genes in intensive aquaculture of hybrid snakehead fish. Front. Mar. Sci. 9:1088176. doi: 10.3389/fmars.2022.1088176
Crossref Full Text | Google Scholar
Lu, J., Zhang, J., Xu, L., Liu, Y., Li, P., Zhu, T., et al. (2018). Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect Control 7:415. doi: 10.1186/s13756-018-0415-0
PubMed Abstract | Crossref Full Text | Google Scholar
Mancilla-Rojano, J., Ochoa, S. A., Reyes-Grajeda, J. P., Flores, V., Medina-Contreras, O., Espinosa-Mazariego, K., et al. (2020). Molecular epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from children at the Hospital INFANTIL de México Federico Gómez. Front. Microbiol. 11:576673. doi: 10.3389/fmicb.2020.576673
PubMed Abstract | Crossref Full Text | Google Scholar
Migliaccio, A., Bray, J., Intoccia, M., Stabile, M., Scala, G., Jolley, K. A., et al. (2023). Phylogenomics of Acinetobacter species and analysis of antimicrobial resistance genes. Front. Microbiol. 14:1264030. doi: 10.3389/fmicb.2023.1264030
PubMed Abstract | Crossref Full Text | Google Scholar
Møller, T. S. B., Overgaard, M., Nielsen, S. S., Bortolaia, V., Sommer, M. O. A., Guardabassi, L., et al. (2016). Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. 16, 1–8. doi: 10.1186/s12866-016-0649-z
PubMed Abstract | Crossref Full Text | Google Scholar
Qian, C., Liu, H., Cao, J., Ji, Y., Lu, W., Lu, J., et al. (2021). Identification of floR variants associated with a novel Tn4371-like integrative and conjugative element in clinical pseudomonas aeruginosa isolates. Front. Cell Infect Microbiol. 11:685068. doi: 10.3389/fcimb.2021.685068
PubMed Abstract | Crossref Full Text | Google Scholar
Somogyi, Z., Mag, P., Simon, R., Kerek, Á., Szabó, P., Albert, E., et al. (2023). Pharmacokinetics and pharmacodynamics of florfenicol in plasma and synovial fluid of pigs at a dose of 30 mg/kgbw following intramuscular administration. Antibiotics 12:12040758. doi: 10.3390/antibiotics12040758
PubMed Abstract | Crossref Full Text | Google Scholar
Trevisi, P., Amatucci, L., Ruggeri, R., Romanelli, C., Sandri, G., Luise, D., et al. (2022). Pattern of antibiotic consumption in two Italian production chains differing by the endemic status for porcine reproductive and respiratory syndrome. Front. Vet. Sci. 9:840716. doi: 10.3389/fvets.2022.840716
PubMed Abstract | Crossref Full Text | Google Scholar
Trif, E., Cerbu, C., Olah, D., Zăblău, S. D., Spînu, M., Potârniche, A. V., et al. (2023). Old antibiotics can learn new ways: a systematic review of florfenicol use in veterinary medicine and future perspectives using nanotechnology. Animals 13:1695. doi: 10.3390/ani13101695
PubMed Abstract | Crossref Full Text | Google Scholar
Veltri, D., Wight, M. M., and Crouch, J. A. (2016). SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44, W41–W45. doi: 10.1093/nar/gkw330
PubMed Abstract | Crossref Full Text | Google Scholar
Verner-Jeffreys, D. W., Brazier, T., Perez, R. Y., Ryder, D., Card, R. M., Welch, T. J., et al. (2017). Detection of the florfenicol resistance gene floR in Chryseobacterium isolates from rainbow trout. Exception to the general rule? FEMS Microbiol. Ecol. 93:fix015. doi: 10.1093/femsec/fix015
PubMed Abstract | Crossref Full Text | Google Scholar
Villalón, P., Ortega, M., Sáez-Nieto, J. A., Carrasco, G., Medina-Pascual, M. J., Garrido, N., et al. (2019). Dynamics of a sporadic nosocomial acinetobacter calcoaceticus-acinetobacter baumanniicomplex population. Front. Microbiol. 10:593. doi: 10.3389/fmicb.2019.00593
PubMed Abstract | Crossref Full Text | Google Scholar
Wang, E., Yuan, Z., Wang, K., Gao, D., Liu, Z., Liles, M. R., et al. (2019). Consumption of florfenicol-medicated feed alters the composition of the channel catfish intestinal microbiota including enriching the relative abundance of opportunistic pathogens. Aquaculture 501, 111–118. doi: 10.1016/j.aquaculture.2018.11.019
Crossref Full Text | Google Scholar
Wang, Q., Zhou, X., Liu, Y., Ding, Q., Wu, Z., Deng, J., et al. (2022). Bacterial community and antibiotic resistance gene profiles of fish gut contents and their aquaculture environment in Tianjin, China. Aquaculture J. 2, 269–284. doi: 10.3390/aquacj2040016
Crossref Full Text | Google Scholar
Wareth, G., Brandt, C., Sprague, L. D., Neubauer, H., and Pletz, M. W. (2021). WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective. BMC Microbiol. 21, 1–13. doi: 10.1186/s12866-021-02270-7
PubMed Abstract | Crossref Full Text | Google Scholar
Wareth, G., Neubauer, H., and Sprague, L. D. (2019). Acinetobacter baumannii- a neglected pathogen in veterinary and environmental health in Germany. Vet. Res. Commun. 43, 1–14. doi: 10.1007/s11259-018-9742-0
PubMed Abstract | Crossref Full Text | Google Scholar
Wasyl, D., Hoszowski, A., Zajac, M., and Szulowski, K. (2013). Antimicrobial resistance in commensal Escherichia coli isolated from animals at slaughter. Front. Microbiol. 4:54851. doi: 10.3389/fmicb.2013.00221
留言 (0)