Acinetobacter baumannii (A. baumannii) is an important Gram-negative pathogen that often causes serious nosocomial infections, especially among immunocompromised and elderly patients in intensive care units (ICUs) (Sarshar et al., 2021). It has been estimated that between 47% and 93% of A. baumannii infections are associated with multi-drug resistance (MDR), which is facilitated through a variety of well-documented mechanisms (β-lactamases, efflux pumps, aminoglycoside-modifying enzymes, permeability defects, and target modifications) (Gallagher and Baker, 2020). In China, the epidemic of A. baumannii in the ICUs is even more severe. Research showed that 71.4% of ICUs were found to be contaminated by carbapenem-resistant A. baumannii strains (Liu et al., 2022). A study reported a 10-fold rise in the incidence of A. baumannii–related bloodstream infections in China during 2009–2018, from 99 to 926 per 1,000,000 ICU populations (Meng et al., 2021). Another research showed that A. baumannii was the most frequent bacterial isolate in ventilator-associated pneumonia in China, and rates were 35.7%−52.7% (Xie et al., 2018).
Carbapenem antibiotics have historically been considered a first-line treatment for A. baumannii infections (Poirel et al., 2011). Data from the Antimicrobial Surveillance Network (CHINET) showed that the proportion of A. baumannii isolates that are resistant to meropenem and imipenem has reached nearly 72% in China during 2022. Several β-lactamases can cause carbapenem resistance in A. baumannii, but the most common are class D β-lactamases, including those belonging to the OXA-23,−24,−51, and−58 families (Poirel and Nordmann, 2006; Walther-Rasmussen and Hoiby, 2006).
Multidrug resistance, especially carbapenem resistance in A. baumannii, is a global healthcare concern. There was a lot of research on MDR A. baumannii in China. However, there was little available data on the phenotypic and genotypic characteristics of MDR A. baumannii isolates in northwest China. To investigate the prevalence and genomic features of MDR A. baumannii transmitted in the first hospital of Lanzhou University, which is a large tertiary hospital, a total of 50 MDR A. baumannii isolates were collected and analyzed by whole-genome sequencing (WGS).
Materials and methods Bacterial isolates and clinical dataWe collected all A. baumannii isolates isolated from people who were hospitalized in the first quarter of 2022 in the first hospital of Lanzhou University, Gansu Province, China. Specimens from the same site in the same patient were removed. We further screened for MDR A. baumannii from these isolates. MDR A. baumannii was defined by resistance to three or more representatives of the following classes of antibiotics: quinolones, extended-spectrum cephalosporins, β-lactam/β-lactamase inhibitor combination, aminoglycoside, and carbapenems (Hujer et al., 2006). In the end, we collected 31 ICU-derived isolates and 19 other department-derived isolates in chronological order. If the patient has a history of ICU exposure within 1 month, we also classify the isolates as ICU-derived. The following clinical data were collected: age, gender, the main indicator of inflammation within 48 h of MDR A. baumannii positive (Zhang et al., 2021), primary disease, hospitalization time duration, outcome at discharge, and administration of antibiotics.
Susceptibility testingAll A. baumannii isolates were isolated from people who were hospitalized in the first hospital of Lanzhou University. A. baumannii isolates were identified by matrix-assisted laser desorption-ionization-time of flight mass spectrometry (MALDI-TOF MS; BioMérieux, Marcy l'Etoile, France). Minimal inhibitory concentrations (MICs) of Piperacillin-tazobactam, Cefoperazone-sulbactam, Ceftazidime, Cefepime, Imipenem, Meropenem, Ciprofloxacin, Levofloxacin, Trimethoprim-sulfamethoxazole, Amikacin, Tobramycin, Tigecycline, Minocycline and Colistin were tested by the VITEK-2 compact automatic microbial analyzer (BioMérieux, Marcy l'Etoile, France). The reference strains, Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922, are being used as quality control. Ampicillin-sulbactam (10/10 μg) was tested for susceptibility using a disk diffusion test. Interpretations of resistance phenotypes follow those of the Clinical and Laboratory Standards Institute (CLSI) guideline document M100-S32. There is no susceptibility breakpoint for Acinetobacter to cefoperazone-sulbactam has been provided by CLSI, we referred to CLSI breakpoints for Enterobacteriaceae susceptibility and resistance to cefoperazone-sulbactam. The criteria for the susceptibility of tigecycline were adapted from the U. S. Food and Drug Administration.
We calculated the multiple antibiotics resistance (MAR) index for all samples. MAR index was calculated as described by Blasco et al. (2008) as follows: MAR = a/b, where a = number of antibiotics to which the isolate was resistant; b = total number of antibiotics against which individual isolate was tested.
Whole genome sequencing and bioinformatics analysisThe total bacterial DNA of 50 clinical MDR A. baumannii isolates was extracted using TIANamp bacteria DNA kit (TIANGEN, Beijing, China), following the manufacturer's instructions. For testing sample qualification, the DNA concentration was measured by NanoDrop 2000 (Thermo Fisher Scientific, The United States of America). The extracted DNA was sent to Zhejiang Tianke Biotechnology Co., Ltd., for whole-gene sequencing. MLST of the test isolates based on the public databases for molecular typing and microbial genome diversity (https://pubmlst.org/) using the Oxford scheme (Jolley et al., 2018). The genetic relatedness of STs was analyzed and visualized using PHYLOVIZ 2.0a goeBURST analysis (Nascimento et al., 2017). For sequence analysis and annotation, the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST) and the RAST server (https://rast.nmpdr.org/) were utilized (Overbeek et al., 2014). The antimicrobial resistance (AMR) genes were identified using the comprehensive antibiotic resistance database (CARD; https://card.mcmaster.ca/) (Alcock et al., 2020). The transposon regions and the integrons were investigated using BacAnt (http://www.bacant.net/BacAnt/) (Hua et al., 2021). Also, the virulence factor genes (VFGs) were detected using the virulence factor database (VFDB; http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi) (Chen et al., 2005).
Phylogenetic analysisOne of the isolates we isolated LZU2 (ST469oxf) was used as the reference strain. Single nucleotide polymorphisms (SNPs) alignment was matched using package Snippy v4.6.0, and phylogenetic trees were constructed by fasttree v2.1.10. Cytoscape v3.7.2 software was used to visualize the phylogenetic tree.
Statistical analysisQuantitative data of normal distribution were presented as mean deviation, while those of abnormal distribution were expressed as the median and quartile. Qualitative data were presented as a percentage (%) and their inter-group comparison was performed with a chi-square test, qualitative data incapable of being analyzed with a chi-square test were compared between two groups using Fisher's exact test. SPSS 27.0 software was used for statistical analysis, and P < 0.05 indicated a statistically significant difference.
Results Patient demographics and clinical profilesIn this study, we collected a total of 50 isolates, including 21 isolates from the ICU, 10 isolates from the emergency intensive care unit (EICU), and 19 isolates from other wards. The cardiac surgery ward had the highest separation rate among wards excluding intensive care units (16.0%).
The clinical profiles showed that, of the 50 patients infected with MDR A. baumannii, 80.0% were male, pulmonary disease was the most common primary disease, these patients had a median age of 59 years, the median total length of hospital stay was 33 days and 44% of cases died. These isolates were collected mostly from sputum and accounted for 75% of all samples (Table 1).
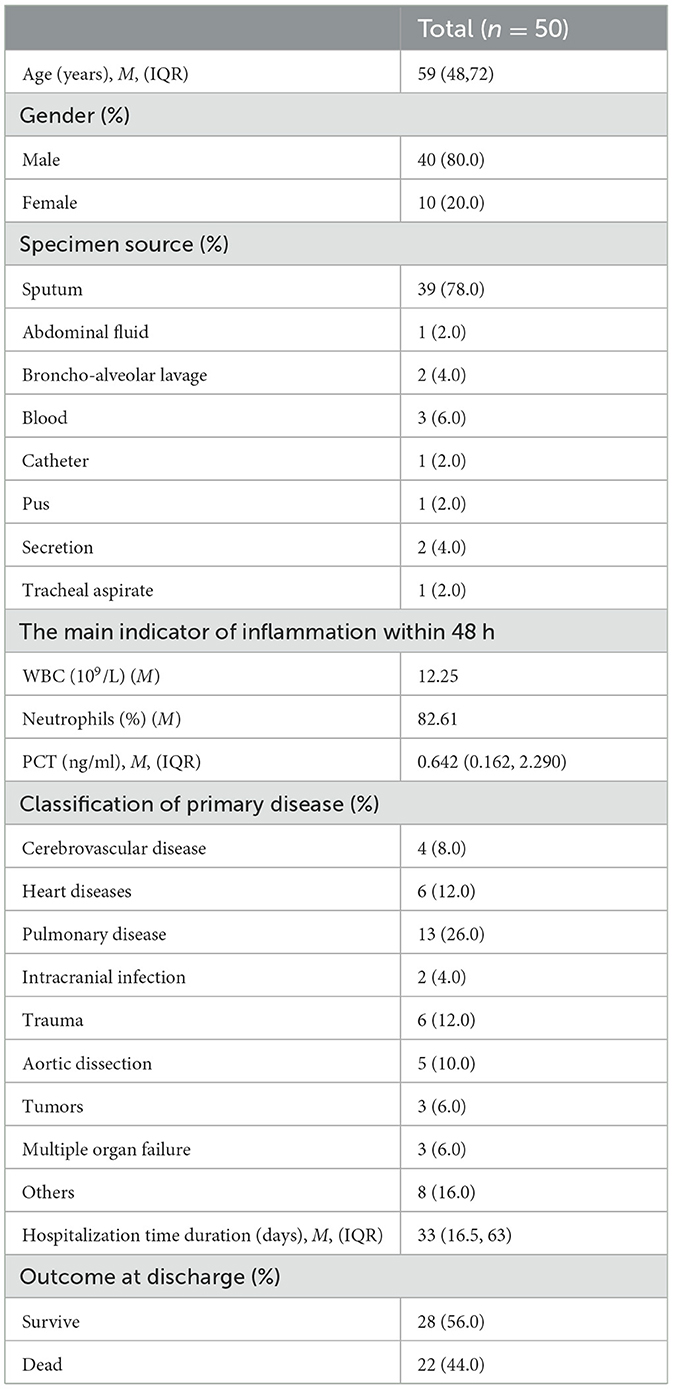
Table 1. General information statistics.
We investigated antibiotic use strategies 1 week before and after MDR A. baumannii was detected in these patients. The survey showed that the top three antibiotic strategies were carbapenem alone, cefoperazone-sulbactam alone, and piperacillin-tazobactam alone.
Susceptibility testingAntibiotic susceptibility testing was performed on the 50 MDR A. baumannii isolates. Just as revealed by drug resistance test results, these isolates were generally resistant to the common β-lactamase antibiotics, the rate of resistance to ampicillin-sulbactam, piperacillin-tazobactam, ceftazidime, and cefepime were 100%, 98.0%, 96.0%, and 86.0%, and 92.0% isolates were found resistance to both imipenem and meropenem. Resistance to Cefoperazone-sulbactam is relatively low, at 20%. Most of the isolates were quinolone resistant, 98.0% of isolates were resistant to ciprofloxacin, and 92.0% of isolates were resistant to levofloxacin. The rate of resistance to trimethoprim-sulfamethoxazole, amikacin and tobramycin were 30%, 44%, and 68%, respectively. Additionally, only 6.0% of isolates were resistant to minocycline, but 28.0% of isolates showed intermediate resistance to minocycline. There were no isolates resistant to tigecycline and colistin. The details are shown in Table 2. We calculated the MAR index for all samples. The MAR index was higher than the 0.2 limit in all our tested isolates (Table 3).
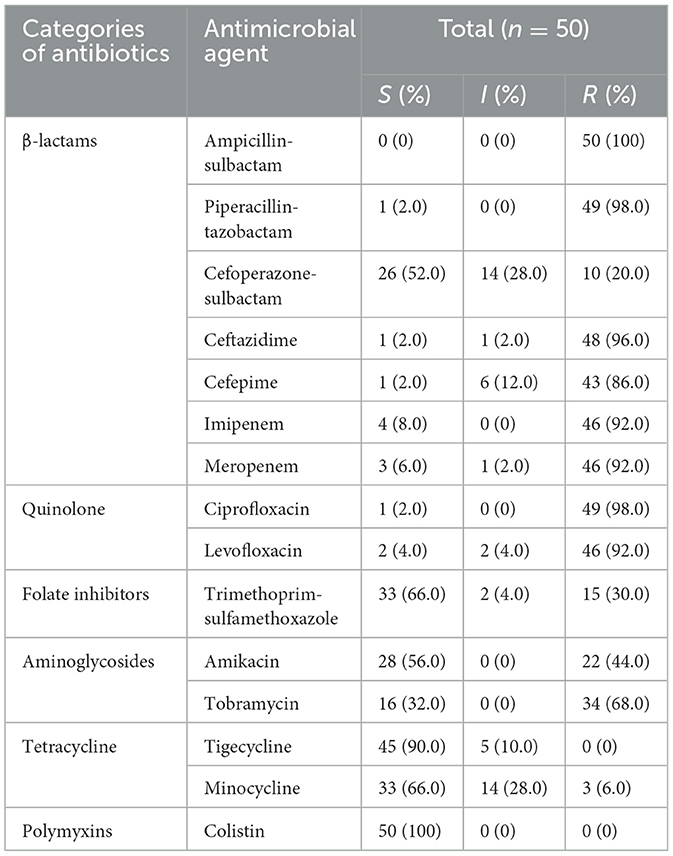
Table 2. The results of drug resistance test and genetic characteristics.
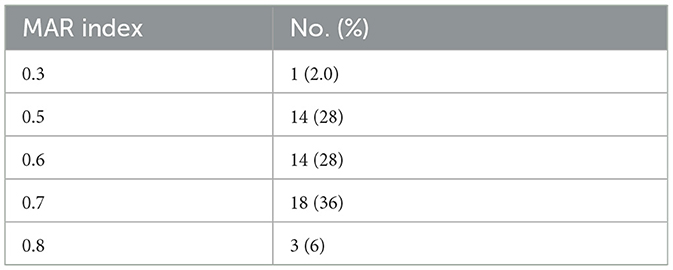
Table 3. Multiple antibiotics resistance (MAR) index of the MDR Acinetobacter baumannii isolates.
MLST analysisIn our study, the gdhB allele in the ST_Oxford scheme had two copies in most isolates except ST931. A total of eight STs were detected for the 50 MDR A. baumannii isolates by MLST, including ST469, ST208/1806, ST195/1816, ST540, ST368/1962, ST369/1837, ST931 and ST2336. ST469 was predominant in all MDR A. baumannii isolates (46.0%), followed by ST208/1806 (18.0%), ST195/1816 (14.0%), ST540 (14.0%), ST368/1962 (2.0%), ST369/1837 (2.0%), ST931 (2.0%) and ST2336 (2.0%).
We analyzed ST types distributed in different departments. We found that ST469 is mainly distributed in the ICU and EICU, and ST195/1816 are almost exclusively present in the ICU. ST208/1806 is mainly found in departments outside the ICU and EICU. Among these isolated clones, ST208/1806, ST195/1816, ST540, ST368/1962, ST369/1837, and ST2336 were SLVs and all these STs belonged to clonal complex 208 (CC208; Figure 1).
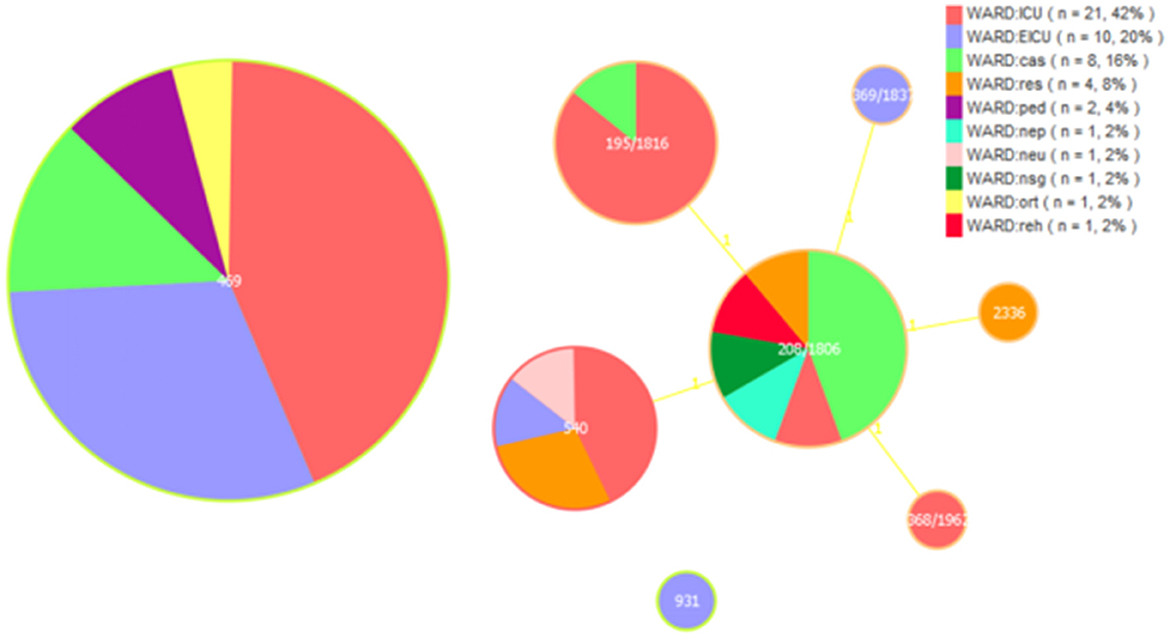
Figure 1. Multi-locus sequence typing (MLST) and minimum spanning tree (MST) population analysis of 50 MDR Acinetobacter baumannii isolates from the different clinical departments. Minimum spanning tree (MST) based on STs profiles of 50 MDR A. baumannii isolates. Each circle represents an ST and the size of the circle is proportional to the number of isolates. Different colors represent different wards. ICU, intensive care unit; EICU, emergency intensive care unit; ped, pediatric ward; ort, orthopedics ward; res, respiratory medicine ward; reh, rehabilitation ward; neu, neurology ward; nsg, neurosurgery ward; nep, nephrology ward; cas, cardiac surgery ward.
Genetic characteristics and phylogenetic analysisWGS analysis demonstrated that most MDR A. baumannii isolates harbored carbapenemase genes blaOXA − 23 (94.0%) and blaOXA − 66 (98.0%). Cephalosporinase genes blaADC30 (36.0%) and blaADC73 (62.0%) are also expressed in most isolates. These isolates tend to carry one of the two genes. The 44% of isolates carried blaTEM − 1, which encodes resistance to β-lactams. A total of 8 genes [AAC(3)-Ia, AAC(6')-Ib9, APH(6)-Id, ANT(3”)-IIc, APH(3')-Ia, APH(3”)-Ib, aadA, armA] encoding aminoglycoside resistance were found in these isolates. The 98.0% of isolates carried APH(6)-Id, ANT(3”)-IIc and APH(3”)-Ib. All of these isolates contained the tet (B) and tetR genes predicting tetracycline resistance and gyrAS81L, parCS84L, parCV104I and parCD105E genes encoding fluoroquinolone resistance. The sul1 and sul2 genes that encode resistance to sulfonamide were observed in 30.0% and 10.0% isolates, respectively. In addition, 84.0% of isolates possessed the mphE and msrE genes encoding macrolide resistance. We also found other genes such as the LpsB (100%) and catB8 (20.0%) genes, which provide resistance to peptide antibiotics and chloramphenicol, respectively. In addition to these genes, the resistance-nodulation-cell division (RND) antibiotic efflux pump and major facilitator superfamily (MFS) antibiotic efflux pump also played an important role in antibiotic resistance.
Overall, the most isolates carried Tn6292, Tn2007, Tn6022-delta-1, Tn6205, Tn6166, the presence is 84%, 94%, 88%, 98%, and 84%, respectively. Twenty-eight percent of isolates possessed Tn2009 and mainly being detected in CC208 isolates. We detected a total of eight integrons in these isolates.
We further analyzed the existence of virulence factor genes. All of the MDR A. baumannii isolates harbored genes adeFGH, bap, csuABCDE and pgaABCD (associated with biofilm formation), plc and plcD (encoding phospholipase), lpsB and lpxABCDLM (associated with immune evasion), barAB, basABCDFGHIJ, bauABCDEF, entE and hemO (related to iron uptake), abaI and abaR (played a role in Quorom sensing), bfmR and bfmS (controlled biofilm formation and cellular morphology), pbpG encoding penicillin-binding protein, OmpA associated with adherence. The 96% of isolates possessed virulence factor katA associated with stress adaption. The distribution of pilE varies from isolate to isolate. Only 46% of isolates expressed the pilE gene which was essential for twitching motility and natural competence and contributed to host cell adherence.
We mapped the heat map of resistance genes, transposons, integrons and virulence factor genes (pilE and katA; Figure 2). Combined with phylogenetic analysis and molecular characteristics, we found that the MDR A. baumannii prevalent in our hospital consists mainly of two clonal populations: ST469, ST208 and its SLVs (CC208). We found that ST469 and CC208 have large differences in genomic characteristics. CC208 isolates carried more resistance genes. Except LZU13, all ST469 isolates possessed the same resistance genes and transposons.
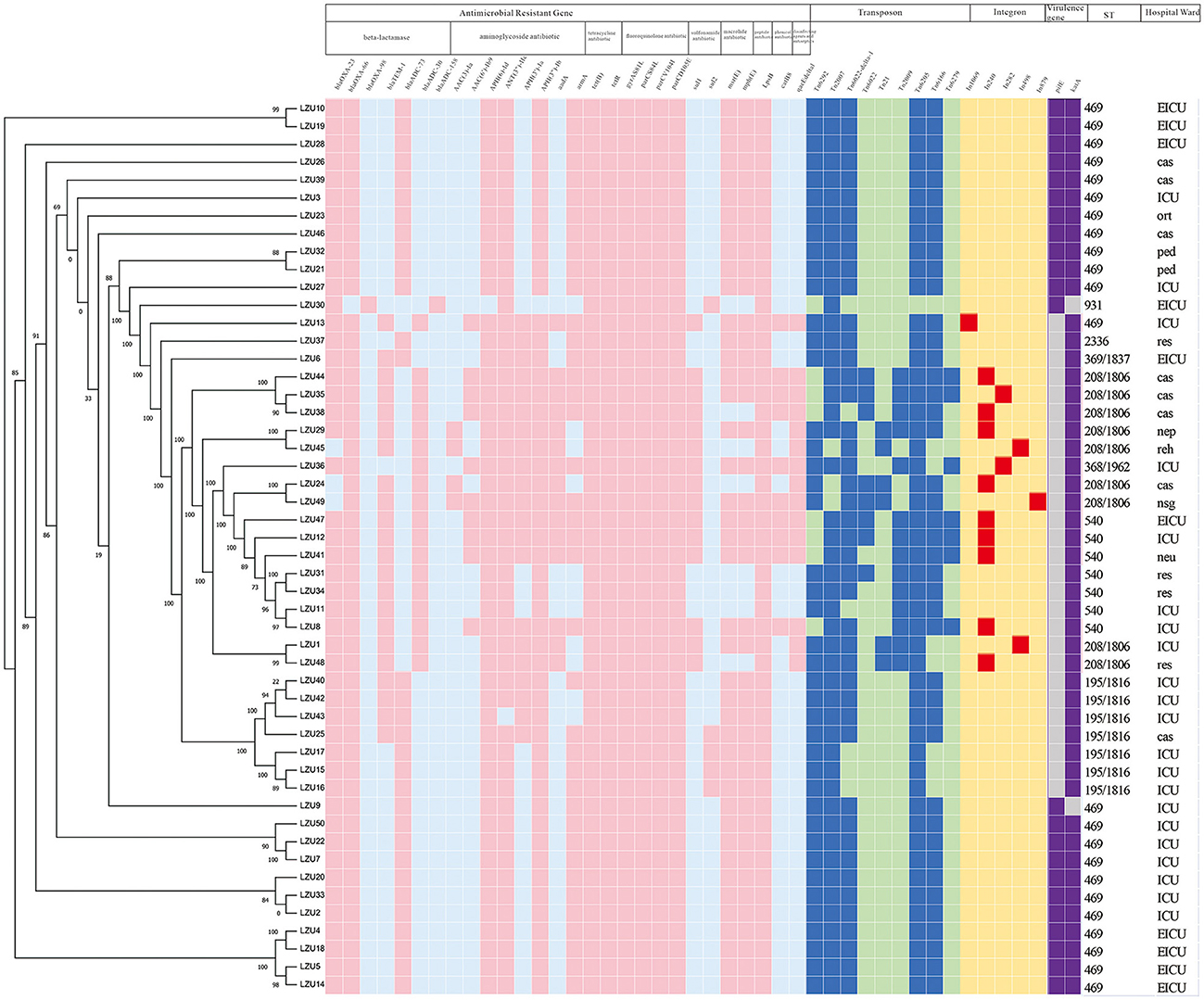
Figure 2. Phylogenetic analysis and heat map of the detection of ARGs, transposons, integrons and VFGs among the 50 MDR Acinetobacter baumannii clinical isolates. The left side is phylogenetic tree constructed by calling SNPs from core gene alignment of the study 50 MDR A. baumannii clinical isolates. Pink, blue, red, purple colors represent express the corresponding gene, respectively. Light blue, light green, yellow, gray colors represent absent of the corresponding gene, respectively. ICU, Intensive care unit; EICU, emergency intensive care unit; ped, pediatric ward; ort, orthopedics ward; res, respiratory medicine ward; reh, rehabilitation ward; neu, neurology ward; nsg, neurosurgery ward; nep, nephrology ward; cas, cardiac surgery ward. Because all of these isolates harbored the following virulence factor genes adeFGH, bap, csuABCDE, pgaABCD, plc, plcD, lpsB, lpxABCDLM, pbpG, bfmRS, abaI, abaR, barAB, basABCDFGHIJ, bauABCDEF, entE, OmpA, we only shown two genes, pilE and katA.
Comparative analysis between the ST469 group and CC208 groupWe compared the differences in resistance gene carriers between the ST469 group (n = 23) and the CC208 group (n = 26; Table 4). We found that the carrying rates of the following genes in CC208 group: blaTEM − 1, blaADC30, AAC(6')-Ib9, APH(3')-Ia, aadA, sul1, catB8 and qacEdelta1 were significantly higher than those in ST469 group, while the carrying rates of blaADC73 and armA in ST469 group were significantly higher than those in CC208 group. Furthermore, the ST469 isolates harbored the same transposon elements, Tn6292, Tn2007, Tn6022-delta-1, Tn6025, and Tn6166. Most CC208 isolates harbored both Tn2007 and Tn2009 elements. We further analyzed the differences in antibiotic susceptibility phenotypes between the two groups (Table 5), and showed that the nonsensitivity rates of cefoperazone-sulbactam and trimethoprim-sulfamethoxazole were significantly higher in the CC208 group than those in the ST469 group. The proportion of CC208 isolates resistant and mediated to cefoperazone-sulbactam was as high as 84.6%.
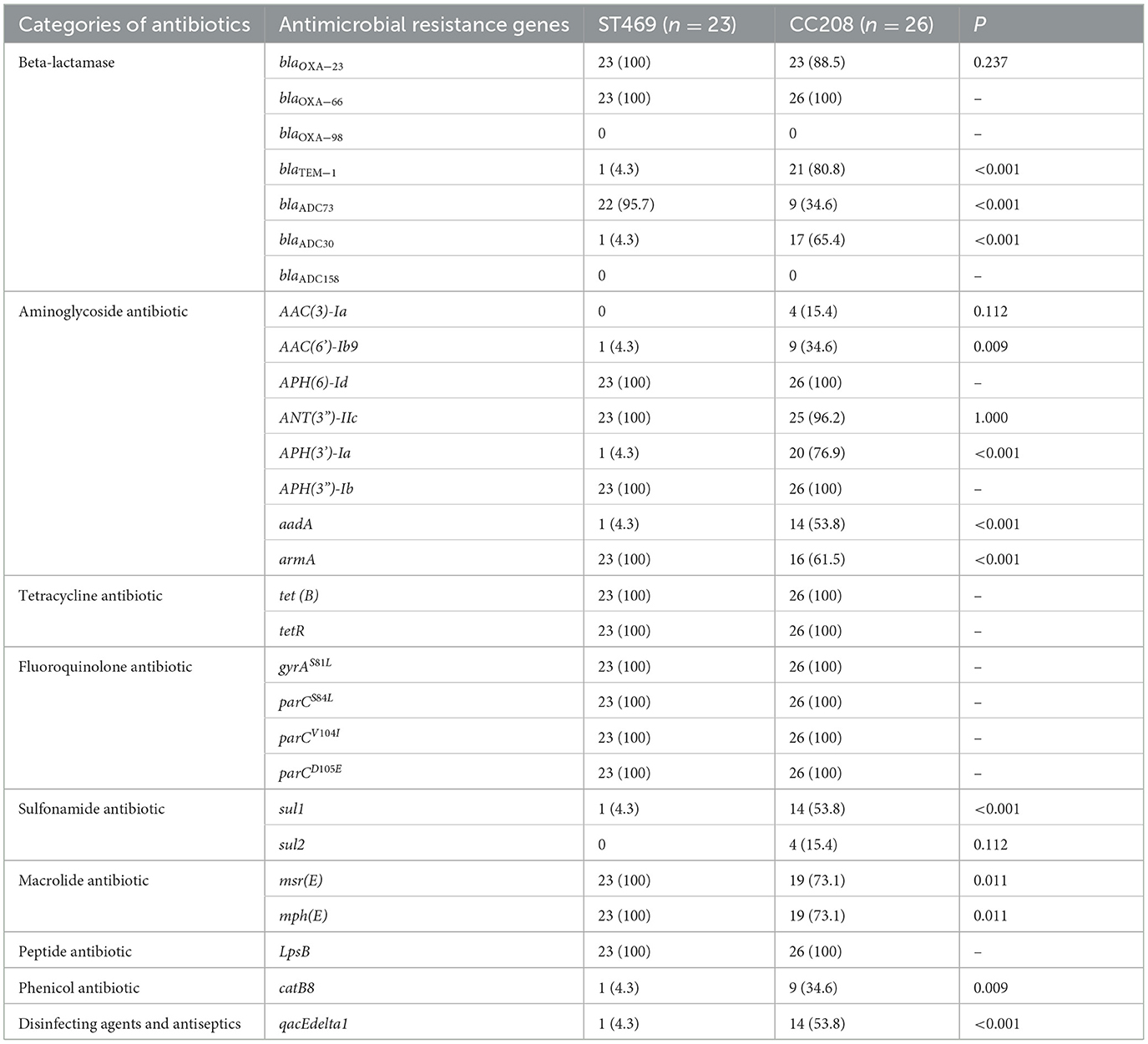
Table 4. Comparative analysis of the antimicrobial resistance genes in ST469 group and CC208 group.
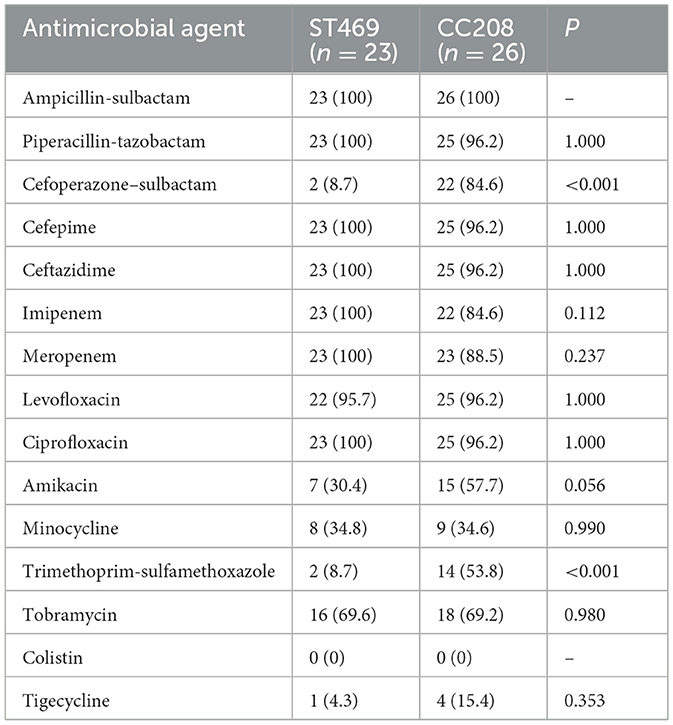
Table 5. Comparative analysis of antibiotic nonsensitivity rates in ST469 group and CC208 group.
Characterization of carbapenemase-encoding genesA variety of carbapenemase encoding genes were found in these MDR A. baumannii isolates, the presence of blaOXA − 23 (94.0%) and blaOXA − 66 (98.0%) was the most frequent determinants for carbapenem resistance. BLASTN indicated that the blaOXA − 23 gene resided exclusively in the Tn2007 and Tn2009 elements. Tn2007 harbors the blaOXA − 23 gene and a single copy of ISAba4 upstream of the blaOXA − 23. The structure of Tn2009 was ISAba1-blaOXA − 23-ATPase-hp-hp-ATPase-hp-hp-DNA helicase-ISAba1, which has two copies of ISAba1 flanking an internal segment containing blaOXA − 23 (Figure 3).
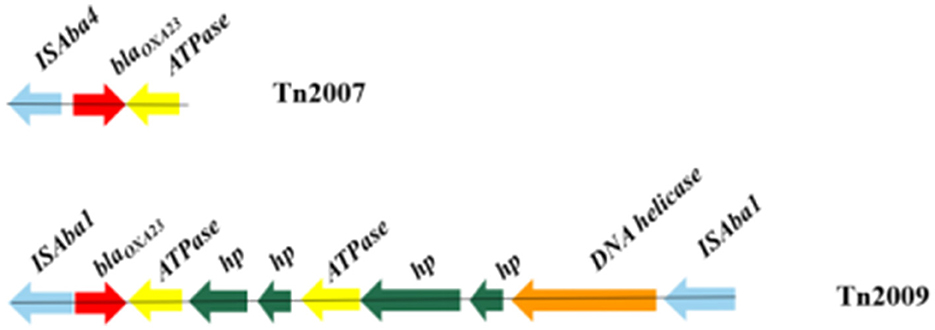
Figure 3. Genotypic characterization of blaOXA − 23 in Acinetobacter baumannii. Carbapenemase genes are represented by red arrows, and other coding sequences are represented with other different colors.
DiscussionA comparatively small number of pathogens are responsible for a substantial burden of hospital-acquired infections globally, A. baumannii is one of the key pathogens (Gallagher and Baker, 2020). Carbapenem antibiotics have historically been considered a first-line treatment for A. baumannii infections. However, according to the literature, increased resistance to carbapenems in the A. baumannii strain has been observed globally over the past decade (Rodríguez et al., 2018; Teerawattanapong et al., 2018). Carbapenem resistance in A. baumannii is medicated by several coexisting mechanisms, while the most prevalent mechanism is associated with the production of oxacillinases (Ambler class D β-lactamases), especially OXA-23 (Li et al., 2019; Nordmann and Poirel, 2019). This work also confirms that the production of the OXA-type carbapenem-hydrolyzing-class D β-lactamase is the key mechanism for carbapenem resistance in A. baumannii. In our study, the detection of resistant genes showed that the most common carbapenemase-resistant gene was blaOXA − 66 (98.0%) in all MDR A. baumannii isolates, followed by blaOXA − 23 (94.0%). Research showed that without the promoter provided by ISAba1, the OXA-23 carbapenemase does not confer clinically relevant levels of resistance to carbapenem antibiotics (Walther-Rasmussen and Hoiby, 2006; Figueiredo et al., 2009). ISAba1 has been shown to provide a strong promoter that drives a high level of expression of blaOXA − 23 (Segal et al., 2007). To date, five transposons containing the blaOXA − 23 gene, including Tn2006, Tn2007, Tn2008, Tn2008B, and Tn2009, have been reported (Nigro and Hall, 2016). In our study, Tn2007 was identified in 94.0% of isolates. However, only 28.0% of isolates possessed Tn2009 and only being detected in CC208 isolates. BLASTN indicated that Tn2009 has two copies of ISAba1 flanking an internal segment containing blaOXA − 23. There is currently no evidence that Tn2007 is a transposon that is capable of self-movement (Nigro and Hall, 2016). Therefore, Tn2009 may played a major role in the horizontal transfer of blaOXA − 23 in the CC208 isolates isolated from our hospital. Gao et al. (2022) described the potential circular intermediate of Tn2009 and suggested that replicative transposition contributes to the evolution and transmission of OXA-23-producing ST208. This structure Tn2009 element has only been reported in China (Zhou et al., 2011; Zhu et al., 2013). Previous research showed that Tn2009 and Tn2008 played dominant roles in the dissemination of blaOXA − 23 in China (Wang et al., 2011; Liu et al., 2015). In this study, Tn2006 and Tn2008 were not detected in these isolates.
There is still no optimal therapeutic regimen for MDR A. baumannii infection, sulbactam-based regimens, tigecycline-based regimens, and polymyxins-based regimens have been recommended as the main treatment for MDR A. baumannii infection in China (Group et al., 2016). Fortunately, resistance to tigecycline and polymyxin has not been found in MDR A. baumannii isolated from our hospital, so combination therapy based on tigecycline and polymyxin is recommended. As for the use of cefoperazone-sulbactam, our study showed that there was a large difference in resistance rates between different clones. The proportion of CC208 isolates resistant and mediated to cefoperazone-sulbactam is as high as 84.6% in our study. Therefore, the use of cefoperazone-sulbactam as empiric treatment regimens in patients with CC208 MDR A. baumannii infection is not recommended. Recently, a research about bloodstream infection caused by MDR A. baumannii showed that the most effective antibacterial regimen was the combination of cefoperazone-sulbactam and tigecycline (Yu et al., 2021). It was worth noting that the MDR A. baumannii isolated in our hospital has a very low resistance rate to minocycline. As the treatment options of MDR A. baumannii infections are considerably limited, reinstituting the use of older antimicrobials has now become a priority. Beganovic et al. (2021) evaluate minocycline alone and in combination with other commonly utilized anti-microbials against CRAB and non-CRAB isolates. They found that triple therapy with high-dose minocycline, continuous-infusion sulbactam, and polymyxin B produced the most significant kill against the CRAB, with no regrowth and minimal resistance development. A growing number of studies have shown that minocycline could constitute an alternative agent for the treatment of MDR A. baumannii infections (Lashinsky et al., 2017; Fragkou et al., 2019).
In our study, ST469 is the predominant sequence types in ICUs (17/31, 54.8%), followed by ST195/1816 (6/31, 19.4%). This differs from the prevalence of MDR A. baumannii in ICUs in other hospitals in China. It has been reported that ST208 and ST195 were the predominant epidemic types of MDR A. baumannii in China (Qu et al., 2016; Jiang et al., 2019). However, ST208 is the predominant circulating strain in out-of-ICUs (8/19, 42.1%) in our study. We found that the popular MDR A. baumannii isolates in our hospital is divided into two main endemic clones, ST469 and CC208. These two endemic clones have large differences in genomic characteristics. CC208 isolates carried more resistance genes. The carrying rates of the following genes in CC208 group: blaTEM − 1, blaADC30, AAC(6')-Ib9, APH(3')-Ia, aadA, sul1, catB8 and qacEdelta1 were significantly higher than those in ST469 group. We further analyzed the differences in antibiotic susceptibility phenotypes between these two groups, and showed that the nonsensitivity rates of cefoperazone-sulbactam were significantly higher in the CC208 group than in the ST469 group. Previous studies have shown that the blaTEM − 1 and blaADC30 could confer sulbactam resistance to A. baumannii (Krizova et al., 2013; Kuo et al., 2015). The current research on the mechanism of sulbactam resistance is very limited, our study provided evidence that blaTEM − 1 and blaADC30 is associated with A. baumannii. resistance to sulbactam. We also found that CC208 isolates had a significantly higher resistance rate to trimethoprim-sulfamethoxazole than ST469 isolates, possibly because most CC208 isolates carry the sul1 gene. Moreover, previous study shown that CC208 isolates was more tolerant to various adverse environments than other isolates (Bian et al., 2021). CC208 isolates has strong resistance to antibiotic and the environment at the same time, so it is very important to rapid identify the CC208 isolates and take some infection control measures to reduce the spread of CC208 MDR A. baumannii.
Recently, discussions arose about the accuracy of the two A. baumannii multilocus sequence typing (MLST) schemes (Castillo-Ramirez and Grana-Miraglia, 2019). Stefano Gaiarsa et al. reported issues of the Oxford scheme regarding gdhB paralogy, recombination, primer sequences, and position of the genes on the genome (Gaiarsa et al., 2019). The same problem was encountered in our study, the gdhB allele in the ST_Oxford scheme had two copies in most isolates except LZU30. However, according to our research, MLST still has its unique advantages in bacterial homologous analysis. The Oxford scheme shows higher concordance with phylogenies. But we think that this approach should be improved. As on multiple occasions, allele sequences resulting from this duplication were incorrectly used to establish new Oxford STs that do not exist, Stefano Gaiarsa et al. suggest that these gdhB-based alleles should be removed from the database (Castillo-Ramirez and Grana-Miraglia, 2019). We maintain the same opinion. The wrong calling of alleles at gdhB locus would artifactually inflated the diversity recorded using the Oxford scheme.
In conclusion, we present a whole-genome-based investigation of the prevalence and genomic features of MDR A. baumannii clinical isolates in a large tertiary hospital in Gansu. The most common carbapenemase-resistant genes still was blaOXA − 66 and blaOXA − 23 in our hospital. Two main endemic clones were identified in our hospital. One clone was predominantly circulating in ICUs and exhibited the same genetic characteristics (ST469), and another clone (CC208) carried more resistance genes and had more widely disseminate. The spread of CC208 isolates may pose a greater challenge to the treatment of MDR A. baumannii infection. We should take timely and effective measures to control the further epidemic of these isolates.
Data availability statementThe data supporting this work are available under NCBI bioproject PRJNA1103124.
Ethics statementThe studies involving humans were approved by the Ethics Committee of LZU No.1 hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributionsMH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. HS: Funding acquisition, Supervision, Writing – review & editing. YX: Investigation, Methodology, Writing – review & editing. XX: Funding acquisition, Supervision, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Gansu Province (No.: 21JR11RA093), the Natural Science Foundation of Gansu Province (No.: 21JR11RA086), the Youth Fund of the first hospital of Lanzhou University (No.: ldyyyn2020-65), and the Education Science and Technology Innovation Fund of Gansu Province (No.: 2022B-022).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAlcock, B. P., Raphenya, A. R., Lau, T. T., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
PubMed Abstract | Crossref Full Text | Google Scholar
Beganovic, M., DaffineeK, E., Luther, M. K., and LaPlante, K. L. (2021). Minocycline alone and in combination with polymyxin B, meropenem, and sulbactam against carbapenem-susceptible and -resistant Acinetobacter baumannii in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 65, e01680-20. doi: 10.1128/AAC.01680-20
PubMed Abstract | Crossref Full Text | Google Scholar
Bian, X., Liu, X., Zhang, X., Li, X., Zhang, J., Zheng, H., et al. (2021). Epidemiological and genomic characteristics of Acinetobacter baumannii from different infection sites using comparative genomics. BMC Genomics 22:649. doi: 10.1186/s12864-021-07942-2
PubMed Abstract | Crossref Full Text | Google Scholar
Blasco, M. D., Esteve, C., and Alcaide, E. (2008). Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J. Appl. Microbiol. 105, 469–475. doi: 10.1111/j.1365-2672.2008.03765.x
PubMed Abstract | Crossref Full Text | Google Scholar
Castillo-Ramirez, S., and Grana-Miraglia, L. (2019). Inaccurate multilocus sequence typing of Acinetobacter baumannii. Emerg. Infect. Dis. 25, 186–187. doi: 10.3201/eid2501.180374
PubMed Abstract | Crossref Full Text | Google Scholar
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33(suppl_1), D325–D328. doi: 10.1093/nar/gki008
PubMed Abstract | Crossref Full Text | Google Scholar
Figueiredo, S., Poirel, L., Croize, J., Recule, C., and Nordmann, P. (2009). In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob. Agents Chemother. 53, 2657–2659. doi: 10.1128/AAC.01663-08
PubMed Abstract | Crossref Full Text | Google Scholar
Fragkou, P. C., Poulakou, G., Blizou, A., Blizou, M., Rapti, V., Karageorgopoulos, D. E., et al. (2019). The role of minocycline in the treatment of nosocomial infections caused by multidrug, extensively drug and pandrug resistant Acinetobacter baumannii: a systematic review of clinical evidence. Microorganisms 7:159. doi: 10.3390/microorganisms7060159
PubMed Abstract | Crossref Full Text | Google Scholar
Gaiarsa, S., Batisti Biffignandi, G., Esposito, E. P., Castelli, M., Jolley, K. A., Brisse, S., et al. (2019). Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front. Microbiol. 10:930. doi: 10.3389/fmicb.2019.00930
PubMed Abstract | Crossref Full Text | Google Scholar
Gallagher, P., and Baker, S. (2020). Developing new therapeutic approaches for treating infections caused by multi-drug resistant Acinetobacter baumannii. J. Infect. 81, 857–861. doi: 10.1016/j.jinf.2020.10.016
PubMed Abstract | Crossref Full Text | Google Scholar
Gao, Y., Li, H., Chen, H., Zhang, J., Wang, R., Wang, Z., et al. (2022). Origin, phylogeny, and transmission of the epidemic clone ST208 of carbapenem-resistant Acinetobacter baumannii on a global scale. Microbiol. Spectr. 10:e0260421. doi: 10.1128/spectrum.02604-21
PubMed Abstract | Crossref Full Text | Google Scholar
Group, C. X. C. W., Guan, X., He, L., Hu, B., Hu, J., Huang, X., et al. (2016). Laboratory diagnosis, clinical management and infection control of the infections caused by extensively drug-resistant Gram-negative bacilli: a Chinese consensus statement. ClinMicrobiol Infect. 22, S15–S25. doi: 10.1016/j.cmi.2015.11.004
PubMed Abstract | Crossref Full Text | Google Scholar
Hua, X., Liang, Q., Deng, M., He, J., Wang, M., Hong, W., et al. (2021). BacAnt: a combination annotation server for bacterial DNA sequences to identify antibiotic resistance genes, integrons, and transposable elements. Front. Microbiol. 12:649969. doi: 10.3389/fmicb.2021.649969
PubMed Abstract | Crossref Full Text | Google Scholar
Hujer, K. M., Hujer, A. M., Hulten, E. A., Bajaksouzian, S., Adams, J. M., Donskey, C. J., et al. (2006). Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50, 4114–4123. doi: 10.1128/AAC.00778-06
PubMed Abstract | Crossref Full Text | Google Scholar
Jiang, L., Liang, Y., Yao, W., Ai, J., Wang, X., Zhao, Z., et al. (2019). Molecular epidemiology and genetic characterisation of carbapenem-resistant Acinetobacter baumannii isolates from Guangdong Province, South China. J. Glob. Antimicrob. Resist. 17, 84–89. doi: 10.1016/j.jgar.2018.11.002
PubMed Abstract | Crossref Full Text | Google Scholar
Jolley, K. A., Bray, J. E., and Maiden, M. C. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
PubMed Abstract | Crossref Full Text | Google Scholar
Krizova, L., Poirel, L., Nordmann, P., and Nemec, A. (2013). TEM-1 beta-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother. 68, 2786–2791. doi: 10.1093/jac/dkt275
PubMed Abstract | Crossref Full Text | Google Scholar
Kuo, S.-C., Lee, Y.-T., Lauderdale, T.-L. Y., Huang, W.-C., Chuang, M.-F., Chen, C.-P., et al. (2015). Contribution of Acinetobacter-derived cephalosporinase-30 to sulbactam resistance in Acinetobacter baumannii. Front. Microbiol. 6:231. doi: 10.3389/fmicb.2015.00231
PubMed Abstract | Crossref Full Text | Google Scholar
Lashinsky, J. N., Henig, O., Pogue, J. M., and Kaye, K. S. (2017). Minocycline for the treatment of multidrug and extensively drug-resistant A. baumannii: a review. Infect. Dis. Ther. 6, 199–211. doi: 10.1007/s40121-017-0153-2
PubMed Abstract | Crossref Full Text | Google Scholar
Li, S., Duan, X., Peng, Y., and Rui, Y. (2019). Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in southern China. BMC Infect. Dis. 19:900. doi: 10.1186/s12879-019-4423-3
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, C., Chen, K., Wu, Y., Huang, L., Fang, Y., Lu, J., et al. (2022). Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg. Microbes Infect. 11, 1730–1741. doi: 10.1080/22221751.2022.2093134
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, L. L., Ji, S. J., Ruan, Z., Fu, Y., Fu, Y. Q., Wang, Y. F., et al. (2015). Dissemination of blaOXA-23 in Acinetobacter spp. in China: main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob. Agents Chemother. 59, 1998–2005. doi: 10.1128/AAC.04574-14
留言 (0)