1Department of Biology, Faculty of Sciences, University of Hail, Hail, Kingdom of Saudi Arabia
2Department of Biology and Chemistry, Faculty of Education, University of Gadarif, El-Gadarif, Sudan
3 Institute of Genetics, Technical University of Dresden, Dresden, Germany
4 Department of Research and training, Research and training station, King Faisal University, Al-Ahsa, Saudi Arabia.
5 Department of Microbiology,MHES College of Sciences and Technology ,Affiliated to the University of Calicut, Calicut, Kerala, India. vajidnv@gmail.com. Orcid : 0000-0002-5194-9051
Corresponding author email: abuelhadi@hotmail.com
Article Publishing HistoryReceived:
Accepted After Revision:
ABSTRACT:Although Gadarif State is an important agricultural area in Sudan, studies must be conducted on rhizobia’s genetic or molecular diversity associated with economically important legumes. Therefore, this study was undertaken to isolate rhizobia related to groundnut (Arachis hypogea), Bambara groundnut (Vigna subterranean), and Cowpea (Vigna unguiculata) in different localities in Gadarif State and study their phylogenetic relationships to make the genetic information of the indigenous rhizobia available and establish a molecular database for monitoring future climate change impact on their diversity. Nodules were collected from 11 localities of the Gadarif state, rhizobia were isolated, DNA was extracted, 16SrRNA, recA, glnII, nifH, and nodA genes were amplified, and data were analyzed.
The results showed that all isolates obtained from six localities were found to be fast-growing. Isolates obtained from groundnut nodules are Rhizobium sp._Haw1 and Rhizobium sp._G6-11, which are found to be related to Rhizobium leguminosarum. These two strains were found to be associated with Rhizobium etli when BLASTN analyzed the sequences. At the same time, Rhizobium sp._UoG27, which was isolated from the same plant, was found to be related to Rhizobium tropici. There is evidence of new species in Rhizobium sp._UoG30, Rhizobium sp._Sab13 (isolated from Bambara groundnut), Rhizobium sp._Umk34 (isolated from Cowpea), and Rhizobium sp._Taw3 (isolated from groundnut). No symbiotic genes (nifH and nod) were found in all isolates except the strain isolated from Alfalfa grown in Gezira state (Central Sudan). The study concluded that fast-growing rhizobia are dominant in Gadarif state soils, characterized by genetic instability, and may play roles other than nitrogen-fixing.
KEYWORDS:Phylogeny, Rhizobium, Isolates, Cowpea, Groundnut, Bambara Groundnut
Download this article as: Copy the following to cite this article:Suleiman A. M. E, Idris A, Gottfert M, Salih Z. A, Veettil V. N. Molecular Biodiversity of Rhizobia Isolated from Root Nodules of Some Economical Important Legumes in Gadarif State – Sudan. Biosc.Biotech.Res.Comm. 2024;17(1).
Copy the following to cite this URL:Suleiman A.M.E, Idris A, Gottfert M, Salih Z.A, Veettil V.N. Molecular Biodiversity of Rhizobia Isolated from Root Nodules of Some Economical Important Legumes in Gadarif State – Sudan. Biosc.Biotech.Res.Comm. 2023;17(1). Available from: <a href=”https://shorturl.at/buILM“>https://shorturl.at/buILM</a>
INTRODUCTION
For decades, many crop inoculation trials were done in the world to increase legume production and soil fertility at the same time. Many of these trials showed no significant increase in production. The failure may be due to the lack of genetic information about the bacteria used as inoculants, which are supposed to fix atmospheric nitrogen in association with legumes. Nitrogen is the most limiting nutrient for plant growth, including leguminous (Howieson and Committee, 2020; Simon et al., 2014). It is essential in plant cells for synthesizing enzymes, proteins, chlorophyll, DNA and RNA, thus essential for plant growth and production of food and feed, (Fahde et al., 2023).
Many times, on-farm activities, such as adjusting crop rotations, boosting the production of legumes in main crops and catch crops, and returning the byproducts to the fields, can fix inadequate N and humus balances. However, as a result, the nutrient cycles may become even more open because of the widespread insufficiency of levels of basic nutrients in the soil that are available to plants. Therefore, deficiencies in the fundamental nutrients P, K, Mg, and S are typically needed for external supplementation by fertilization with organic fertilizers and, in some circumstances, also with mineral fertilizers (Kolbe, 2022).
Rhizobia are Gram-negative bacteria (Proteobacteria) that live in the root nodules of legume plants. They are renowned for being able to fix nitrogen for their hosts’ legume species in return for carbon. Members of the family proteobacteria and the genera Rhizobium, Bradyrhizobium, Ensifer, Phyllobacterium, Mesorhizobium, Devosia, Allorhizobium, Azorhizobium, and Microvirga make up the majority of the bacteria that fix nitrogen in the root nodules of leguminous plants. Rhizobia, or members of the Burkholderiaceae family of proteobacteria, also nodulate legumes (Mukhtar et al., 2020).
According to Vanlauwe et al. (2019), the common bean (Phaseolus vulagaris L.), soybean (Glycine max), pigeon pea (Cajanuscajan), broad bean (Viciafaba), chickpea (C. arietinum), and cowpea (Vigna unguiculata L. Walp.) are the most important legumes in Africa. In the majority of African nations, subsistence farmers grow legumes primarily for food, however any surplus can be sold to generate revenue (Kawaka et al., 2014). Farmers in rural Africa cannot afford nitrogenous fertilizers because of the extreme poverty there; as a result, nitrogen requirements for cultivation are mostly met by native rhizobia. Low legume yields with those isolates have been consistently recorded in East Kenya, albeit certain native isolates there are ineffective at fixing nitrogen (West Africa (Binde et al., 2009), South Africa, as well (Biro et al., 2010 Abubakar, and Yusuf, 2016).
Commercial rhizobia inoculants have had some success in some areas (FAO, 2015), however multiple studies also show that they were ineffective in many agricultural areas in terms of promoting plant growth and final production. [Lack of successful adaptation of the isolates to the local soil conditions may be the cause of the commercial rhizobia’s failure in African farms. The majority of formulations (Kawaka et al., 2014) use strains that have been obtained from different continents that may not be well enough acclimated to the local environment. Therefore, it is essential to identify, study, and employ the rhizobial strains in the local soils as commercial inoculants in certain areas of Africa. However, formulations including local elite isolates from the specific localities must be developed and commercialized.
The diversity and biotechnological potential of symbiotic bacteria are high in tropical soils. Nevertheless, tremendous strains’ phylogenetic relationships are still poorly understood (Biro et al., 2010). In Sudan, many inoculation trials were achieved, and some of these trials showed that some local isolates have a potential effect on nitrogen fixation. However, few studies concentrate on the genetic characterization of the microbes associated with legumes and responsible for nitrogen fixation.
Gadarif state is located in the eastern part of Sudan; it consists of 12 localities. The people’s main job is agriculture, one of the most important agricultural areas in Sudan (Food Net of Sudan). Legumes like groundnut (Arachis hypogaea), Bambara groundnut (Vigna subterranean), and Cowpea (Vigna unguiculata) are grown and used for direct local consumption, and the shoot system residuals are used for animal feeds. Groundnut is also used in oil production and groundnut cake for animal feeding after processing. Despite this, genetic information on bacteria associated with leguminous plants is scarce in the Gadarif State. Although the soil’s organic matter and nitrogen content are low in the Gadarif State (FAO, 2015), it necessitates looking for indigenous efficient nitrogen-fixing bacteria to be used in inoculation.
The aims of this study are to: Investigate the molecular differences of rhizobia associated with groundnut, Bambara groundnut, and Cowpea in different localities in Gadarif State. Study the phylogenetic relationship of the isolates associated with the same plants and obtained from different sites. Contribute to the availability of genetic information on the indigenous rhizobia to help in efficient inoculants preparation for restoring and conserving soil fertility. Establish a molecular database to be used as a baseline to monitor future climate change’s impact on the molecular biodiversity of rhizobia in Gadarif State.
MATERIALS AND METHODS
Study area: The Gadarif State is located in eastern Sudan between Latitudes 12º N and 13º N and 33º E and 37º E. It covers a total area of approximately 78,000 km². The annual rainfall in the northern part is less than 500 mm. The mean monthly temperature ranges from 26º – 32º C, while the mean maximum temperatures rise to 41º C. Soils are heavy dark-cracking clays; the clay content is very high, 70 % to 80 %. The soil’s organic matter and nitrogen content are low, but as there is no deficiency of other plant nutrients, the soil is moderately fertile (FAO, 2015).
The state has 12 localities: the Gadarif, Middle locality, Eastern Galabat, Western Galabat, Alrahad, Alfashaga, Almafaza, Albotana, Alfaw, Galaalnahal, Algoraesha, and Basonda. Agriculture is the main activity in the state; the total agricultural area is 8602600 acres. It contributes 54.8% of the state’s gross domestic product (GDP).
The main crops are maize, sesame, sorghum, and sunflower, which grow in rain-fed areas. Groundnut was grown in irrigated regions on about 72000 acres in 2014, and the area is increasing (Vincent, 1970); sometimes, it is grown in rain-fed areas. Besides groundnut, leguminous crops like Bambara groundnut and Cowpea are grown in minimal areas for local consumption.
Collecting nodules: Nodules were collected from different localities of the Gadarif State in the autumn (September 2016). The plant roots were carefully removed from the soil, as nodules will be dislodged easily if the plant is pulled from the soil. The nodules were stored and preserved in screw-capped plastic tubes containing silica gel, with a cotton plug separating nodules from the desiccant in the bottom; the tube was marked with a permanent marker to record the location of the collection sites (Howieson and Committee, 2020).
Bacteria isolation: Bacteria were isolated in the Biofertilization lab, department of Biofertilization, National Center for Research, Sudan. Collected nodules were washed with sterile water, and surface sterilization was done using 70% ethanol and 20 times diluted sodium hypochlorite solution and repeatedly washed with sterile water. After surface sterilization, the nodules were crushed. The resulting suspension was streaked onto yeast extract mannitol agar (YEMA) with or without Congo Red or Bromothymol blue at pH 6.8. The medium contains (g / l): mannitol, 10; K₂HPO₄, 0.5; MgSO₄.7H₂O, 0.2; NaCl, 0.4; yeast extract, 1; agar (Sulieman et al., 2022).
DNA extraction: A colony of bacteria was grown in AG broth medium in an incubator shaker (150 rev/minutes) at 28°C for two days. Centrifugation was done to collect about 20 ml of the bacterial culture. Bacteria were resuspended in 300 µl TE buffer after washing the bacterial biomass once with TE buffer (10 mM tris, one mM EDTA, pH 8). After that, 100 µl of 5% SDS (Sodium dodecyl sulfate) and 100 µl pronase E (2.5 mg/ml in TE buffer pre-incubated for 90 minutes at 37°C) were added and mixed. The resulting solution was incubated overnight, and the DNA was sheared thoroughly using a syringe. The DNA was purified by two extractions with 300 µl of Tris-buffered phenol and one extraction with methylene chloride. DNA obtained was precipitated with 2.5 volumes of ethanol (Wekesa et al., 2022).
Amplification of the different genes: 16SrRNA: To amplify 16SrRNA, PCR reaction was set with the following: 5 µl of 10x dream Taq buffer, 1 µl dNTPs, 1 µl forward primer (16Sa 5-cgctggcggcaggcttaaca-3), 1 µl reverse primer (16Sb 5-ccagccgcaggttcccct-3), 1 µl template DNA, 0.5 µl dream taq polymerase, and 40.5 µl double distilled water total volume of 50 µl. To obtain full-length 16SrRNA, it was amplified again with the same reaction using forward primer (16SLoa 5-aacgcattaaacattccgcctgg-3) and reverse primer (16Slob 5-ttaatcttgcgaccgtactcc-3). PCR conditions were initial denaturation at 95°C for 5 minutes, 30 cycles of denaturation at 95°C for 30 seconds, annealing at 58°C for the 30s, extension at 72°C for 1.5 minutes, and final extension at 72°C for 10 minutes.
recA: PCR reaction for recA amplification contains 5 µl of 10x dream Taq buffer, 1 µl dNTPs, 1 µl forward primer (recA-a 5- gacgaccttgacgcgsgtctgrttg-3 for all strains and MrecAAI2f 5-ccgaacatgacgccgatcttcatgc-3 for strain Gez1), 1 µl reverse primer (recA-b 5- aaatcggtggayaaaagcaargc-3 for all strains and MrecAAI1r 5-tgtatcatggctcagaattctttgc-3 for strain Gez1), 1 µl template DNA, 0.5 µl dream taq polymerase, and 40. 5 µl double distilled water total volume of 50 µl. PCR conditions were: initial denaturation at 95°C for 3 minutes, 30 cycles of denaturation at 95°C for 30 seconds, annealing at 53°C for the 30s, extension at 72°C for 45 seconds, and final extension at 72°C for 5 minutes. \
glnII: PCR reaction for glnII amplification contains 5 µl of 10x dream Taq buffer, 1 µl dNTPs, 1 µl forward primer ( glnIIetF 5 atgacaaaatataagctcgagtatatttggc-3 for strains Haw1, G6-11, UoG27, and Gez; glnIIAbdF2 5-atcaaccacgaaggcatcaacg-3 for strain Taw3; glnII3for 5-tacaaggacggycgcccgctcggcttcc-3 for strains Sab13, UoG30 and Umk34), 1 µl reverse primer (glnIIetR 5-tcgatgtcgatgccgtatttttcggtcag-3 for strains Haw1, G6-11, UoG27 and Gez; glnIIAbdR 5-gtaggagaacttgttccacgg-3 for strain Taw3; glnII4rev 5- cgcggtctcgtgcttgccgg-3 for strains Sab13, UoG30, and Umk34), 1 µl template DNA, 0.25 µl dream taq polymerase, and 40.75 µl double distilled water total volume of 50 µl. PCR conditions were: initial denaturation at 95°C for 5 minutes, 30 cycles of denaturation at 95°C for 30 seconds, annealing at 58°C for the 30s, extension at 72°C for 1.5 minutes, and final extension at 72°C for 10 minutes. To amplify the strain Umk34 glnII gene PCR conditions were initial denaturation at 95°C for 30 seconds, 25 cycles of denaturation at 95°C for 1 minute, annealing at 53°C for 30 seconds, extension at 72°C for 45 seconds, and final extension at 72°C for 5 minutes.
For other strains, gradients PCR was done as follows: initial denaturation at 95°C for 3 minutes, 25 cycles of denaturation at 95°C for minute, annealing (6 cycles at 60 – 55°C for 30 seconds for strains Hw1, G6-11, and UoG27; 6 cycles at 58 – 50℃ for 30 seconds for strains Sab13 and UoG30), extension at 72°C for 30 seconds and final extension at 72°C for 30 seconds.
nifH: PCR reactions for nifH contain 5 µl of 10x dream Taq buffer, 1 µl dNTPs, 1 µl forward primer (MnifHAI1f 5- atcggcaagtccaccacctcycaaa-3), 1 µl reverse primer (MnifHAI2r 5- ctccatggtratyggggtcgggatg-3), 1 µl template DNA, 0.5 µl dream Taq polymerase, 1.5 µl DMSO and 39.5 µl double distilled water total volume of 50 µl. PCR conditions were: initial denaturation at 95°C for 5 minutes, 25 cycles of denaturation at 95°C for one minute, annealing at 55°C for 1 minute, extension at 72°C for 45 seconds, and final extension at 72°C for 5 minutes.
nodA: PCR reaction for nodA contained 5 µl of 10x dream Taq buffer, 1 µl dNTPs, 1 µl forward primer (nodA-Sin-F 5- tgtccttaaamgtgcagtggaag-3 for strain 37), 1 µl reverse primer (nodA-Sin-R 5- caatgtacctggcggccattcgt-3 for strain 37), 1 µl template DNA, 0.25 µl dream taq polymerase, and 40.75 µl double distilled water total volume of 50 µl. For amplification, gradient PCR was done as follows: initial denaturation at 95°C for 3 minutes, 25 cycles of denaturation at 95°C for 1 minute, annealing (6 cycles at 58 – 53 ℃ and 25 cycles at 52 ℃ for), extension at 72°C for 30 seconds and final extension at 72°C for 30 seconds.
All molecular characterization experiments were done in the Molecular Genetic lab, Institute of Genetics, Faculty of Science, TU Dresden, Germany (Table 1 and 2).
PCR products were purified and sequenced, and then the sequences were analyzed by the algorithm BLASTN to identify similarities.
Nucleotide Accession Numbers
Table 1. Different gene sequences accession numbers
GenesStrain
16SrRNA recA glnII nodA Rhizobium sp Haw1 MN211542 MN218348 MN218340 – Rhizobium sp Taw3 MN211543 MN218349 MN218341 – Rhizobium sp G6-11 MN211544 MN218350 MN218342 – Rhizobium sp Sab13 MN211545 MN218351 MN218343 – Rhizobium sp UoG27 MN211546 MN218352 MN218344 – Rhizobium sp UoG30 MN211547 MN218353 MN218345 – Rhizobium sp Umk34 MN211548 MN218354 MN218346 – Rhizobium sp Gez1 MN211549 MN218355 MN218347 MN218356
RESULTS
Table 2. GC% of the different rhizobia isolated from different legumes
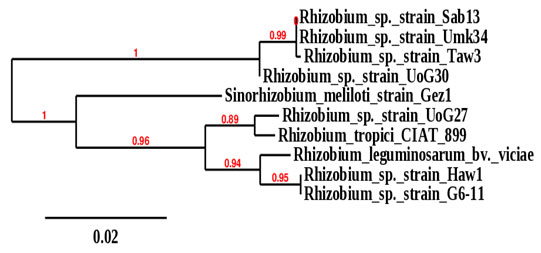
recA glnII 16SrRNA Plants Isolates 61 62 56 Groundnut Rhizobium sp._Haw1 59 59 Groundnut Rhizobium sp._Taw3 61 62 56 Groundnut Rhizobium sp._G6-11 61 59 55 Bambara groundnut Rhizobium sp._Sab13 61 61 55 Groundnut Rhizobium sp._UoG27 61 58 55 Bambara groundnut Rhizobium sp._UoG30 61 58 55 Cowpea Rhizobium sp._Umk34 61 63 55 Alfalfa SinorhizobiummelilotiAccording to the 16srRNA phylogeny tree, the isolates obtained from groundnut, Bambara groundnut, and cowpea root nodules were clustered on two main phylogeny branches or main well-defined clusters (Figure 2). One branch was divided into two subclusters comprised of rhizobia isolated from nodules of the three plants with bootstrap support of 99%. These include Rhizobium sp._Sab13, which was isolated from Bambara groundnut grown in Sabarna (Western Galabat locality), Rhizobium sp._Umk34, which was isolated from nodules of Cowpea grown Umkhareet (Basonda locality), and Rhizobium sp._Taw3 isolated from nodules of groundnut grown in Tawareet (Eastern Galabat locality). The second subcluster included Rhizobium sp._UoG30, isolated from Bambara groundnut grown in the Gadarif locality.
The second branch was also divided into two subclusters with bootstrap support of 96%; one subcluster comprised only Sinorhizobiummeliloti isolated from nodules of Alfalfa grown in Gazira state (Central Sudan). The second subcluster branched to two more subclusters; the first contains Rhizobium sp._UoG27 isolated from groundnut grown in Gadarif locality, which was clustered with Rhizobium tropici _CIAT_899 (sequences obtained from the gene bank database). The second subcluster contains Rhizobium sp._Haw1 isolated from root nodules of groundnut grown in Al-Hawata (Al-Rahad locality) and Rhizobium sp._G6-11 isolated from groundnut grown in Garia6 (Al-fashaga locality). These last two isolates were found clustered with Rhizobium leguminosarum_bv._viciae, whose sequences were also obtained from the gene bank database (Figure 1).
Figure 1: Phylogeny tree analysis of 16srRNA of Rhizobia
isolated from different legumes.
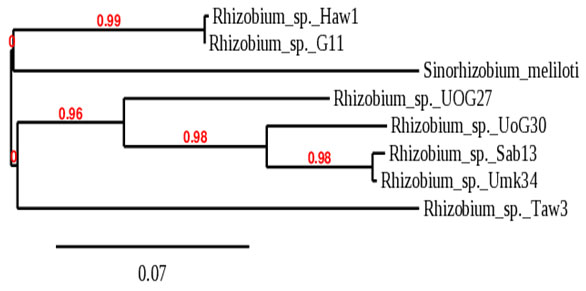
Like16srRNA phylogeny tree, the phylogeny tree of the recA gene also clustered the different isolates into two main groups with bootstrap-support of 99%, and group one branched to two subclusters, one contains Rhizobium sp._Haw1 and Rhizobium sp._G6-11 which were clustered with Rhizobium leguminosarum_bv._viciae in 16srRNA phylogeny tree. The second subcluster contains Sinorhizobiummeliloti only. Group two also branched into two subclusters with bootstrap support of 96%, the first subcluster represented by Rhizobium sp._UoG27. The second subcluster branched to two additional subclusters with bootstrap support of 98% containing Rhizobium sp._UoG30 as one cluster, while another subcluster contains Rhizobium sp._Sab13 and Rhizobium sp._Umk34. The second subcluster of group two contains only Rhizobium sp._Taw3, as shown in (Figure 2).
Figure 2: Phylogeny tree analysis of recA of Rhizobia isolated
from different legumes
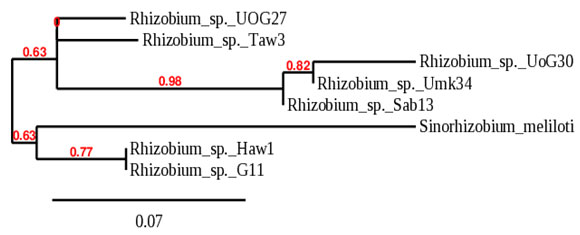
The phylogeny tree built with glnII split the isolates into two large groups with bootstrap support of 63%. The first group comprised three subgroups, subgroup (I) included Rhizobium sp._UoG27 only, subgroup (II) included Rhizobium sp._Taw3 only, both isolated from groundnut, and subgroup (III) included isolates branched to two subclusters with bootstrap support of 82%; one subcluster contains two isolates, Rhizobium sp._UoG30, and Rhizobium sp._Umk34. The second subcluster in group (III) contains Rhizobium sp._Sab13 only. The second group of the phylogeny tree built with glnII comprised two subgroups with bootstrap support of 77%, the first subgroup included Sinorhizobium meliloti only, and the second subgroup included Rhizobium sp._Haw1 and Rhizobium sp._G6-11 as illustrated in (Fig. 3).
Figure 3: Phylogeny tree analysis of glnII of Rhizobia isolated
from different legumes
The phylogeny tree of 16SrRNA, recA, and glnII confirmed that Rhizobium sp._Haw1 and Rhizobium sp._G6-11 are the same. However, they were isolated from nodules of groundnut from different localities (Al-Rahad and Al-Fashaga), located in the Western and Eastern parts of the Gadarif State, respectively. Rhizobium sp._Sab13 and Rhizobium sp._Umk34 were also found the same according to 16SrRNA and recA phylogeny trees despite their different hosts (Bambara groundnut and Cowpea, respectively).
However, the glnII phylogeny tree confirmed that they belong to different species. Other isolates related to these last two isolates were Rhizobium sp._Taw3 according to 16SrRNA and Rhizobium sp._UoG30 according to to16SrRNA, recA, and glnII phylogeny trees which were isolated from groundnut and Bambara groundnut, respectively. Accordingly, these results indicate that Rhizobium sp._Haw1 and Rhizobium sp._G6-11 are related to Rhizobium leguminosarum, although BLASTN analysis showed that they are between Rhizobium etli and Rhizobium sp. N324 depending on the gene sequence analyzed. While Rhizobium sp._UoG27 is Rhizobium tropici typically used as BLASTN analysis.
On the other hand, Rhizobium sp._UoG30, Rhizobium sp._Sab13 (isolated from Bambara groundnut), and Rhizobium sp._Umk34 (isolated from Cowpea) may be new species as supported by BLASTN analysis which revealed that they are related to Rhizobium sp. IRBG74 and found in one subcluster with some differences in the phylogenies built with the three genes analyzed in this study.
In addition, Rhizobium sp._Taw3 (isolated groundnut) may be a new species because it was clustered with Rhizobium sp._Sab13 and Rhizobium sp._Umk34 in 16SrRNA phylogeny; it appears in a separate group in recA and gene phylogenies. In addition, it is classified between Rhizobium etli and Rhizobium sp. S41in BLASTN analysis. We did not find 16SrRNA, recA, and glnII sequences in the gene bank for fast-growing rhizobia isolated from Bambara groundnut and Cowpea to use in a phylogeny tree to estimate the relationship between already sequenced genes and Rhizobium sp._UoG30, Rhizobium sp._Sab13, Rhizobium sp._Umk34, and Rhizobium sp._Taw3.
DISCUSSION
The isolates obtained from the Gadarif State were isolated from different localities with different environmental conditions. Therefore, they can be used to solve poor nitrogen fixation problems, look for strains well-adapted for stresses, use in legume domestication programs, for biodiversity studies, or genetic studies of nitrogen fixation (Howieson and Committee, 2020; Lindström and Mousavi, 2020; Dai et al., 2012). The study of these isolates also can enrich the understanding of the phylogenetic relationships of the different strains, as they are still poorly understood (Biro et al., 2010).
In this study, Rhizobium sp._Haw1 and Rhizobium sp._G6-11 were the same despite their different site directions (Western and Eastern parts of Gadarif State), respectively. Moreover, Rhizobium sp._Sab13 and Rhizobium sp._Umk34 were found the same despite their different hosts (Bambara groundnut and Cowpea, respectively), although they appeared as different species in the glnII phylogeny tree. These results are supported
partially by other findings, which show that the symbiotic association between rhizobia and legumes may be subject to environmental factors, interactions among rhizobia, legumes, and biogeography. It was found that rhizobia can form nodules in legumes in distinctive geographic regions, and the same rhizobia may form nodules in different legume species (Grönemeyer et al., 2014). Therefore, we expect that each of the isolates mentioned above, which clustered in the same groups, may associate symbiotically with different hosts in different sites due to the minor influence of the plant origin on the relatedness of the isolates (Flores et al., 2005).
The Phylogeny trees built with 16SrRNA and recA were found to be nearly the same, with some exceptions. The 16S rRNA sequence analysis agreement with recA was reported before (Grönemeyer et al., 2014). However, glnII phylogeny shows inconsistency with 16SrRNA and recA phylogenies, which agrees with the previous finding that “discordant phylogenies within different loci of rhizobia can result in different phylogenetic tree topologies for rhizobia species.
The results in this study also revealed that groundnut is associated with different Rhizobium species because the isolates were found clustered with Rhizobium leguminosarum and Rhizobium tropici simultaneously sequence analyses of BLASTN indicate that Rhizobium etli.Rhizobium leguminosarum and Rhizobium etli are “the symbionts of the common bean plant Phaseolus vulgaris” (Angelini et al., 2011). This supports that “groundnut nodules are a reservoir for different rhizobial lineages (Roughley, 1970).
This may be an advantage in inoculants manufacturing in which the inoculants produced from groundnut Rhizobia can be helpful. However, the performance of rhizobia under conditions dissimilar to the original habitat is poor (Law et al., 2007; Botha et al., 2004), and their effectiveness depends on the interaction of environmental factors (Pule-Meulenberg et al., 2010) and host plant variety or genotypes (Keyser et al., 2002 Martínez-Hidalgo and Hirsch, 2017).
Therefore, inoculants of effective indigenous rhizobia adapted to local conditions may perform better (Flores et al., 2005). However, all isolates obtained from groundnut, Bambara groundnut, and Cowpea in this study were found to induce nodules in all three legumes, even in cross-inoculation performed under laboratory conditions directly after the isolates were obtained. In addition, the shoot dry weight of the inoculated plants showed a significant increase (data not shown). After about one year, all isolates failed to form nodules even in their original hosts.
To interpret these findings, we exclude the assumption that these isolates are non-symbionts, and we assume that they lost their symbiotic genes may be due to the storage conditions and repeated sub-culturing, which means that these isolates are genetically unstable. However, maintaining genetic stability is essential in determining the isolate’s validity as an inoculant (Zhang et al., 2008).
Even though symbiotic genes were not found in our isolates, the role of rhizobia is not restricted to nitrogen fixation; they also provide plants with plant growth-promoting factors like hormones, which we expect our isolates to play also. Besides that, we isolated rhizobia from six localities of the eleven localities of the Gadarif State surveyed. These findings necessitate conducting more studies to obtain isolates from other localities, searching for the reasons leading to the loss of nitrogen-fixing (nifH) and nodulation (nod) genes, and studying these isolates’ roles may play in promoting legumes and growth.
For Cowpea, we obtained only one isolate (Rhizobium sp._Umk34), which was found to be related to Rhizobium sp._UoG30, Rhizobium sp._Sab13 isolated from Bambara groundnut. These isolates may be due to Cowpea and Bambara groundnut affiliation to the same plant genus (Vigna). We assumed these isolates were new species because they clustered in one group in the phylogeny tree built with all three genes. We did not find sequences for 16SrRNA, recA, and glnII deposited in the gene bank related to Cowpea rhizobia. Despite its importance as a legume crop, cowpea rhizobia have yet to be well characterized (Ibny et al., 2019).
More than that, recently, it was reported that different isolates obtained from nodules of Bambara groundnut from South Africa, Ghana, and Mali were found to be closely related to different species of Bradyrhizobium (Herridge et al., 2002; Keyser et al., 2002), of their isolates belong to fast-growing Rhizobium. In addition, BLASTN analyses of these genes classified them between different species. More than that, rhizobia isolated from cowpea showed unstable relationships. The relationship between Cowpea and Bambara groundnut is a normal phenomenon because they belong to the same plant genus (Vigna) as mentioned above. Many studies reported that Cowpea is considered promiscuous, which is modulated by a wide range of rhizobia (Toolarood et al., 2012 Ibny et al., 2019).
Besides groundnut, Cowpea, and Bambara groundnut isolates, we isolated Sinorhizobium meliloti from nodules of Alfalfa grown in the Gezira state, which was identified by BLASTN analyses of 16SrRNA, recA, glnII, nifH, and Noda genes. It was found in a separate branch in the phylogeny tree built with 16SrRNA, recA, glnII. No relationship exists between this isolated and the others isolated from the Gadarif State. Previously, “Sinorhizobium meliloti is the dominant genus in alfalfa nodules with a relatively high genetic diversity” (Aloo et al., 2022).
In contrast to the other isolates in this study, symbiotic genes were found in the isolate obtained from Alfalfa nodules. However, it was maintained in the same storage conditions as the isolates obtained from the other three plants. This indicates that the isolate obtained from Alfalfa is genetically stable and supports our assumption that the other isolates are characterized by genetic instability.
In all isolates obtained in these fast-growing rhizobia, no slow-growing Bradyrhizobium was isolated, although many studies reported fast-growing Rhizobium and slow-growing Bradyrhizobium from nodules of Cowpea (Flores et al., 2005; Ibny et al., 2019). The advantages of fast-growing rhizobia are that they require a shorter time in inoculants production, contamination occurs at a lower rate during the industrial process, more accessible establishment in the soil, and gene manipulation is easier. However, fast-growing rhizobia lacks competitiveness against Bradyrhizobium, which limits their recommendation to use as a commercial inoculant (Kolbe, 2022). Therefore, more studies are required to look for slow-growing rhizobia and its advantages over the fast-growing in the Gadarif State soil.
Finally, studying the diversity and characteristics of soil rhizobia has practical importance for ecology and agriculture. These studies help to enrich agricultural microbial genetic resources (AMiGRs) (Howieson and Committee, 2020), select effective combinations of Rhizobium-legume genotypes to increase nitrogen fixation (Grönemeyer et al., 2014), and study the indigenous rhizobia populations adapted to the local environmental conditions contribute to the general understanding of regional species abundance. In addition, they provide a basis for the formulation of a rhizobial inoculant matching local settings (Flores et al., 2005). Moreover, the “variety of rhizobia is a valuable bioresource for the exploitation of bacterial selection in attempts to find bacterial strains with desirable traits that maximize legume crop productivity” (Biro et al., 2010).
CONCLUSION
Fast-growing Rhizobium dominates the Gadarif State soils, lose symbiotic genes (unstable genetically), and is related to each according to the host and regardless of the site from which they were isolated. Different species of these fast-growing Rhizobium are associated with groundnut; evidence of new species is found in those associated with Bambara groundnut and Cowpea. Thus, more studies are required to isolate both fast-growing and slow-growing rhizobia, characterize those that seem to be new species and search for symbiotic genes and factors affecting the isolates.
Acknowledgments
This work was supported by Ministry of Higher Education and Scientific Research.
Conflict of Interest: The authors have no financial conflicts of interest to declare.
Authors contribution: A.S., A.I., M.G, V.N.V developed the concept, A.S., A.I, Z.S designed the experiment and; A.S., A.I. collected data and performed the analyses; A.S., A.I., V.N.V wrote the manuscript.
Data Availability: The data will be made available on request.
REFERENCES
Abubakar, F.J.; Yusuf, A.A. (2016). Relative efficiency and response of promiscuous soybean to rhizobia inoculant in Savanna region of Nigeria. Afr. J. Microbiol. Res,10: 1187–1193.
Aloo, BN.; Tripathi, V.;Makumba, B.A.;Mbega, E.R. (2022). Plant growth-promoting rhizobacterial biofertilizers for crop production: The past, present, and future. Front. Plant Sci, 13: 1002448.
Angelini, J.; Ibáñez, F.; Taurian, T.; Tonelli, M. L.; Valetti, L. and Fabra, A. (2011). A study on the prevalence of bacteria that occupy nodules within single peanut plants. Current microbiology, 62:1752-1759.
Binde, D.R.;Menna, P.; Bangel, E.V.; Barcellos, F.G.; et al. (2009). rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl Microbiol Biotechnol,83: 897-908.
Biro, K.;Biswajeet, P.; Manfred, B.;Franz,M.(2010).The effects of different land use types on soil compaction and infiltration rate in the dry lands vertisol of Gadarif Region, Sudan. International Research on Food Security, Natural Resource Management and Rural Development ETH Zurich.
Botha,W.J,\.; Jaftha, J.B.;Bloem, J.F.;Habig, J.H.; Law, I.J. (2004). Effect of soil Bradyrhizobia on 605 the success of soybean inoculant strain CB 1809. Microbiol Res, 159:219-231.
Dai, J.; Liu, X.; Wang, Y. (2012). Genetic diversity and phylogeny of rhizobia isolated from Caragana microphylla growing in desert soil in Ningxia, China. Genet Mol Res, 11 (3): 2683-2693.
Fahde, S.;Boughribil, S.;Sijilmassi, B.;Amri, A. (2023). Rhizobia: A Promising Source of Plant Growth-Promoting Molecules and Their Non-Legume Interactions: Examining Applications and Mechanisms. Agricult,13 (7): 1279.
FAO. (2015). AQUASTAT Country Profile – Sudan. Food and Agriculture Organization of the United Nations (FAO). Rome, Italy.
Flores, M.; Morales, L.; Avila, A.;Gonza´lez, V.; Bustos, P.;Garcı´a, D.; Mora, Y.;Guo, X.;Collado-Vides, J.;Pin˜ero, D.;Da´vila, G.; Mora, J.; Palacios, R. (2005). Diversification of DNA Sequences in the Symbiotic Genome of Rhizobium et. Jour Bacter,187 (21): 7185–7192.
Grönemeyer, J. L.; Kulkarni, A.;Berkelmann, D.;Hurek, T., & Reinhold-Hurek, B. 2014. Rhizobia indigenous to the Okavango region in Sub-Saharan Africa: diversity, adaptations, and host specificity. Applied and environmental microbiology, 80(23): 7244-7257.
Herridge, D.; Gemmell, G.; Hartley, E. (2002). Legume Inoculants and Quality Control. In: Inoculants and Nitrogen Fixation of Legumes in Vietnam edited by Herridge, D., (2002): pp 105 – 115.
Howieson, J.; Committee, G.R.P. (2020). Technical Issues Relating to Agricultural Microbial Genetic Resources (AMiGRs), including Their Characteristics, Utilization, Preservation and Distribution: Draft Information Paper.
Ibny, F. Y.; Jaiswal, S. K.; Mohammed, M. and Dakora, F.D. (2019). Symbiotic effectiveness and ecologically adaptive traits of native rhizobial symbionts of Bambara groundnut (Vigna subterranea L. Verdc.) in Africa and their relationship with phylogeny. Scient rep, 9 (1): 12666.
Kawaka, F.; Dida, M.M.;Opala, P.A.; Ombori, O.; Maingi, J.; Osoro, N.; Muthini, M.; Amoding, A.; Mukaminega, D.; Muoma, J. (2014). Symbiotic efficiency of native rhizobia nodulating common bean (P. vulagaris L.) in soils of Western Kenya. Int. Sch. Res. Not, 2014: 258497
Keyser, H.H.;Somasegaran, P.;Bohlool, B.B. (2002). Rhizobial ecology and technology. In: Herridge, D.; Gemmell, G. and Hartley, E. (2002). Legume Inoculants and Quality Control. In: Inoculants and Nitrogen Fixation of Legumes in Vietnam edited by D. Herridge, 2002: pp 105 – 115.
Kolbe, H. (2022). Comparative Analysis of Soil Fertility, Productivity, and Sustainability of Organic Farming in Central Europe—Part 2: Cultivation Systems with Different Intensities of Fertilization and Legume N2 Fixation as well as Perspectives for Future Development. Agronom,12: 2060.
Law, I.J.; Botha, W.J.;Majaule, U.C.;Phalane, F.L. (2007). Symbiotic and genomic diversity of ‘cowpea’ bradyrhizobia from soils in Botswana and South Africa. Biol FertilSoi,43:653- 663.
Lindström, K.; Mousavi, S.A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol,13: 1314–1335.
Martínez-Hidalgo, P.;& Hirsch, A. M. (2017). The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiom Jourl, 1 (2): 70-82.
Mukhtar, S.;Ann, M.; Hirsch,N.K.; Kauser, A.; Malik,E.A.; Humm,M.P.;Baochen.S.;Leah, B.;Marcel, H.; Alicia, C.; et al. (2020). Impact of Soil Salinity on the Cowpea Nodule-microbiome and the Isolation of Halotolerant PGPR Strains to Promote Plant Growth under Salinity Stress. Phytob Jour,4: 364-374.
Pule-Meulenberg, F.; Belane, A. K.; Krasova-Wade, T. and Dakora, F.D. (2010). Symbiotic functioning and bradyrhizobial biodiversity of cowpea (Vigna unguiculata L. Walp.) in Africa. BMC Microbiol, 10: 1-12.
Roughley, R.J. (1970). The influence of root temperature, Rhizobium strain, and host selection on the structure and nitrogen-fixing efficiency of the root nodules of Trifolium subterraneum. Ann Bot. (Lond.), 34: 631-646.
Simon, Z.; Kelvin, M.; Amare, G.; Patrick, A.N.;(2014). Isolation and Characterization of Nitrogen Fixing Rhizobia from Cultivated and Uncultivated Soils of Northern Tanzania. Ameri jour plascien,5:4050-4067.
Sulieman, A.E.;Abdelmalik, O.A.;Alshammari, N.A.;Naimah, A.;Alanazi,N.A.; Al-Azmi, A.A.;Hamadou, W.S.;Ebbadri, G.A.;Khamisabad, H. (2022). Molecular Biodiversity of Bacteria Isolated from Medicago sativa Rhizosphere in Haʼil
District, Saudi Arabia. Cell Molec Biol,68 (2): 1-7.
Toolarood, A.S.;Alikhani, H.A.; Salehi,G.h.;Asadi-Rahmani, H.;Khavazi, K.;Poorbabaee, A.A.;Lindström, K. (2012). Molecular diversity of rhizobia isolated from root nodules of Alfalfa evaluated by analysis of IGS and 16SrRNA. Ann. of Biol. Res,3 (5): 2058-2063.
Vanlauwe, B.;Hungria, M.;Kanampiu, F.;Giller, K.E. (2019). The role of legumes in the sustainable intensification of African smallholder agriculture: Lessons learnt and challenges for the future. Agric. Ecosyst. Environ,284: 106583.
Vincent, J. M. (1970). A manual for the practical study of the root-nodule bacteria. A manual for the practical study of the root-nodule bacteria.
Wekesa, C.; Jalloh, A.A.;Muoma, J.O.;Korir, H.;Omenge, K.M.;Maingi, J.M.;Furch, A.C.U.;Oelmüller, R. (2022). Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa. International Journal of Molecular Sciences,23 (12): 6599.
Zhang, Y.F.; Wang, E.T.; Tian, C.F.; Wang, F.Q.; Han, L.L.; Chen, W.F.; Chen, W.X. (2008). Bradyrhizobiumelkanii, Bradyrhizobiumyuanmingense, and Bradyrhizobium japonicum are the main rhizobia associated with Vigna unguiculata and Vigna radiata in the subtropical region of China. FEMS Microbiol Lett, 285: 146–154.
留言 (0)