Leptospirosis, a zoonotic disease caused by pathogenic spirochaetes of the genus Leptospira, is constantly present in some parts of the world and holds significant relevance in both veterinary and public health contexts due to its ability to cross over between humans, domestic animals, wildlife and even environment (water). Reported cases of leptospirosis are global with over one million cases annualy, leading to approximately 60,000 fatalities (Costa et al., 2015). To date, a minimum of 64 distinct Leptospira species have been validated worldwide using the average nucleotide identity (ANI) values of their genomes (Vincent et al., 2019). While rats are traditionally known as the primary reservoirs for pathogenic Leptospira species, there have been numerous reports on various vertebrate and invertebrate hosts as excreting this pathogen through their urine. Wild and domestic mammals (Arent et al., 2017; Vieira et al., 2017), livestock (Shiokawa et al., 2019; Zhang et al., 2019), amphibians (Dezzutto et al., 2017), reptiles (Rodamilans et al., 2020) and bats (Mateus et al., 2019) also appear to play significant roles in the spread of Leptospira sp. Human infections typically result from exposure to soil or water contaminated with Leptospira, mostly from the urine of reservoir animals (Adler and de la Peña Moctezuma, 2010). Detecting Leptospira through traditional growth on media can be problematic due to their slow growth, making it impractical for timely diagnoses. To address this, molecular diagnostic methods, such as the real-time PCR of the lipL32 gene, have been developed (Ferreira et al., 2014; Wu et al., 2014). PCR-based amplification of secY and ompL1 genes using species-specific primers and probes has been used to identify Leptospira species directly from clinical samples. These assays can identify common pathogenic Leptospira species when combined with a lipL32 assay, including L. borgpetersenii, L. interrogans, L. kirschneri, and Leptospira noguchii (Victoria et al., 2008). Furthermore, sequence-based typing schemes utilizing gene targets like 16S rRNA rrs2, secY, and lfb1, or adk, icdA, lipL32, lipL41, rrs2 and secY have been developed for Leptospira (Ahmed et al., 2006; Morey et al., 2006). For example, a ∼ 435-bp fragment of the secY gene shows good phylogenetic discrimination between pathogenic Leptospira species. Sequence-based methods can also be applied directly to clinical samples to determine the infecting species and genotype, as well as investigate links between human and animal Leptospira infection (Hamond et al., 2015). In Serbia, the presence of pathogenic Leptospira sp. has been documented in various animals including small wild mammals (Blagojević et al., 2019), however most of the studies in Serbia have been focused on seroprevalence and seroepidemiological detection of antibodies in samples from cats (Obrenović et al., 2014), dogs (Vojinović et al., 2022) and humans (Svirčev et al., 2009). To the best of our knowledge this is the first study to perform molecular and multilocus sequence typing analysis of Leptospira species in Serbia. Moreover, this study revealed the presence of Leptospira kirshneri serovar Mozdok in Serbia for the first time.
2 Materials and methods 2.1 Animal collectionThe research was conducted in accordance with ethical principles and was approved by the Ministry of Agriculture, Forestry and Water Management (Republic of Serbia) - Veterinary Directorate (No. 323-07-04943/2020-05/2, 29.05.2020 and 323-07-04155/2023-05/2, 16.05.2023). During 2020, 2021 and 2022, a total of 344 (186 female and 158 male) carcasses of Norway rats (Rattus norvegicus) were collected in the broad environs of Belgrade City, predominantly in their urban and suburban 163 habitats. With the aim of collecting material that would be of good quality for further analysis of the presence of bacteria in the tissues, animals were collected by trapping (snap traps with fish mixture with peanut butter and oat flakes as a bait) and carcasses were collected daily and kept at 4°C during transportation to the laboratory where they were measured, followed by necropsy, during which the kidneys were removed and kept at −20°C until further processing and analysis. The average body mass of all used animals (± SD) was 236.97 ± 99.31 g (range 30–498 g). The average body length of the individuals was 201.79 ± 40.09 cm (range 91–374 cm), while the average tail length was 168.14 ± 30.48 cm (range 80–230 cm). Carcasses were collected predominantly in their urban and suburban habitats. The largest number of individuals was collected after the implementation of control measures or the implementation of monitoring measures. The collected carcasses were kept in a freezer at −20°C for a short time, until further processing. During autopsy, the kidneys were separated for further analysis and the morphological data, body weight and sex of the animals were recorded.
2.2 DNA extraction, molecular detection, sequencing, and MLST analysisDNA was extracted from the kidney using the Quick-DNA MiniPrep kit (Zymo Research, USA, Cat. no. D3024) according to manufacturers’ instructions. To validate the extraction processes and all downstream steps, nuclease-free water and DNA extracted from Leptospira positive samples were used as negative and positive controls, respectively. DNA extracted from each sample was stored at −20°C until downstream use. To distinguish between pathogenic and non-pathogenic Leptospira, we performed qPCR targeting the lipL32 partial target genes. Specifically, we used primers LipL32F (5′-GGA TCC GTG TAG AAA GAA TGT CGG-3′) and LipL32R (5’-GTC ACC ATC ATC ATC ATC GTC C-3′) to amplify a 101 bp fragment of the lipL32 gene, which was detected by the probe LipL32P (6-carboxyfluorescein [FAM]-5′-ATG CCT GAC CAA ATC GCC AAA GCT GCG AAA-3’-Black Hole Quencher 1 [BHQ1]) (Wu et al., 2014). An internal control, represented by exogenous DNA added before the extraction phase, representing simultaneously the extraction and PCR amplification control (qPCR Extraction Control RED, Meridian Bioscience, UK) was also included. The qPCR was carried out in a 12 μL reaction mixture containing 3 μL of Leptospira spp. genomic DNA, 0.5 μL (concentration of 20 pmol/μL) of forward and reverse primer and probe and 5 μL (concentration of 10 pmol/μL) of FastGene 2x PROBE Universal (Nippon Genetics, Germany) and 2.5 μL of PCR water. All reactions were conducted in duplicates using a 7,500 Fast Real-Time PCR System (Applied Biosystems, ThermoFisher, USA) with the following conditions: initial denaturation at 95°C for 2 min, followed by 45 cycles of denaturation at 95°C for 20 s, and annealing/elongation at 65°C for 50 s. Each PCR test included a negative control (DNA extracted from water) and a positive control (DNA extracted Leptospira spp. positive samples). Among the positive samples obtained through qPCR, only those with threshold cycle (Ct) values lower than or equal to 30 underwent further analysis. Specifically, 27 kidney samples and 27 Leptospira DNAs were subjected to PCR using a set of primers amplifying adk, icdA, lipL32, lipL41, rrs2 and secY partial genes (Table 1) (Ahmed et al., 2006). PCR reagents and their volumes, as well as PCR cycling conditions are shown in Tables 2, 3, respectively. The PCR products were visualized by electrophoresis on a 1.5% agarose gel and examined under UV transillumination.
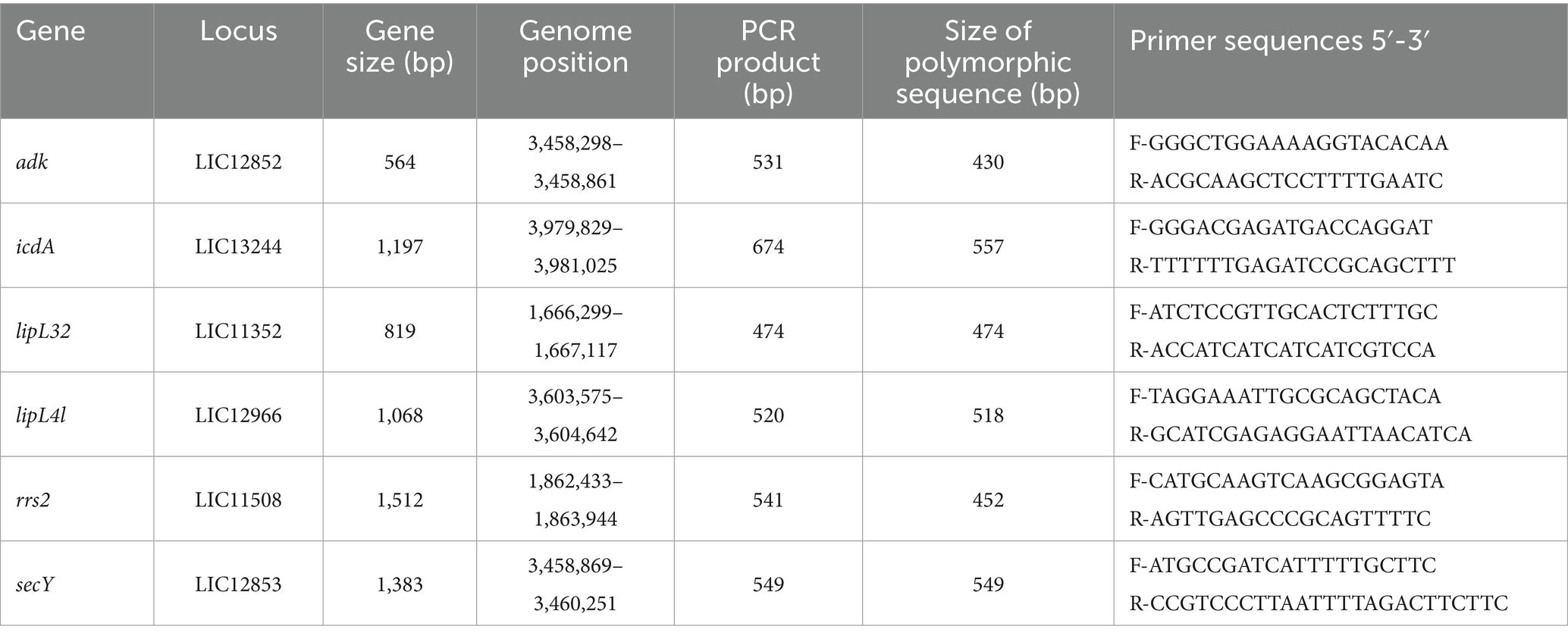
Table 1. Details of gene loci and the corresponding primer sequences used for MLST analysis.
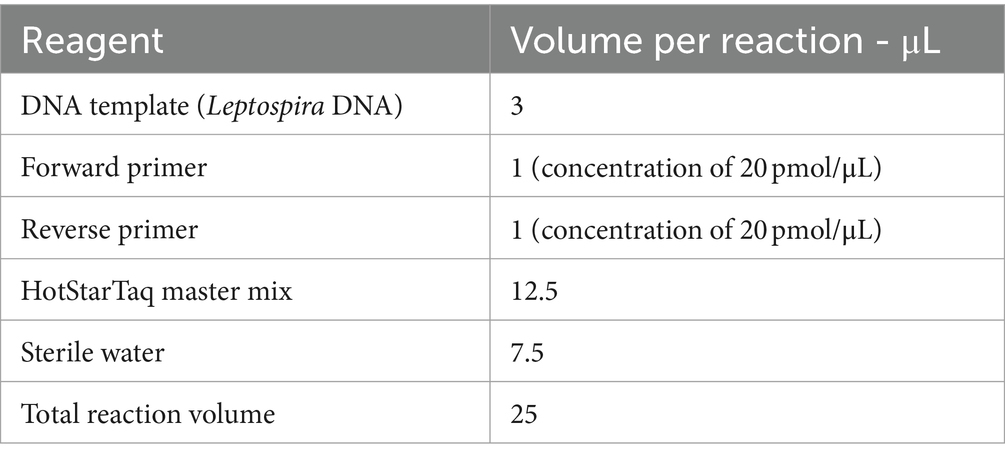
Table 2. Reagents and volumes.
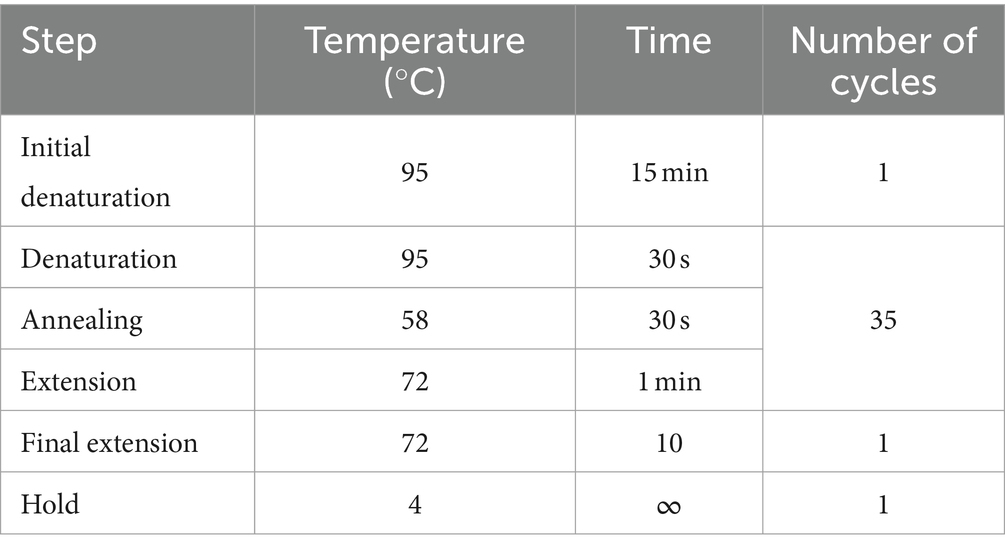
Table 3. PCR cycling conditions.
We purified the amplicons using the GeneJET PCR Purification Kit (ThermoFisher Scientific, USA, cat. no. K0702) and sent them to Macrogen Europe (Amsterdam, Netherlands) for Sanger sequencing. Sequences were analyzed and edited using the Staden package (Staden et al., 2003). Consensus sequence validation was performed against a custom Leptospira database using nucleotide blast (BLASTn) (Altschul et al., 1990). Each allele and the allelic profiles (adk-icdA-lipL32-lipL41-rrs2-secY) were submitted to the Leptospira database (Jolley et al., 2018) for ST assignment. Sequence similarity of our samples was performed with a custom reference database using Biopython (Cock et al., 2009). All sequences were submitted to NCBI’s GenBank under the following accession numbers: OR920389 - OR920523 for adk, icdA, lipL32, LipL41 and secY, while for rrs2 OR912477-OR912503.
2.3 Statistical analysisMean prevalence and confidence intervals (95% CI) for Leptospira spp. were determined using the Clopper and Pearson method.
3 ResultsAll 344 samples were analyzed for the presence of pathogenic Leptospira species. In kidney tissues, Leptospira spp. was detected in a total of 103 out of 344 individuals (29.94, 95% CI: 25.15–35.09) upon amplification by qPCR (Table 4). A total of 27 out of 103 positive samples (with Ct values between 20 and 28) were used in this study. Among all samples, the BLASTn analysis indicated that 26 sequences were affiliated with the L. interrogans, and 1 sequence exhibited the closest resemblance to the L. kirschneri (with 100% identity). The calculated sequence similarity of our samples with a cutoff value of 95% performed with Biopython was in concordance with the BLASTn results and for some of the samples it was possible to determine the serovar. For the final and definite characterization of our samples we determined the allele profile using the MLST scheme 3 from the PubMLST database. The MLST analysis yielded the following results: 11 of our samples belong to L. interrogans serovar Copenhageni, 12 to L. interrogans serovar Icterohaemorrhagiae (all belong to the serogroup Icterohaemorrhagiae) and one to L. kirschneri serovar Mozdok (serogroup Pomona). For the rest 3 of our samples, we were only able to determine the taxonomy to the level of species (L. interrogans) due to lower sequence quality.
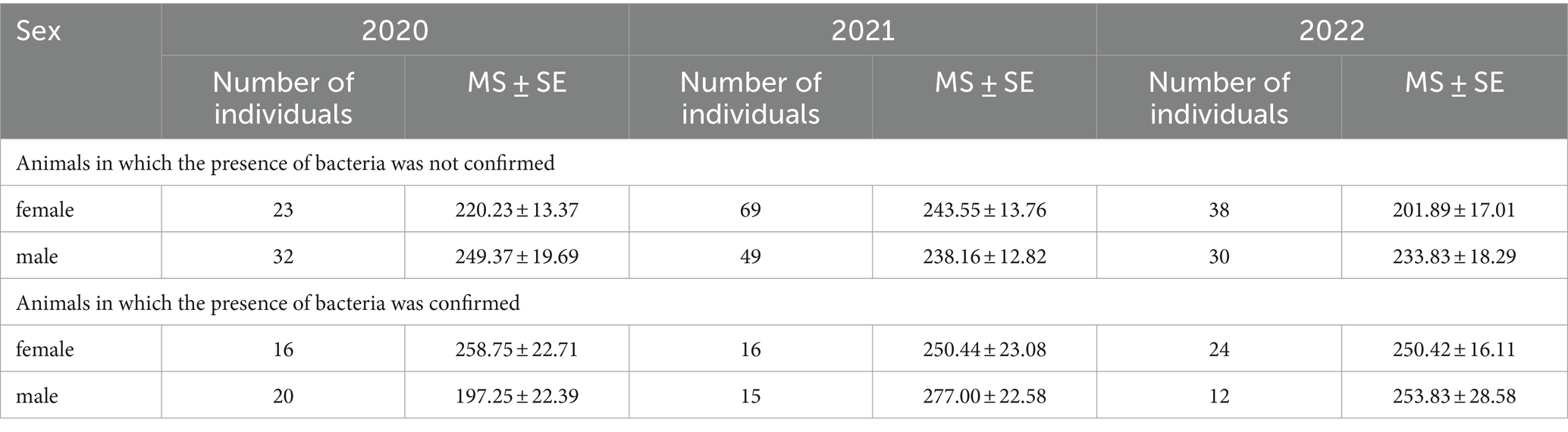
Table 4. The average weight (g) ± standard error (MS ± SE) and presence of Leptospira ssp. in Norway rat kidney tissues, collected in the period 2020–2021 in the Belgrade, Serbia.
4 DiscussionThere is a growing interest in the surveillance of Leptospira spp. hosts, and investigations into the prevalence of this pathogen in wildlife and synanthropic faunaacross Europe are on the rise and the significance of rodents as reservoirs for various Leptospira serovars has been extensively explored worldwide with various results. It is well-established that wild rats (Rattus spp.) are the principal sources of Leptospira infection, particularly in urban and peri-domestic environments (Boey et al., 2019). Rattus norvegicus is known as the primary host of L. interrogans related to the serogroup Icterohaemorrhagiae, which is responsible for the most severe forms of the disease in humans (Haake and Levett, 2014). Our study aimed to examine the circulating Leptospira strains in wild rats, utilizing qPCR for initial detection of pathogenic Leptospira and MLST analysis for molecular characterization. Our findings confirm that wild rats harbor different serovars of pathogenic Leptospira spp. which pose threat to both animal and public health, highlighting the importance of continuous monitoring the presence and diversity of these bacteria in wild animals. The identification of L. interrogans serovar Icterohaemorrhagiae and L. interrogans serovar Copenhageni aligns with studies from all over Europe: in Sicily the bacteria has been detected in stray dogs and cats (Grippi et al., 2023). In Sardinia authors have reported pathogenic Leptospira in hedgehogs, mustelids and wild rodents (Piredda et al., 2021). In Germany, researchers in one study reported that 6% of the tested animals (various small mammals) exhibited positive results for L. kirschneri and L. interrogans (Obiegala et al., 2016), while L. interrogans serovar Icterohaemorrhagiae has been reported in wild rats all over the world (Boey et al., 2019) which is not surprising given that it represents the most common serovar in animals and humans. Additionally, this study relied on the utilization of the adk, icdA, lipL32, lipL41, rrs2 and secY partial genes as a means for molecular typing and differentiating Leptospira serovars. The results obtained using these genes align with those obtained from other MLST analyses. Although Leptospirosis has been the subject of numerous studies across various geographical regions, this present investigation in Serbia marks a significant contribution to the field. Prior research in Serbia had mainly focused on seroprevalence and seroepidemiological studies (Svirčev et al., 2009; Obrenović et al., 2014; Vojinović et al., 2022). However, our study distinguishes itself as the first in Serbia to employ molecular and multilocus sequence typing analysis for Leptospira species. This unique approach has yielded in discovering the presence of L. kirshneri serovar Mozdok in Serbia. This marks the first documented occurrence of this serovar in the country. Similar reports have been documented in Croatia (Majetić et al., 2014). The comprehensive and systematic testing conducted in our study, which included various Leptospira genes, facilitated the detailed characterization of positive samples. The sequencing and BLASTn analysis unveiled a predominance of L. interrogans in our samples, reinforcing its role as a common pathogenic Leptospira species. Further analysis, including the calculation of sequence similarity and allele profiling using the PubMLST database, refined our understanding of the Leptospira strains present. Notably, our findings unveiled specific serovars, such as L. interrogans serovar Copenhageni and L. interrogans serovar Icterohaemorrhagiae, underscoring the diversity of Leptospira strains within the Belgrade region. The significance of our discovery of L. kirshneri serovar Mozdok in Serbia extends beyond the confines of our study. This novel serovar presence has potential implications for vaccine strategies and epidemiological studies in both human and veterinary epidemiology. Serovars play a crucial role in vaccine formulation, as they determine the specific Leptospira strains that the vaccine should target. Consequently, our findings serve as a starting point for a more comprehensive and continuous Leptospira sp. surveillance in order to detect new serovars and accordingly adapt management strategies (presently the present vaccine strategies in Serbia include preparations for different animals which contain L. interrogans serovar Icterohaemorrhagiae, Canicola, Copenhageni and Bratislava, L. kirshneri serovar Grippotyphosa) However, it should be noted that this finding does not imply immediate change in vaccine strategy. To date, in Serbia, this serovar has not been confirmed as the causative agent of leptospirosis, most probably due to the lack of testing in patients infected with this bacterium. On the other hand, multiple research has proved that this serovar is clinically relevant since it has been implicated in human and animal leptospirosis (Cunha et al., 2016; Bertasio et al., 2020a,b). The presence of a novel serovar implies the need further explore and determine the presence of this serovar in other animals and/or humans and perform serological tests to screen for seropositive individuals that may have come into contact with this specific bacterium. Regarding leptospiros epidemiology, the identification of L. kirshneri serovar Mozdok opens doors to a more comprehensive understanding of the disease’s distribution and dynamics in the region. The serovar’s presence highlights the complexity of Leptospira populations in Serbia and warrants further investigation into its reservoir hosts and transmission dynamics. While the detection of a single serovar in one rat does not definitively establish that species as a reservoir it should be noted that while our findings do not conclusively determine the rat as a carrier, they are supported by other research [that has identified the same strain in additional rats, suggesting a potential role as carriers (Majetić et al., 2014; Obiegala et al., 2016)]. Epidemiological studies must now consider the unique characteristics of this serovar, as it may exhibit distinct patterns of host adaptation and disease transmission. Understanding the prevalence and distribution of this serovar is crucial for developing effective control measures, both in terms of prevention and treatment. Moreover, the discovery emphasizes the importance of continued surveillance and monitoring of Leptospira diversity in the region, as new serovars may continue to emerge over time. In conclusion, our study has provided valuable insights into the presence and diversity of Leptospira species in Serbia. The discovery of L. kirshneri serovar Mozdok serves as a pivotal point for advancing management strategies and epidemiological research in the region. By adapting our approaches to the unique characteristics of this novel serovar, we can better address the challenges of leptospirosis and work toward more effective prevention and control measures for both human and veterinary health. Furthermore, the presence of L. kirshneri serovar Mozdok opens new avenues for epidemiological research in Serbia. Further research is essential to unveil the full implications of this discovery and to refine our understanding of the epidemiological landscape in Serbia.
Data availability statementThe original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/nuccore/; OR920389-OR920523, OR912477-OR912503.
Ethics statementThe animal study was approved by Ministry of Agriculture, Forestry and Water Management (Republic of Serbia) - Veterinary Directorate (No. 323-07-04943/2020-05/2, 29.05.2020 and 323-07-04155/2023-05/2, 16.05.2023). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributionsVG: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. GJ: Conceptualization, Investigation, Supervision, Writing – original draft, Writing – review & editing. SS: Conceptualization, Formal analysis, Supervision, Writing – review & editing. MZ: Investigation, Methodology, Validation, Writing – original draft. TB: Data curation, Investigation, Resources, Writing – original draft. MR: Data curation, Formal analysis, Writing – original draft. TP: Conceptualization, Investigation, Methodology, Validation, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by Ministry of Science, Technological Development and Innovation of Republic of Serbia by the Contract of implementation and funding of research work of NIV-NS in 2023, Contract No: 451-03-47/2023-01/200031.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1379021/full#supplementary-material
Footnotes ReferencesAdler, B., and de la Peña Moctezuma, A. (2010). Leptospira and Leptospirosis. Vet. Microbiol. 140, 287–296. doi: 10.1016/j.vetmic.2009.03.012
Crossref Full Text | Google Scholar
Ahmed, N., Devi, S. M., Valverde Mde, L., Vijayachari, P., Machang'u, R. S., Ellis, W. A., et al. (2006). Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 5:28. doi: 10.1186/1476-0711-5-28
PubMed Abstract | Crossref Full Text | Google Scholar
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Crossref Full Text | Google Scholar
Arent, Z., Gilmore, C., Ayanz, J. M. S.-M., Neyra, L., and García-Peña, F. J. (2017). Molecular epidemiology of Leptospira serogroup Pomona infections among wild and domestic animals in Spain. EcoHealth 14, 48–57. doi: 10.1007/s10393-017-1210-8
PubMed Abstract | Crossref Full Text | Google Scholar
Bertasio, C., Boniotti, M. B., Lucchese, L., Ceglie, L., Bellinati, L., Mazzucato, M., et al. (2020a). Detection of new Leptospira genotypes infecting symptomatic dogs: is a new vaccine formulation needed? Pathogens 9:484. doi: 10.3390/pathogens9060484
PubMed Abstract | Crossref Full Text | Google Scholar
Bertasio, C., Papetti, A., Scaltriti, E., Tagliabue, S., D’Incau, M., and Boniotti, M. B. (2020b). Serological survey and molecular typing reveal new Leptospira serogroup Pomona strains among pigs of northern Italy. Pathogens 9:332. doi: 10.3390/pathogens9050332
PubMed Abstract | Crossref Full Text | Google Scholar
Blagojević, J., Šekler, M., Rajičić, M., Pejić, B., Budinski, İ., Jovanović, V., et al. (2019). The prevalence of pathogenic forms of Leptospira in natural populations of small wild mammals in Serbia. Acta Vet. Hung. 67, 338–346. doi: 10.1556/004.2019.035
PubMed Abstract | Crossref Full Text | Google Scholar
Boey, K., Shiokawa, K., and Rajeev, S. (2019). Leptospira infection in rats: a literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 13:e0007499. doi: 10.1371/journal.pntd.0007499
PubMed Abstract | Crossref Full Text | Google Scholar
Cock, P. J. A., Antão, T., Chang, J. T., Chapman, B., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Crossref Full Text | Google Scholar
Costa, F., Hagan, J. E., Calcagno, J. I., Kane, M. J., Torgerson, P. R., Martinez-Silveira, M. S., et al. (2015). Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl. Trop. Dis. 9:e0003898. doi: 10.1371/journal.pntd.0003898
PubMed Abstract | Crossref Full Text | Google Scholar
Cunha, C. E., Félix, S. R., Neto, A. C. P. S., Campello-Felix, A., Kremer, F. S., Monte, L. G., et al. (2016). Infection with Leptospira Kirschneri Serovar Mozdok: first report from the southern hemisphere. Am. J. Trop. Med. Hyg. 94, 519–521. doi: 10.4269/ajtmh.15-0505
PubMed Abstract | Crossref Full Text | Google Scholar
Dezzutto, D., Barbero, R., Canale, G., Acutis, P. L., Biolatti, C., Dogliero, A., et al. (2017). Detection of Leptospira Spp. in water turtle (Trachemys Scripta) living in ponds of urban parks. Vet. Sci. 4:51. doi: 10.3390/vetsci4040051
PubMed Abstract | Crossref Full Text | Google Scholar
Ferreira, A., Costa, P., Rocha, T., Amaro, A., Vieira, M. L., Ahmed, A., et al. (2014). Direct detection and differentiation of pathogenic Leptospira species using a multi-gene targeted real time PCR approach. PLoS One 9:e112312. doi: 10.1371/journal.pone.0112312
PubMed Abstract | Crossref Full Text | Google Scholar
Grippi, F., Cannella, V., Macaluso, G., Blanda, V., Emmolo, G., Santangelo, F., et al. (2023). Serological and molecular evidence of pathogenic Leptospira Spp. in stray dogs and cats of Sicily (South Italy), 2017–2021. Microorganisms 11:385. doi: 10.3390/microorganisms11020385
PubMed Abstract | Crossref Full Text | Google Scholar
Hamond, C., Pestana, C. P., Medeiros, M. A., and Lilenbaum, W. (2015). Genotyping of Leptospira directly in urine samples of cattle demonstrates a diversity of species and strains in Brazil. Epidemiol. Infect. 144, 72–75. doi: 10.1017/s0950268815001363
PubMed Abstract | Crossref Full Text | Google Scholar
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
PubMed Abstract | Crossref Full Text | Google Scholar
Majetić, Z. Š., Galloway, R. L., Sabljić, E. R., Milas, Z., Perko, V. M., Habuš, J., et al. (2014). Epizootiological survey of small mammals as Leptospira Spp. reservoirs in eastern Croatia. Acta Trop. 131, 111–116. doi: 10.1016/j.actatropica.2013.12.009
PubMed Abstract | Crossref Full Text | Google Scholar
Mateus, J. E., Gómez, N., Herrera-Sepúlveda, M. T., Hidalgo, M., Pérez-Torres, J., and Cuervo, C. (2019). Bats are a potential reservoir of pathogenic Leptospira species in Colombia. J. Infect. Dev. Ctries. 13, 278–283. doi: 10.3855/jidc.10642
Crossref Full Text | Google Scholar
Morey, R. E., Galloway, R. L., Bragg, S. L., Steigerwalt, A. G., Mayer, L. W., and Levett, P. N. (2006). Species-specific identification of Leptospiraceae by 16S RRNA gene sequencing. J. Clin. Microbiol. 44, 3510–3516. doi: 10.1128/jcm.00670-06
PubMed Abstract | Crossref Full Text | Google Scholar
Obiegala, A., Woll, D., Karnath, C., Silaghi, C., Schex, S., Eßbauer, S., et al. (2016). Prevalence and genotype allocation of pathogenic Leptospira species in small mammals from various habitat types in Germany. PLoS Negl. Trop. Dis. 10:e0004501. doi: 10.1371/journal.pntd.0004501
PubMed Abstract | Crossref Full Text | Google Scholar
Obrenović, S., Radojičić, S., Stević, N., Bogunović, D., Vakanjac, S., and Valčić, M. (2014). Seroprevalence of cat leptospirosis in Belgrade (Serbia). Acta Vet-Beogr. 64, 510–518. doi: 10.2478/acve-2014-0047
Crossref Full Text | Google Scholar
Piredda, I., Ponti, M. N., Palmas, B., Noworol, M., Pedditzi, A., Rebechesu, L., et al. (2021). Molecular typing of pathogenic Leptospira species isolated from wild mammal reservoirs in Sardinia. Animals 11:1109. doi: 10.3390/ani11041109
PubMed Abstract | Crossref Full Text | Google Scholar
Rodamilans, G. M., Fonseca, M. S., Paz, L. N., Fernandéz, C. C., Biondi, I., Lira-Da-Silva, R. M., et al. (2020). Leptospira Interrogans in wild Boa Constrictor snakes from Northeast Brazil Peri-urban rainforest fragments. Acta Trop. 209:105572. doi: 10.1016/j.actatropica.2020.105572
PubMed Abstract | Crossref Full Text | Google Scholar
Shiokawa, K., Welcome, S., Kenig, M., Lim, B., and Rajeev, S. (2019). Epidemiology of Leptospira infection in livestock species in saint Kitts. Trop. Anim. Health Prod. 51, 1645–1650. doi: 10.1007/s11250-019-01859-5
PubMed Abstract | Crossref Full Text | Google Scholar
Svirčev, Z., Marković, S. B., Vukadinov, J., Stefan-Mikić, S., Ružić, M., Doder, R., et al. (2009). Leptospirosis distribution related to freshwater habitats in the Vojvodina region (republic of Serbia). Sci. China C Life Sci. 52, 965–971. doi: 10.1007/s11427-009-0124-2
PubMed Abstract | Crossref Full Text | Google Scholar
Victoria, B., Ahmed, A., Zuerner, R. L., Ahmed, N., Bulach, D., Quinteiro, J., et al. (2008). Conservation of the S10-spc-α locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One 3:e2752. doi: 10.1371/journal.pone.0002752
PubMed Abstract | Crossref Full Text | Google Scholar
Vieira, A. S., Pinto, P. S., and Lilenbaum, W. (2017). A systematic review of leptospirosis on wild animals in Latin America. Trop. Anim. Health Prod. 50, 229–238. doi: 10.1007/s11250-017-1429-y
PubMed Abstract | Crossref Full Text | Google Scholar
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, È., Thibeaux, R., et al. (2019). Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 13:e0007270. doi: 10.1371/journal.pntd.0007270
PubMed Abstract | Crossref Full Text | Google Scholar
Vojinović, D., Žutić, J., Vasić, A., Stanojević, S., Spalević, L., and Sapundžić, Z. Z. (2022). A serological survey of canine leptospirosis in the City of Belgrade, Serbia. Vet. Glas. 76, 47–55. doi: 10.2298/vetgl210708001v
Crossref Full Text | Google Scholar
Wu, Q., Prager, K. C., Goldstein, T., Alt, D. P., Galloway, R. L., Zuerner, R. L., et al. (2014). Development of a real-time PCR for the detection of pathogenic Leptospira Spp. in California Sea lions. Dis. Aquat. Org. 110, 165–172. doi: 10.3354/dao02752
PubMed Abstract | Crossref Full Text | Google Scholar
Zhang, C., Junjun, X., Zhang, T., Qiu, H., Li, Z., Zhang, E., et al. (2019). Genetic characteristics of pathogenic Leptospira in wild small animals and livestock in Jiangxi Province, China, 2002–2015. PLoS Negl. Trop. Dis. 13:e0007513. doi: 10.1371/journal.pntd.0007513
留言 (0)