Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Kadir Gonzalez, Santiago S. De León, Vanessa Pineda, Franklyn Samudio, Zeuz Capitan-Barrios, José Antonio Suarez, Adriana Weeden, Betsi Ortiz, Margarita Rios, Brechla Moreno, Nathan D. Gundacker, Juan M. Pascale, Sandra López-Vergès, Néstor Sosa, Azael SaldañaAuthor affiliations: Gorgas Memorial Institute for Health Studies, Panama City, Panama (K. González, S.S. De León, V. Pineda, F. Samudio, Z. Capitan-Barrios, J.A. Suárez, A. Weeden, B. Ortíz, M. Ríos, B. Moreno, J.M. Pascale, S. López-Vergès, N. Sosa, A. Saldaña, L.E. Ábrego); University of Panama, Panama City (K. Gonzalez, S.S. De León, Z. Capitan-Barrios, A. Saldaña, L.E. Ábrego); Medical College of Wisconsin—Zablocki VA Medical Center, Milwaukee, Wisconsin, USA (N.D. Gundacker); University of New Mexico, Albuquerque, New Mexico, USA (N. Sosa)
Leishmania RNA virus 1 (LRV1) belongs to the Totiviridae family, Leishmaniavirus genus, and infects different Leishmania lineages. This virus is not enveloped and is composed of a viral capsid ≈40 nm in diameter and a double-stranded RNA (dsRNA) of 5,280 nt (1,2). The genome has 3 open reading frames (ORF), 2 of which are coding. The orf2 codes for the capsid protein and the orf3 codes for an RNA-dependent RNA polymerase (RdRp). orf1 has been described in other members of the family, but its function is unknown (1,3). This virus has been categorized in LRV1 and LRV2, according to the subgenuses of Leishmania in which they have been identified (4,5). The presence of LRV1 has been reported more frequently in specific regions of South America associated with cases of cutaneous leishmaniasis (CL) and mucocutaneous leishmaniasis (MCL) (6,7). L. (Viannia) panamensis is the predominant species and is responsible for most cases of CL in Panama (8,9) and the presence of LRV1 has been reported in 2 isolates of L. (V.) panamensis from Ecuador and Costa Rica (7,10).
We analyzed Leishmania spp. parasite isolates from clinical samples from 2014–2018 that were cryopreserved at Gorgas Memorial Institute’s parasitology research department (Panama City, Panama). The Bioethics Committee of the Gorgas Memorial Institute for Health Studies submitted and approved this study (protocol no. 056/CBI/ICGES/19). We extracted clinical and epidemiologic data such as sex, age, clinical classification (location, severity, and number of lesions), and province of origin from the database. The disease was classified as nonsevere or severe according to Infectious Disease Society of America guidelines (11). We activated the isolates at 26°C by using Schneider’s medium enriched with 25% fetal bovine serum until reaching exponential growth (2–3 ×107 parasites/mL) (9). We centrifuged this concentration of parasites for 10 minutes at 3,500 rpm and divided it into 2 pellets; we used 1 pellet to extract DNA from Leishmania spp. for characterization and confirmation and the other to extract RNA and detect LRV1. We characterized the isolates as L. (V.) panamensis by the RFLP/PCR-Hsp70 methodology (12). For the detection of LRV1, we amplified 245 nucleotides corresponding to the orf1 gene region using the primers described by Ito et al. (6,13) and sequenced the product by the Sanger method.
Figure 1
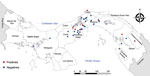
Figure 1. Distribution of Leishmania (Viannia) panamensis isolates analyzed for RNA virus 1, Panama, 2014–2018.
We recovered parasite isolates from 56 patients. Of those isolates, 11 (20%) were positive for LRV1, 63.3% from female patients and 36.4% from male patients. Patient age range was 8–59 years; mean (+SD) age was 34 (+5.4) years (Appendix Table 1). All the patients came from leishmaniasis-endemic areas in Panama: 36.4% from Panama Oeste, 18.2% from Panama, 18.2% from Colón, 18.2% from Darién, and 9.0% from Coclé (Figure 1). Most of the patients had single lesions (7/11 [63.6%]); mean (+SD) was 1 (+0.2) and range 1–3 lesions per patient. Mean (+SD) time of evolution of the lesion was 50 (+9.6) days and range was 21–120 days. Most (6/11 [54.5%]) patients had an evolution time of 30 days. All the lesions were CL and were classified as nonsevere; lesions consisted of a crusty, moist ulcer with raised margins and a clean base (Table) (11). The lesions were distributed mainly on the arms (9/11: 81.8%); only 2 were visible elsewhere, on the leg (1/11: 9.1%) and face (1/11: 9.1%).
Figure 2
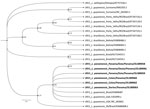
Figure 2. Phylogenetic analysis of Leishmania (Viannia) panamensisisolates positive for RNA virus 1 from Panama (boldface) and reference isolates from GenBank. A phylogenetic tree reconstruction of RNA virus...
We performed data analysis using GraphPad Prism 5.0 software (GraphPad, https://www.graphpad.com). We performed the Kolmogorov-Smirnov test to assess the normality of the samples. To analyze the differences between groups, we performed a t test for Gaussian distribution data. We considered differences statistically significant when p was <0.05. We found no significant difference to suggest that those with LRV1-positive parasites developed more severe diseases (data not shown). From 10 sequences downloaded from GenBank (accession nos. OL389058–67), we selected 6 sequences based on phylogenetic analysis quality (Appendix Table 2); those sequences clustered within the phylogenetic group of LRV1 sequences detected in the species of the subgenus Viannia, close to those found in isolates of L. (V.) guyanensis (Figure 2).
We detected LRV1 in 11/56 (20%) of L. (V.) panamensis–evaluated isolates, all of them in patients with CL, consistent with the preliminary description of the presence of LRV1 in 2 isolates of L. (V.) panamensis from clinical samples from Ecuador and Costa Rica, countries geographically close to Panama (7). The prevalence of LRV1 has been reported as higher in Leishmania spp. isolates from the New World (39.1%) than in those from the Old World (8.4%); prevalence also is higher in isolates from patients with severe skin forms of leishmaniasis, such as disseminated leishmaniasis and MCL, than from patients with CL (14).
The use of Leishmania spp. isolates could be a limitation for the analysis because we were able to analyze only the parasites that grew in medium. To avoid this bias, future studies analyzing the presence of the virus directly from clinical samples are needed. In South American countries, prevalence of ≈25% of LRV1 has been described in isolates of L. (V.) braziliensis and L. (V.) guyanensis from Peru (7), Bolivia (14), and Brazil (15). The presence of LRV1 in L. (V.) panamensis in this study (20%) indicates circulation of this virus in Panama, suggesting LRV1 is likely widespread across the Americas and in different Leishmania (V.) species. Future analysis using a higher number of samples is necessary to estimate LRV1 prevalence in Panama.
In this study, we found no evidence that correlates the presence of LRV1 with severe clinical forms of leishmaniasis caused by L. (V.) panamensis, which was consistent with previous findings of no predisposition of the Th2 response induced by LRV1 for the favorable survival of the parasite for L. (V.) panamensis (7). In addition, previous studies described a general decrease in the expression of virulence factor transcription in L. (V.) panamensis (7) compared with an earlier study of L. (V.) braziliensis (10). It is possible that L. (V.) panamensis strains infected with LRV1 have low expression of virulence factor, which would be reflected in the presence of uncomplicated symptoms of CL cases in the analyzed samples.
The role of LRV1 and its subtypes modulating the immune response in infection caused by L. (V.) panamensis is unclear. It is important to carry out studies of the virus subtypes that are circulating in the country and analyze whether the differences in the modulation of the immune response reflected in the clinical manifestations are because of intrinsic factors of the virus, the Leishmania species that it infects, or both.
In conclusion, the data we obtained show the presence of LRV1 in isolates of L. (V.) panamensis from Panama from different years and locations, suggesting wide spread of the virus in this species. In addition, the recent documented circulation of L. (V.) guyanensis and L. (V.) braziliensis in Panama (9) and the proposed association of LRV1 presence in these species with severity of disease highlight the necessity of future studies on the presence of LRV1 in non–L. (V.) panamensis species in Panama. The role of Leishmania in disease severity may depend on the species infected and the role of viral, parasite, and human host factors in pathogenesis.
Dr. González is a medical technologist and senior health researcher at Gorgas Memorial Institute, Panama City, Panama. Primary research interests are immunopathology of cutaneous leishmaniasis and molecular characterization of Leishmania spp., and in vitro studies of Leishmania (V.) panamensis infection.
We thank all the members of the Department of Research in Virology and Biotechnology, the Department of Research in Parasitology, and the clinical research unit of Gorgas Memorial Institute for Health Studies. We thank Alberto Cumbrera for map creation.
K.G., F.S., J.A.S., J.M.P., S.L.V., A.S., L.E.A. are members of the Sistema Nacional de Investigación of SENACYT, Panama.
This study was possible thanks to the support of the Sistema Nacional de Investigación (SNI-SENACYT), Panama, awarded to J.A.S., J.M.P., S.L.V., N.S., A.S., K.G., and L.E.A. This work also received administrative and financial support from Gorgas Memorial Institute for Health Studies.
留言 (0)