Despite the availability of many interventions in India, children (<15 yrs) accounted for 12% of new HIV infections in 2016-17, and mother to child transmission rate was 6.4%.1 Among host genetic factors, human leukocyte antigen molecules have been consistently associated with HIV transmission.2 The extensive polymorphism of human leukocyte antigen results in a broad diversity of potential immune responses against HIV-1 thereby affecting disease outcomes.3
Studies in various cohorts around the world have reported that individuals with certain human leukocyte antigen types may be more or less susceptible to HIV-1 infection in a population.4-6 HLA-B*57 and -B*27 have been associated with slower progression to AIDS whereas HLA-B*35 alleles with more rapid disease progression.7 Specifically, B*57:01 has been reported to be directly involved in providing virus-specific immunity in individuals.8 Cytotoxic T lymphocyte responses elicited towards viruses are specific and enhanced in presence of certain human leukocyte antigen alleles as compared to others and changes in even a single amino acid may have a substantial effect on disease progression.9,10 A few studies have also reported alleles such as HLA-B*13 and HLA-B*58:01 to be protective in groups of HIV controllers when compared to HIV progressors.11 Apart from alleles, haplotypes have also been found to be associated with HIV transmission. One study done in the Scottish population reported an increased frequency of HLA-A1-B8-DR3 haplotype among HIV-infected children whereas HLA-A3-B7-DR2 haplotype was associated with protection against HIV-1 infection.12 A similar observation was made by another group among white American infants wherein the HLA-DR2 allele (DRB1*1501) was associated with seroreversion while the HLA-DR3 (DRB1*03011) allele was positively associated with the occurrence of HIV-1 infection.13 In an East African population HLA-A2 was associated with a nine-fold reduction in the risk of perinatal HIV infection.14 Mackelprang in 2009 reported that the presence of A*2301 was associated with higher mother to child transmission whereas B*1801 had a protective effect for the infants.15
Not many studies have been performed on Indian children exposed to HIV through their mothers. Hence this study aimed to evaluate the association of human leukocyte antigen class I and class II alleles with vertical transmission of human immunodeficiency virus type 1 (HIV-1) infection in the Indian population.
Methods Study subjectsEnrolment of subjects for this exploratory study was done at a tertiary care hospital from Mumbai, India. Enrolment and follow-up of subjects were carried out between January 2010 and December 2014. Approvals from the ethics committees of the institute and collaborating centre were obtained. HIV seropositive women, who had registered at the prevention of parent to child transmission (PPTCT) centre, under the department of obstetrics & gynaecology, and had delivered a live infant were contacted within 48 hours of delivery. Also, HIV seropositive mothers who had delivered a live infant at the peripheral hospital and attended the integrated counselling and testing centre under the department of microbiology, with their babies for further testing and treatment were informed about the study. These parents were counselled and motivated to follow up regularly at specified intervals for HIV screening of the infant. Only those mothers who gave consent for enrolment and follow-up of their babies were included. A detailed questionnaire was prepared to obtain demographic data. Infants who were in neonatal intensive care and infants who did not complete the follow-up were excluded from the study.
Maternal characteristicsPlasma HIV-1 viral load was estimated in the mother’s samples, obtained within 48 hours of delivery. This could be done only in mothers who were enrolled through the prevention of parent to child transmission centre. The procedure included isolation of total nucleic acid using the MagNa Pure Compact Nucleic Acid Automated System (Roche Diagnostic, Mannheim, Germany). Subsequently, the viral load was measured by Cobas Taqman Real time PCR (Roche Molecular Systems, Branchburg, NJ) according to the manufacturer’s instructions. CD4 count was done using BD FACS count CD4 reagents from BD Biosciences (Becton Dickinson and Company, San Jose, CA) following the instruction manual provided by the manufacturer. During the study period, a triple-drug antiretroviral therapy was initiated in pregnant women only if their CD4 count fell below 350 cells/mm3 or were classified as WHO stage III or IV (according to national guidelines during the study period). Women who maintained a CD4 count above 350 cells/mm3 were only given a single dose of nevirapine (200 mg) at the onset of labour as prophylactic treatment. Antiretroviral prophylaxis given to the infant included a single dose of nevirapine syrup (2 mg/kg) within two hours of delivery followed by zidovudine syrup (4 mg/kg) twice daily for six weeks.16
Infants’ HIV screeningProviral DNA assay in babies was done to detect in-utero HIV-1 transmission, from whole blood samples collected within 48 hours of birth. Briefly, a nested PCR was used to amplify a specific part of the gp41 region of the ‘env’ gene.17 The amplicons of the PCR reaction were then sequenced and confirmed as being HIV-1 subtype C using the NCBI-BLAST software. Dried blood spots were collected during the follow-up visits for DNA PCR. In cases where the status was indeterminate whole blood was collected and PCR was repeated. A confirmation by serology (ELISA) was done after the completion of 18 months of age for all the infants. These infants were then divided into two groups: 1st group with infected children (positive) and 2nd group with uninfected children (negative).
DNA isolation and quantitationDNA from the whole blood specimen was extracted manually by salting out method or using Qiagen DNA extraction mini kit (QIAGEN, GmbH, Hilden). The absence of inhibitory factors was checked by PCR amplification of housekeeping gene, β-globin, using the following primer pair: 5’ ACA CAA CTG TGT TCA CTA GC 3’ and 5’ GAA ACC CAA GAG TCT TCT CT 3’.
HLA typing using polymerase chain reaction-sequence-specific oligonucleotide probe/sequence-specific primers methodTwo methods namely polymerase chain reaction-sequence-specific oligonucleotide probe and sequence-specific primers were used for human leukocyte antigen typing. The polymerase chain reaction-sequence-specific oligonucleotide probe and sequence-specific primers method consists of three steps namely, PCR amplification, hybridization reaction and detection. Reactions and interpretations were carried out according to the manufacturer’s protocol (Invitrogen, USA).
Sequence-specific primer (SSP) method was used for DNA-based tissue typing. The assignment of alleles consists of visualization and detection on agarose gel electrophoresis. The interpretation was done using the AllSet uniMatch software, available along with the reagents, for specific amplification patterns using the manufacturer’s protocol (AllSet Gold SSP kit, Life technologies, USA).
Quality controlRandomly selected specimens were tested using both methods and also reassessed by another co-author to check for the reproducibility of the analysis.
Statistical analysisThe population genetics package, PyPop, developed by the biostatistics core for workshop18 was used for the analysis of pairwise linkage disequilibrium and estimation of haplotype frequencies of the different studied loci. Statistical analysis was done using SPSS software version 19. As many alleles were uncommon, odds ratios were computed only for alleles found in 10% of infants to limit spurious associations. For allele, genotype and haplotype analysis, false discovery rate was calculated and applied to determine true significance.19 ‘Power for cohort studies’ module available online in the OpenEpi software was used for post hoc power determination.20
In silico analysisSequences obtained from positive samples were compared to the HIV sequence database available online. GenCutter alignment tool was then used for multiple sequence alignment and also to confirm the position of the sequences in the HIV genome. These aligned sequences were then run in the consensus maker software to construct a consensus sequence for the ‘env’ protein that we had sequenced.21 Peptides specific to significant alleles were predicted from the consensus sequence of ‘env’ protein generated using ProPred-I for major histocompatibility complex (MHC) Class I.22 The threshold value was set at 4%. The peptides identified as epitopes by this method was used for molecular modelling. Short-listed epitopes were modelled with the HLA-B*40 (PDB: 5IEH) using the CABS-Dock web server which enables docking of peptides to the surface of flexible proteins.23
ResultsDetails of patient enrolment and follow-up have been mentioned in [Figure 1]. Human leukocyte antigen typing and analysis was done in 30 HIV positive and 60 HIV negative children. In a few samples, human leukocyte antigen typing could not be done due to insufficient DNA or non-amplification.
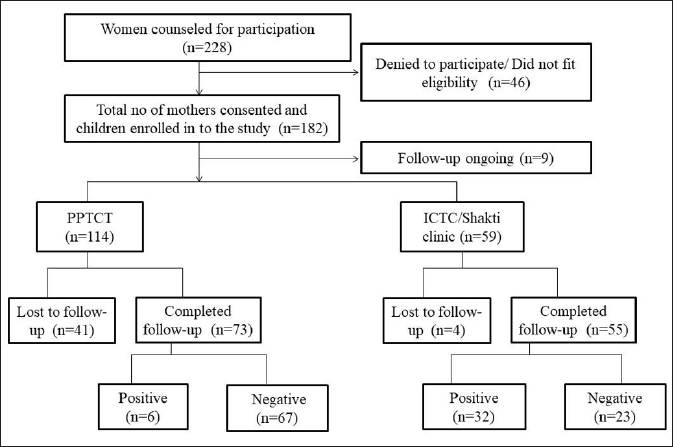
Figure 1:: Enrollment and follow-up details of women counselled and their infants enrolled. HLA typing was done in 30 seropositive and 60 seronegative children whose DNA was available for typing. The positive numbers do not indicate the prevalence rate of the studied population. PPTCT: Prevention of Parent to Child Transmission; ICTC: Integrated Centre for Testing and Care.
Export to PPT
HLA class I and class II allele frequencies in positive vs. negative childrenFifteen HLA-A alleles and 21 HLA-B alleles were observed in our study population. HLA-B*07, HLA-B*35 that are known to be enriched in HIV progressors were together present in 21.7% of the positive children. HLA-A*01 was marginally significant with a P-value of 0.05. The frequency of HLA-B*37 was high in the infected children (8.3% in positives vs. 1.7% in negatives, P = 0.033) [Table 1]. In the negative children, HLA-A*02, -A*11 and -A*24 were present with a frequency >10% and together constituted more than 62% of the total alleles but were not statistically significant. The known protective alleles such as HLA-B*13, -B*18, -B*27, -B*57 together constituted only 10.1% of the HLA-B alleles in the negative children; and HLA-B*27 was exclusively present only in the negative children. HLA-B*40 was present significantly more frequently in exposed uninfected children (20.3% in negatives vs. 6.7% in positives, P = 0.018) [Table 1].
Table 1:: Significant alleles of HLA class I and class II in positive vs. negative children
Alleles Positive Negative Power (2n = 54) % (2n = 110) % P-value A*01 10 18.5 9 8.9 0.05 3.3% A*02 8 14.8 29 26.3 0.09 86.9% Positive Negative (2n = 60) (2n = 118) B*13 4 6.7 2 1.7 0.08 11.9% B*37 5 8.3 2 1.7 0.032 19.6% B*40 4 6.7 24 20.3 0.018 92.3% Positive Negative (2n = 60) (2n = 112) DRB1*01 1 1.7 9 8.0 0.09 54.5% DRB1*09 3 5 0 0 0.017 66.8%Thirteen alleles were observed for the DRB1 locus. Frequencies of the alleles were similar in both groups. DRB1*09 was only present in the positive children and was also significantly associated (P = 0.017). Although DRB1*01 was present more frequently in negative children, it was not statistically significant (P = 0.09) [Table 1]. Seven DQB1 alleles were observed in the population ([Table 4]). The frequencies of all the alleles of the HLA-DQB1 locus was similar in both groups; except for the alleles DQB1*08 and DQB1*15.
High resolution HLA typingFurther, the allelic distribution was studied in both groups at a higher resolution. Only those sub-types that had a frequency greater than 5% in either of the groups were considered for comparison [Table 2]. HLA-A*01:01 was the only subtype that was present with an increased frequency in positive children and was associated with transmission.
Table 2:: High-resolution analysis of alleles of HLA class I and class II in positive vs. negative children
Alleles Positive Negative (2n = 54) % (2n = 110) % P-value A*01:01 10 18.5 8 7.1 0.035 A*02:01 2 3.7 12 10.7 0.149 A*11:01 4 7.4 12 10.7 0.586 A*24:02 6 11.1 13 11.6 1.0 A*32:01 3 5.5 2 1.8 0.331 A*33:01 5 9.3 7 6.3 0.529 Positive Negative (2n = 60) (2n = 118) B*13:01 3 5 2 1.6 0.333 B*35:01 2 3.3 6 4.9 1.000 B*40:01 0 0 7 5.7 0.097 B*40:06 4 6.6 10 8.1 1.0 B*44:02 5 8.3 8 6.5 0.761 B*51:01 5 8.3 4 3.2 0.158 B*52:01 6 10 4 3.2 0.083 Positive Negative (2n = 60) (2n = 112) DRB1*01:01 1 1.7 8 7.0 0.166 DRB1*07:01 5 8.3 8 7.0 0.767 DRB1*07:11 3 5 1 0.8 0.119 DRB1*09:07 3 5 0 0 0 DRB1*10:02 3 5 0 0 0 DRB1*12:38 3 5 0 0 0 DRB1*15:01 10 16.7 23 20.2 0.686 Positive Negative (2n = 60) (2n = 118) DQB1*02:01 7 12.1 14 11.9 1.00 DQB1*03:01 3 5.1 9 7.6 0.753 DQB1*03:02 3 5.1 5 4.2 0.720 DQB1*05:01 12 20.7 30 25.4 0.574 DQB1*06:01 9 15.5 25 21.2 0.422 DQB1*06:02 9 15.5 5 4.2 0.615 DQB1*06:03 0 0 8 6.7 0.054 Haplotypes in HIV-1 positive and negative childrenHaplotypes of the two groups involving various combinations of class I and class II loci were determined using PyPop. Only those haplotypes that had a frequency greater than 5% or were exclusively present were considered for analysis. All the haplotypic combinations that were found to be significantly associated with infection transmission or protection have been outlined in [Table 3].
Table 3:: Haplotypes found to be significantly associated with vertical transmission in positive vs. negative children
Haplotypes Positive Negative (2n = 60) % (2n = 120) % P-value* A: B 24:40 0 0 9 7.5 0.03 02:15 0 0 8 6.7 0.041 24:52 5 8.3 0 0 0.001 33:35 4 6.7 1 0.8 0.025 A:B:DRB1 24:52:14 3 5 0 0 0.014 33:35:07 3 5 0 0 0.014 33:35:02 4 6.7 0 0 0.004 A: B: DQ 24:52:05 3 5 0 0 0.014 01:51:05 3 5 0 0 0.014 A: DR: DQ 01:15:05 6 10 0 0 0.000 B:DRB1:DQB1 40:14:05 0 0 7 5.8 0.05 52:14:05 4 6.7 0 0 0.004 13:15:06 3 5 0 0 0.014 A:B:DRB1: DQB1 01:51:15:05 3 5 0 0 0.014 33:35:07:02 3 5 0 0 0.014 False discovery rate correctionsThe false discovery rate is a method of conceptualizing the rate of type I errors in null hypothesis testing when conducting multiple comparisons.23 All the other haplotypes that were found to be associated lost their significance except for the haplotypes A: DR: DQ - 01: 15: 05; A: B - 02: 15 and A: B: DQ - 33: 35: 02 [Table 4].
Table 4:: FDR corrected significant P-values
Allele/ haplotype P q* P<q* A: DR: DQ - 01: 15: 05 0.0000 0.0011 TRUE A: B - 02: 15 0.0010 0.0023 TRUE A: B: DQ - 33: 35: 02 0.0040 0.0045 TRUE Confounding factorsA non-parametric chi-square test was performed for confounding factors. The type of treatment given to the mother at the time of delivery influenced HIV transmission in children (P = 0.001). The feeding option selected for the baby (P = 0.908) and sex of the child (P = 0.442) were not associated with transmission. All the confounding factors along with the significant human leukocyte antigen alleles were tested for multivariate logistic regression. After performing logistic regression, only HLA-B*40 (P = 0.01) and DRB1*09 (P = 0.011) remained significant [Table 5]. Mann-Whitney U test was performed for viral load (P = 0.05) and CD4 count (P = 0.462). Logistic regression further confirmed that viral load (P = 0.01) affected the disease outcome in children.
Table 5:: Effect of confounding factors
Alleles Unadjusted P-value Adjusted P-value HLA-A*01 0.05 0.053 HLA-B*37 0.032 0.273 HLA-B*40 0.018 0.010 HLA-DRB1*09 0.017 0.011 Viral Load 0.050 0.010 In silico analysisWe next attempted to identify epitopes from the ‘env’ region of the sequences from our study population. Using the available HIV databases, we could identify a consensus sequence in our population. We then used the ProPred1 software to generate epitopes of this region that specifically binds to HLA-B*40, since this was the most significant allele obtained. We also planned to study HLA-DRB1*09 but this allele was not available in the ProPred database. The three-dimensional structure of HLA-B*40 available in the UniProt database was used to dock epitope. The epitope was found to bind to the peptide-binding cleft of the HLA-B*40 groove implying a protective response in the host.
DiscussionOf all new HIV infections in the Asia and Pacific region, India accounted for 38%.24 In our study, 15 HLA-A, 21 HLA-B, 13 HLA-DRB1and 7 HLA-DQB1 alleles were identified in children exposed to HIV through their mothers, in Mumbai city from Maharashtra, India.
We found a significant association of one allele each of HLA-A & HLA-DRB1 and three alleles of HLA-B with vertical HIV transmission. HLA-A*01 was marginally significant and was associated with susceptibility to the transmission of the infection. Although HLA-A*02 was not significantly associated with protection from infection, it was present more frequently in negative children. Both these alleles have not been reported by any other group to be associated with vertical HIV transmission.25 It is now well established that HLA-B alleles bear the major burden of HIV-specific Cytotoxic T lymphocytes activity in chronic HIV infection, and these alleles are principally associated with diverse outcomes from HIV infection in various populations.26 HLA-B alleles have also been strongly associated with mother to child transmission of HIV irrespective of the viral load.27 In the current analysis, HLA-B*40 showed a protective effect towards the acquisition of the disease whereas HLA-B*37 was associated with susceptibility to disease transmission. HLA*A01-B*37-C*06 is a well-known haplotype and the associations observed could reflect an LD relationship between the two alleles. The fact that HLA-B*40 remained significant after performing a logistic regression and had a power greater than 90% indicates its strong association with protection from HIV, similarly, DRB1*09 was associated with transmission.
We did not find any similarity in protective or susceptible alleles between the reported data and our observations. There could be two possible explanations; one of them being that the HIV clade circulating in the Indian population is clade C; most of the studies have been reported in clade B HIV-1. Another reason could be the ethnicity of the studied population; almost all the studies have been done in either Caucasian, African or American populations.
In an earlier study in HIV discordant couples, it was reported that HLA-B*35 was associated with susceptibility whereas HLA-B*18 and *40 were associated with protection.28 HLA-B*40 was the only allele that was common in both studies. A similar finding was observed in commercial blood donors of China who were highly exposed to HIV but were persistently seronegative.29 Farquhar et al. reported a protective association of HLA-B*18 against the acquisition of HIV infection from the mothers but this group did not find any association of HLA-A locus with HIV infection.30 Though no report was available on HLA-B*37 association with HIV infection, our results indicated its possible association with susceptibility to HIV transmission.
Among the class II alleles studied, we found a significant association of HLA-DRB1*09 with susceptibility to the infection. Similar to our results, one study from India reported the DRB1*09 allele to be associated with disease transmission.31 We were unable to find any other reports with a similar observation.
It is known that maternal-infant human leukocyte antigen concordance increases the risk of perinatal HIV transmission since children are haploidentical to their mothers.32 Hence, we did not do the allele typing of the mothers. The main aim of our study was to identify which human leukocyte antigen alleles are enriched in the positive or negative population conferring different clinical outcomes. Other important factors such as viral load, CD4 count and treatment regimen of the mothers at the time of delivery along with feeding options given to the baby also affect disease outcome in children. As reported in our earlier study, high viral load at the time of delivery is the strongest predictor of disease transmission to children.17
HIV is a highly mutative virus and it continues to evolve and manifest itself in various conditions due to this nature. We could identify peptides in the ‘env’ region of the viral sequences in our population. The binding capacity of the peptide to the HLA-B*40 molecule was evident as obtained by docking analysis. HLA-B*40 was consistently associated with protection from HIV transmission even after correcting for multiple factors. This is in agreement with a previous study that had reported HLA-B*40 to be protective in HIV sero-negatives.28
LimitationsOne major limitation of our study is the small sample size. This could be attributed to the fact that participation and follow-up of the study subjects were voluntary. Also, many parents were hesitant to enroll newborn children into the research study. To maintain confidentiality of the HIV positive individuals, personal contact was avoided. Viral load analysis in positive children could not be done due to insufficient sample amount.
ConclusionOur study could identify specific HLA-B alleles’ association with vertical HIV transmission. The frequencies and distribution of human leukocyte antigen class I alleles obtained from our study add to the existing knowledge of the genetic makeup of the population. This may help in deciding intervention strategies and treatments given to the exposed children. Peptide interactions reported can help in vaccine design strategies for the Indian population.
留言 (0)