Nontuberculous mycobacteria (NTM) are environmental bacteria that can cause progressive, chronic and sometimes fatal pulmonary disease. Population-based investigations indicate that the prevalence of NTM-related pulmonary disease (NTM PD) continues to increase globally, with Mycobacterium avium complex (MAC) species and Mycobacterium abscessus and its subspecies (MABS) being the two most common NTM recovered from respiratory specimens (Prevots and Marras, 2015; Park and Olivier, 2015). In immunocompetent patients with predisposing lung conditions, such as cystic fibrosis (CF) and chronic obstructive pulmonary disease, the presentations of NTM PD share striking similarities with tuberculosis (TB) in immunocompetent subjects, including necrotizing and non-necrotizing granulomas and cavitation. These lesions tend to be paucibacillary with organisms seen primarily in the areas of necrosis (O’Connell et al., 2012; Tomashefski et al., 1996; Jain et al., 2017; Choi et al., 2021). These shared pathologic manifestations suggest that Mycobacterium tuberculosis (Mtb) and MAC/MABS are probably exposed to similar stresses during host infection. Whether or not Mtb and NTM have evolved the same strategies to overcome these stressors and establish a chronic lung infection is a question of considerable interest when designing dual anti-tuberculosis (TB)/NTM therapies.
NTM and Mtb are thought to share the ability to persist within lung lesions in a slow- or non-replicating state, which contributes to their drug tolerance and to treatment failure in chronically infected individuals (Wu et al., 2018). Low oxygen tension and respiratory competitors of oxygen, such as nitric oxide (NO) and carbon monoxide (CO), in avascular necrotic regions of granulomas and in the CF airway likely signal mycobacterial pathogens to adopt a non-replicating state in the absence of aerobic respiration. In Mtb, the initial response to hypoxia is controlled by the three-component regulatory system, DosRST. Upon activation, the transcriptional regulator DosR drives the physiologic adaptation of Mtb required for survival in the absence of oxygen through the induction of a 48-gene “dormancy regulon” (Park et al., 2003; Rustad et al., 2008; Leistikow et al., 2010; Reichlen et al., 2017). The finding that macaques infected with Mtb mutants harboring dosR, dosS and/or dosT null mutations displayed reduced infection levels and granulomatous inflammation during the persistent stage of infection compared to animals infected with wild-type Mtb supports the notion that the DosR regulon is important for persistence in a human-like model of TB infection (Mehra et al., 2015). Accordingly, inhibitors of DosRST are being sought for their potential to shorten TB treatment and lower relapse rates when used in combination with standard-of-care antibiotics (Zheng et al., 2017, 2020).
Homologs of the DosR and DosS proteins have been identified in the sequenced genomes of many Mycobacterium spp. as well as in a number of other environmental Actinomycetes, suggestive of a broad role of this two-component regulator in the physiologic adaptation of Actinomycetes to hypoxic conditions beyond those encountered in the infected host (Bartek et al., 2009; Gerasimova et al., 2011). Recent work by our group and others confirmed that, similar to the situation in Mtb, DosRS plays an important role in the adaptation of MABS to hypoxia (Belardinelli et al., 2022; Simcox et al., 2023). Importantly, our work further revealed that a deficiency in DosRS expression significantly impairs the ability of MABS to form biofilms, a phenotype likely attributable to the reduced ability of oxygen-deprived bacilli located within the biofilms to remain viable in the absence of a functional DosRS system (Belardinelli et al., 2022). Biofilm formation is a common strategy used by pathogens of the airway to colonize the host while enhancing the bacterium’s drug tolerance and resistance to host defense mechanisms, and could help explain why DosS inhibitors can reverse the tolerance of MABS bacilli to some antibiotics in a mouse model of MABS infection not known to develop hypoxic lesions (Belardinelli et al., 2022).
Like Mtb and MABS, MAC species are endowed with a DosRS system and further possess a second kinase (annotated as MAV_2508 in the genome of M. avium 104) thought to function with DosR (Gerasimova et al., 2011). In the context of our search for novel therapeutic strategies to address the growing issue of multidrug-resistant NTM infections, we here sought to experimentally characterize the DosR regulon of MAC and its activation by DosS and/or MAV_2508, and to assess its contribution to survival under hypoxia, drug tolerance, biofilm formation and pathogenicity.
2 Materials and methods2.1 Strains and culture mediaM. avium subsp. hominissuis MAH11 (Dragset et al., 2019) was grown under agitation at 37°C in Middlebrook 7H9 medium supplemented with 10% albumin-dextrose-catalase (ADC) (BD Sciences) and 0.05% Tween 80, in Synthetic CF medium (SCFM) (Belardinelli et al., 2021), or on Middlebrook 7H11 agar supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) (BD Sciences). Kanamycin and hygromycin were added to the culture media at a final concentration of 25 μg mL-1 and 50 μg mL-1, respectively. For growth under microaerophilic and hypoxic conditions, MAH11 strains were grown in Dubos-Tween albumin broth at a final pH of 7.3 or 5.7, either in standing T25 vented tissue culture flasks (microaerophilic conditions) (Zheng et al., 2017) or in 16x100 mm glass tubes with tightly sealed screw caps with rubber septa under constant stirring using Teflon-coated magnetic bars (hypoxic Wayne model) (Wayne and Hayes, 1996). Decolorization of methylene blue (1.5 μg mL-1 final concentration) in control tubes served as a visual indication of oxygen depletion in the Wayne model.
2.2 M. avium dosRS knock-out mutant and complemented mutant strainsA dosRS deletion mutant was generated in MAH11 by allelic replacement using the Ts-sacB system (Pelicic et al., 1997). Briefly, an allelic exchange substrate consisting of the kanamycin-resistance cassette (kan) bracketed by ~ 500 bp of upstream and downstream DNA immediately flanking the dosRS operon of MAH11 (genes MAV_4109-MAV_4108 based on the M. avium 104 genome annotation) was cloned in pPR27xylE. The resulting plasmid, pPR27xylE-dosRS::kan, was electrotransformed in MAH11 and a transformant selected on kanamycin-containing agar plates at 32°C. Upon a culturing step at 32°C in liquid broth, the transformant was plated on agar with kanamycin and 2% sucrose at 39°C to select for allelic exchange mutants. Allelic replacement at the dosRS locus was verified by PCR and sequencing.
For complementation, 220-bp of the promoter region of dosR and the entire coding sequence of dosRS were PCR-amplified from MAH11 genomic DNA and cloned into the integrative plasmid pMV306H, yielding pMV306H-dosRS. Alternatively, 220-bp of the promoter region of dosR and the entire coding sequence of dosR only were PCR-amplified and cloned into the integrative plasmid pMV306H, yielding pMV306H-dosR. The sequences of the primers used to generate the different constructs are available upon request. A mutant carrying a transposon insertion in the second putative sensor kinase associated with DosR (gene MAV_2508 based on the M. avium 104 genome annotation; position of insertion = bp 220), and a mutant carrying a transposon insertion in dosR (position of insertion = bp 549) were obtained from Dr. M. Dragset (Norwegian University of Science and Technology, Trondheim, Norway) (Dragset et al., 2019).
2.3 Biofilm assayBiofilm formation in SCFM was monitored by crystal violet staining as described by Belardinelli et al. (2021) except that MAH11 strains were allowed to grow in poly-D-lysine-coated microtiter plates for 14 days (instead of 5 days for MABS).
2.4 NO susceptibility assaysFor in vitro NO susceptibility assays, MAH11 strains and M. abscessus ATCC 19977 grown in 7H9 supplemented with albumin-dextrose-NaCl (ADS) and 0.05% Tween 80 to an OD600nm of 0.1 were added 50 or 500 μM DETA/NO (Cayman Chemical) or Spermine/NO (Cayman Chemical) and incubated for 24 h at 37°C under agitation. Viable CFUs were enumerated by plating serial dilutions of the cell suspensions on 7H11-OADC agar.
2.5 Susceptibility to metronidazole, nitrofurans and other antibiotics under hypoxiaThe susceptibility of MAH11 strains to metronidazole, nitrofurans and clinically used antibiotics under hypoxic conditions was determined using the Wayne model (pH 7.3) as described above. When cultures reached hypoxia (as determined by decolorization of methylene blue), metronidazole (ThermoFisher), 2-nitrofuran (Sigma-Aldrich), nitrofurazone (Sigma-Aldrich), rifampicin (Sigma-Aldrich), amikacin (Sigma-Aldrich), ethambutol (Sigma-Aldrich), clarithromycin (Sigma-Aldrich), bedaquiline or clofazimine (Sigma-Aldrich) were added with a syringe through a rubber septum, to avoid introducing oxygen. After 7 days of incubation under hypoxia at 37°C, tubes were opened and serial dilutions of the cultures plated to enumerate CFUs.
2.6 Metabolic labeling of lipidsMetabolic labeling of MAH11 cells with [1,2-14C]acetic acid (0.5 μCi mL-1; specific activity, 54.3 Ci/mol, PerkinElmer) was performed for 16 h at 37°C in Dubos-Tween albumin broth under microaerophilic conditions in standing T25 vented flasks, or under well aerated conditions in Erlenmeyer flasks. [1,2-14C]acetic acid-derived lipids extracted from whole bacterial cells with a mixture of chloroform and methanol (1:2 and 2:1, by vol.) were analyzed by thin-layer chromatograph (TLC) on aluminum-backed silica gel 60-precoated plates F254 (E. Merck) and revealed by PhosphorImaging.
2.7 RNA extraction and RT-qPCRTwo independent cultures of bacteria grown under microaerophilic conditions in Dubos-Tween albumin broth (or aerophilic conditions for 24 h) were used for transcriptomics analyses. RNA extraction with the Direct-zol™ RNA Miniprep kit (Zymo Research), reverse transcription reactions using the Superscript IV First-Strand Synthesis System (Thermo Fisher) and RT-qPCR using the SYBR Green PCR Master Mix (Sigma-Aldrich) were conducted as per the manufacturers’ protocols and analyzed on a CFX96 real-time PCR machine (Bio-Rad). PCR conditions: 98°C (30 s; enzyme activation), followed by 40 cycles of 98°C (10 s; denaturation) and 60°C (30 s; annealing/extension). Mock reactions (no reverse transcription) were done on each RNA sample to rule out DNA contamination. The target cDNA was normalized internally to the sigA cDNA levels in the same sample. For RT-qPCR of MAH11-infected macrophages, infections were conducted as described below and, at the indicated time points, 1 mL TRIzol reagent (Thermo Fisher) was added to triplicate wells. Cells were scraped, and RNA extracted and processed as described above. The following primers were used: sigA Fw (5’-CCTACCTCAAGCAGATCGGT-3’); sigA Rv (ATCTCCGACATCAGCTGGG); dosR Fw (5’-GATGCTGACGTCGTTCACC-3’); dosR Rv (5’-TCCATGCCCTTGATGTCCTT-3’); MAV_1793 Fw (5’-GCCCAAGGACCTGACTAACC-3’); MAV_1793 Rv (5’-TCCACTCCTTGAACTTCGCC-3’); MAV_2505 Fw (5’ATGCAAATGACCGCGGATAC-3’); MAV_2505 Rv (5’-GATTTCACTGTTCGGCGCG-3’); MAV_2507 Fw (5’TTGCTCGGTTCGGTCAGTTC-3’); and MAV_2507 Rv (5’-GGTAGGGCATCATCGGATCC-3’).
2.8 RNAseq library preparation and data analysisRNA-seq libraries preparation and data processing was conducted as described previously (Belardinelli et al., 2022). Gene expression and differential expression analysis was completed in R (version 3.6.0) using DESeq2 (version 1.26.0) (Love et al., 2014). Genes were identified as differentially expressed if they had a log2 fold change greater than 2 and a Benjamini-Hochberg multiple testing correction adjusted P-value of 0.05 or less. Venn diagrams were designed and analyzed using InteractiVenn (Heberle et al., 2015).
2.9 Data availabilityThe sequencing data described in this publication have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1191895, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1191895.
2.10 Macrophage infectionsMurine RAW 264.7 cells were grown in Dulbecco’s modified Eagle medium (DMEM) (Corning) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin and seeded in 24-well or 96-well plates for RNA extraction and CFU enumeration, respectively. After a 24 h incubation period to allow the cells to attach to the wells, cells were washed with warm PBS and triplicate wells were infected with well-dispersed suspensions of the WT and mutant strain in antibiotic-free DMEM at an MOI of 1 for 2 h at 37°C. Cells were then washed three times with warm PBS and the wells were replenished with DMEM containing 250 μg mL-1 amikacin to kill extracellular bacteria. After an hour of incubation, cells were washed three more times with PBS and incubated in DMEM for the remainder of the experiment. Two, 24, 48, 72 and 120 hours post-infection, viable intracellular bacteria were assessed by lysing the cells in sterile water containing 0.1% Triton X-100, and plating serial dilutions on 7H11-ADC agar plates to enumerate CFUs.
2.11 Mouse infectionsAll protocols and use of these animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Colorado State University. Studies were performed in accordance with recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Six to eight-week-old female BALB/c mice were purchased from Jackson laboratories. Animals were rested for a week, weighed, and divided into randomized groups for the study. Mice were infected with fresh cultures of MAH11 WT and MAH11ΔdosRS (grown to OD600 nm 0.6 – 0.8 in 7H9-ADC-Tween 80). To this end, mice were anesthetized with a mix of isoflurane and oxygen (1.5- 2% at a flow rate of 0.4-0.8 L/min) and two doses of fifty microliters each of inoculum were delivered intratracheally as an intrapulmonary spray instillation to each animal using a high-pressure syringe device (PennCentury), for a targeted dose of 1x106 CFU/lung. To confirm the actual bacterial deposition in the lungs, mice (n=5) were sacrificed 16 h after instillation and the whole lung prepared for viable bacteria quantification. Tissues were homogenized using the Precellys Tissue Homogenizer (Precellys Lysing Kit, 220325-830) and serial dilutions of each homogenate were applied to 7H11-OADC agar supplemented with carbenicillin (Sigma-Aldrich) and cycloheximide (GoldBio). The plates were cultured for 2-3 weeks at 37°C until visible CFUs could be enumerated. Following infection, the mice were monitored daily for indications of weight loss or abnormal behavior requiring pre-endpoint euthanasia. The remaining groups of mice (n=5) were euthanized at each defined timepoint, and the left lobe lung and spleen were enumerated for CFUs while the right lung lobe was fixed and permeabilized in 4% para-formaldehyde (PFA) for staining and histological analysis. Mice were euthanized via narcosis with CO2 (5.8L, flow rate of 3.0 (1.7-4) L/min). Standard histological protocols for sectioning and staining with Hematoxylin-Eosin (H&E) and Ziehl-Neelsen acid fast stain were used. Slide were scanned at 40X magnification using a multispectral automated PhenoImager (Akoya Biosciences) and analyzed as described previously (Cooper et al., 2024), and final edits performed by the reviewing pathologist.
2.12 Statistical analysisStatistical tests were performed as indicated in the figure legends. Calculations were performed using Graphpad Prism version 9.5.1 for Windows (San Diego California USA).
3 Results3.1 Transcriptional response of MAC to growth under microaerophilic conditionsThe M. avium complex consists of a growing number of species as described in recent reviews (Daley, 2017; Busatto et al., 2019). However, the three most important human pathogens are M. avium, M. intracellulare and M. chimaera. Of these, M. avium subsp. hominissuis is the most frequent species isolated from patients with pulmonary infections and was thus chosen for the purpose of this study. Isolate MAH11 was chosen for its amenability to genetic manipulations, genome sequence availability, and ability to establish an infection in the lungs of mice (Dragset et al., 2019).
RNAseq transcriptional profiling was used to determine differential gene expression between wild-type (WT) M. avium subsp. hominissuis MAH11 grown for 16 h in Dubos-Tween-albumin broth under well-aerated (normoxic) conditions and microaerophilic conditions (Figure 1A). The list of differentially expressed (DE) genes (log2 fold change (FC) > 2 with a false discovery rate adjusted P < 0.05), which is presented in Table 1, indicated that 16 genes were upregulated under microaerophilic conditions. Eleven of them were upregulated more than 5 log2-fold. No genes were found to be significantly downregulated per the log2 FC and Padj cut-off values set for this study. Upregulated genes mapped to seven different locations of the M. avium genome with the largest cluster of induced genes encompassing MAV_2494 through MAV_2508 (based on the M. avium 104 genome annotation). The products of these genes included four universal stress family proteins (USPs), three NAD(P)H nitroreductases, a sensor histidine kinase (MAV_2508) previously proposed to function with DosR (Gerasimova et al., 2011), a putative fatty acyl desaturase, a truncated hemoglobin, oxidoreductases, and hypothetical proteins of unknown function (Table 1). dosS (MAV_4108) and dosR (MAV_4109) were not part of the list despite the finding of a putative DosR-binding motif in the promoter region of M. avium dosR suggestive of autoregulation. RT-qPCR analyses conducted on three arbitrarily chosen DE genes (MAV_1793, MAV_2505 and MAV_2507) confirmed the RNAseq results, showing a 630 to 17,800-fold upregulation of these genes, and revealed a comparatively mild (~ 4-fold) induction of dosR under microaerophilic conditions (Figure 1B). The same three upregulated genes under microaerophilic conditions were also induced ~ 160 to 1,954-fold upon exposure of M. avium MAH11 to NO (Figure 1C). dosR expression, in contrast, was not significantly induced by NO (Figure 1C).
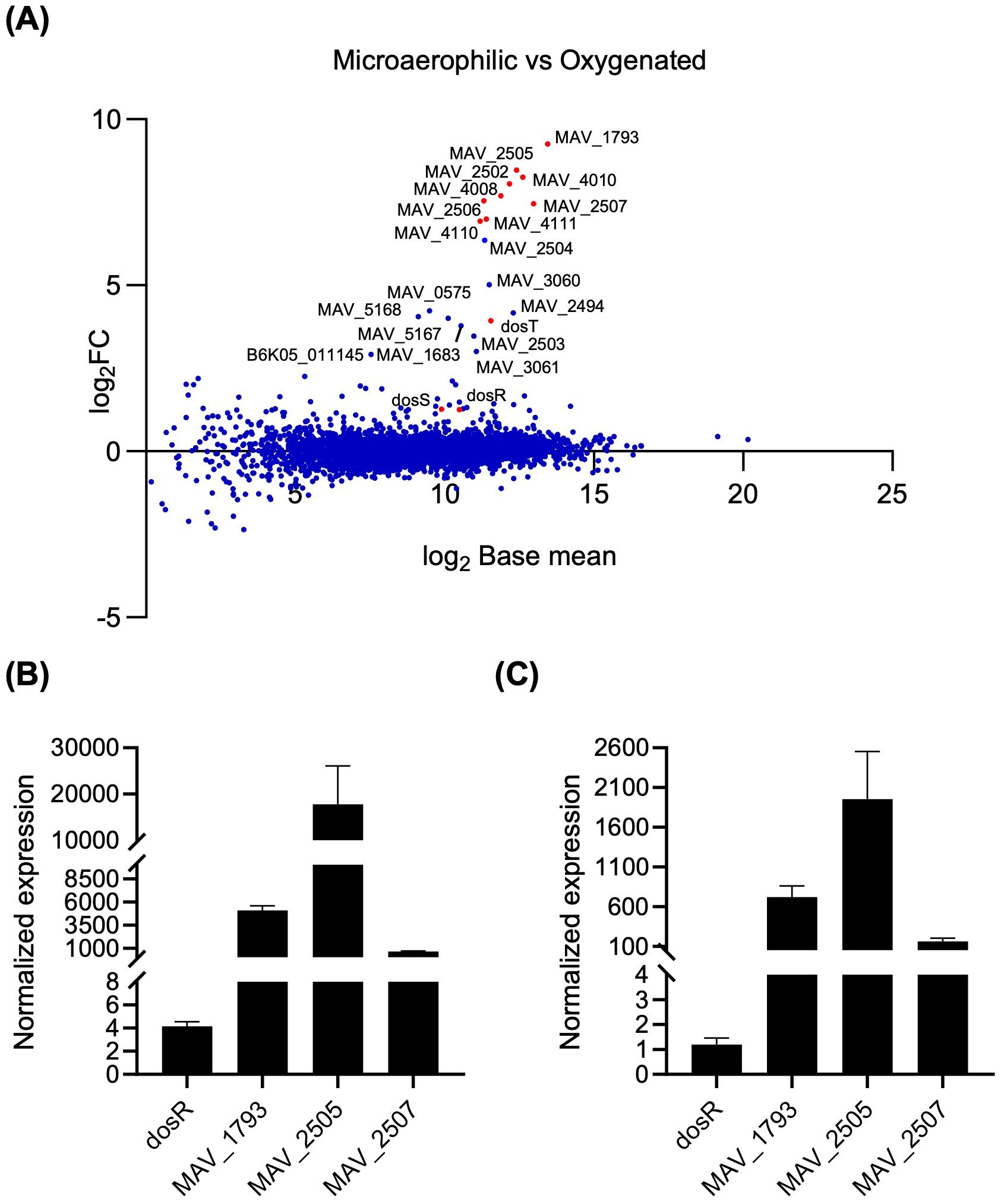
Figure 1. Transcriptional response of wild-type M. avium MAH11 to oxygen depletion. (A) Differentially expressed genes in WT M. avium MAH11 grown in Dubos-Tween albumin broth for 24 h under microaerophilic conditions in standing T25 vented tissue culture flasks compared to well-aerated shaking flasks (oxygenated). Values are expressed as log2-fold change (FC) versus log2 base mean. Positive log2FC values indicate upregulation under microaerophilic conditions. Genes predicted bioinformatically to belong to the DosRS regulon of M. avium, including dosR and dosS, are shown in red (Gerasimova et al., 2011). (B) RT-qPCR showing the upregulation of dosR and the MAV_1793, MAV_2505 and MAV_2507 genes in WT MAH11 grown under microaerophilic conditions compared to oxygenated conditions as described above. cDNA was normalized internally to sigA cDNA in the same samples. For each gene, cDNA levels under microaerophilic conditions are expressed relative to cDNA levels under oxygenated conditions arbitrarily set to 1. The results shown are means ± SD of biological triplicates (n = 3 RNA extractions and RT-qPCR reactions). (C) RT-qPCR showing the upregulation of MAV_1793, MAV_2505 and MAV_2507 in WT MAH11 grown in 7H9-ADS-Tween 80 to an OD600nm of 0.2 and treated with 500 μM DETA/NO for 1 h. cDNA was normalized internally to sigA cDNA in the same samples. cDNA levels in NO-treated samples are expressed relative to cDNA levels measured in the untreated control arbitrarily set to 1. The results shown are means ± SD of biological triplicates (n = 3 RNA extractions and RT-qPCR reactions).
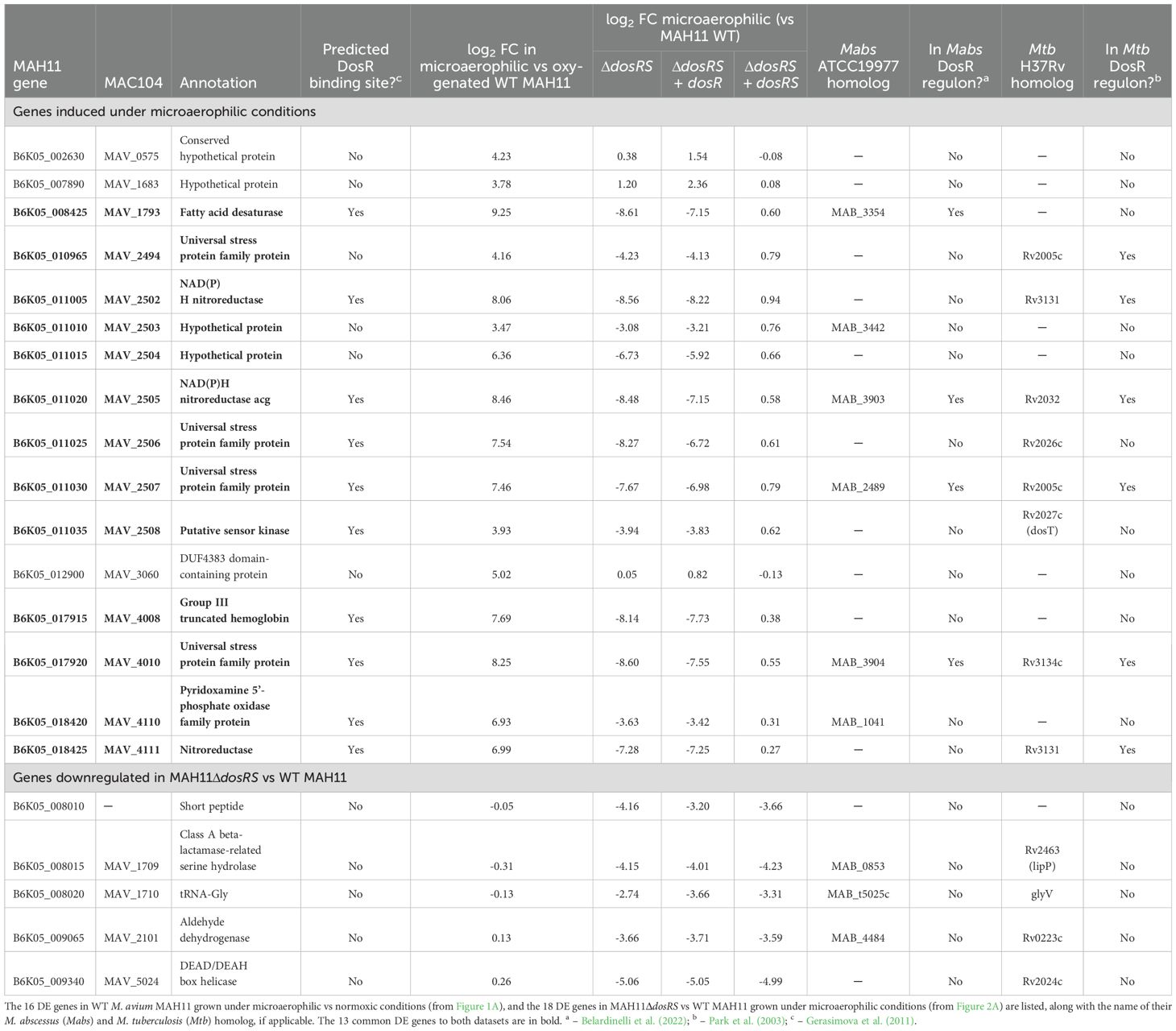
Table 1. DosRS-dependent and -independent response of M. avium MAH11 to microaerophilic conditions.
3.2 Role of DosRS in the transcriptional adaptation of M. avium to anaerobiosisWith the goal to determine how the loss of a functional DosRS regulatory system might impact the transcriptional response of M. avium to oxygen depletion, a dosRS knock-out mutant was generated by allelic replacement in MAH11 (Supplementary Figure S1), and the gene expression of the mutant under microaerophilic conditions was compared to that of the similarly grown WT parent using RNAseq transcriptional profiling. Two complemented mutants, one fully complemented with rescue copies of dosR and dosS, and one solely rescued with dosR, were included in the study.
In the absence of DosRS, 18 genes were expressed at lower levels relative to WT MAH11 (log2 fold change > 2 with a false discovery rate adjusted P < 0.05) (Figure 2A; Table 1). Thirteen of these genes were identical to those reported to be upregulated in WT MAH11 grown under microaerophilic conditions compared to normoxic conditions (Figure 2B; Table 1). We conclude from these results that the upregulation of these 13 genes in response to oxygen depletion is directly or indirectly dependent on the two-component regulator DosRS. No genes were expressed at higher levels in the mutant relative to the WT strain (per the log2 FC and Padj cut-off values set for this study) suggesting that M. avium DosRS solely acts as an activator under microaerophilic conditions. Complementation of dosRS knock-out with a WT copy of the dosRS genes restored WT gene expression in the mutant (Table 1). In contrast, complementation of the mutant with the sole dosR gene did not restore the expression of any of the 13 genes (Table 1). It follows that the sensor kinase encoded by dosS is required for the upregulation of these genes under microaerophilic conditions, and that no other sensor kinase can compensate for its activity. While the putative sensor histidine kinase encoded by MAV_2508 has previously been proposed to function with M. avium DosR (Gerasimova et al., 2011), an analysis of its primary sequence indicated that it is in fact devoid of a functional heme-binding (GAF-A) domain (Sivaramakrishnan and de Montellano, 2013) (data not shown). Collectively, these results thus exclude a participation of MAV_2508 in the DosR-mediated response of M. avium to hypoxia. RT-qPCR analyses conducted on three DE genes (MAV_1793, MAV_2505 and MAV_2507) confirmed the RNAseq results (Figure 2C).
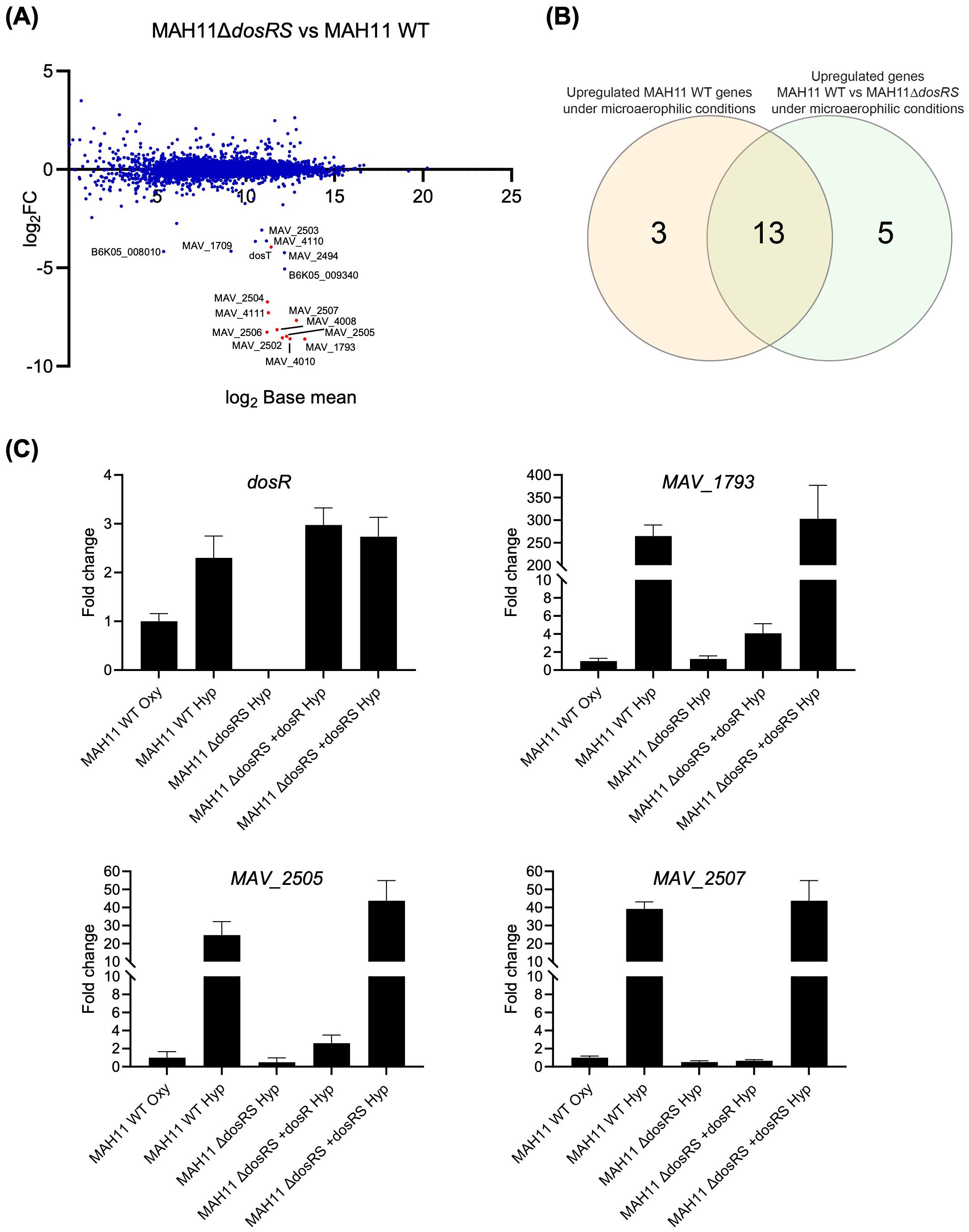
Figure 2. Effect of disrupting dosRS on the transcriptional profile of M. avium MAH11 under microaerophilic conditions. (A) Differentially expressed genes comparing M. avium MAH11 WT to MAH11ΔdosRS grown in Dubos-Tween albumin broth for 24 h under microaerophilic conditions in standing T25 vented tissue culture flasks. Values are expressed as log2FC versus log2 base mean. Negative log2FC values indicate reduced expression in MAH11ΔdosRS compared to WT MAH11. Genes predicted bioinformatically to belong to the DosRS regulon of M. avium (not including dosR and dosS) are shown in red (Gerasimova et al., 2011). (B) Venn-diagram showing the overlap between genes induced under microaerophilic conditions in WT MAH11 (from Figure 1A; log2FC > 2, padj < 0.05) and genes downregulated in MAH11ΔdosRS under microaerophilic conditions from (A) (log2FC < -2, padj < 0.05). (C) An aliquot of the RNA used for RNA-seq in (A) was reverse transcribed to cDNA followed by qPCR of dosR and DosRS regulon genes, MAV_1793, MAV_2505 and MAV_2507. All cDNAs were normalized internally to sigA cDNA. Results are expressed as fold changes over the level of expression of the same genes under well-aerated conditions (“oxy”). The results shown are means ± SD of biological triplicates (n = 3 RNA extractions and RT-qPCR reactions).
A prior bioinformatics study identified a putative DosR-binding motif in the promoter region of 12 M. avium 104 genes (Gerasimova et al., 2011). Ten of the 13 DosRS-dependent DE genes identified in our RNAseq study harbored this DosR-binding motif in their promoter (Table 1). In contrast, none of the 5 genes found to be differentially expressed between WT MAH11 and MAH11ΔdosRS, but not between normoxic vs microaerophilic WT MAH11 (Figure 2B), harbored this motif (Table 1). Moreover, WT expression of these five genes was not restored in MAH11ΔdosRS upon complementation with dosRS raising doubts as to their direct control by the two-component system regulator (Table 1).
In line with the fact that no triglyceride synthase gene appears to be under control of DosRS in MAH11 (Figure 2A; Table 1) [in contrast to the situation in Mtb (Rustad et al., 2008)], the metabolic labeling of WT MAH11, MAH11ΔdosRS and the two complemented mutants with [1,2-14C]acetate failed to reveal any differences in the de novo synthesis of triglycerides under microaerophilic conditions between the four strains (Supplementary Figure S2).
3.3 Effect of DosRS inactivation on the viability of MAH11 under hypoxic conditionsTo next determine whether the inability of the dosRS mutant to induce a subset of genes in response to oxygen depletion negatively impacted the survival of MAH11 under hypoxia, we resorted to the well-established Wayne model to culture MAH11 for up to 35 days under hypoxic conditions, and monitored the viability of the WT, mutant and complemented mutant strains over time. This experiment was conducted at both pH 7.3 and pH 5.7 because acidic conditions, which prevail in the phagolysosome of activated macrophages, have been shown to drastically decrease the anaerobic survival of an Mtb dosR mutant (Reichlen et al., 2017). The results, which are shown in Figure 3A, revealed a significant survival defect of the MAH11 dosRS mutant at both pHs, though the defect was more pronounced at acidic pH. WT survival was restored in the mutant complemented with dosRS but not with dosR only, highlighting the critical role of DosS in the ability of M. avium to sense and respond to hypoxic stress.
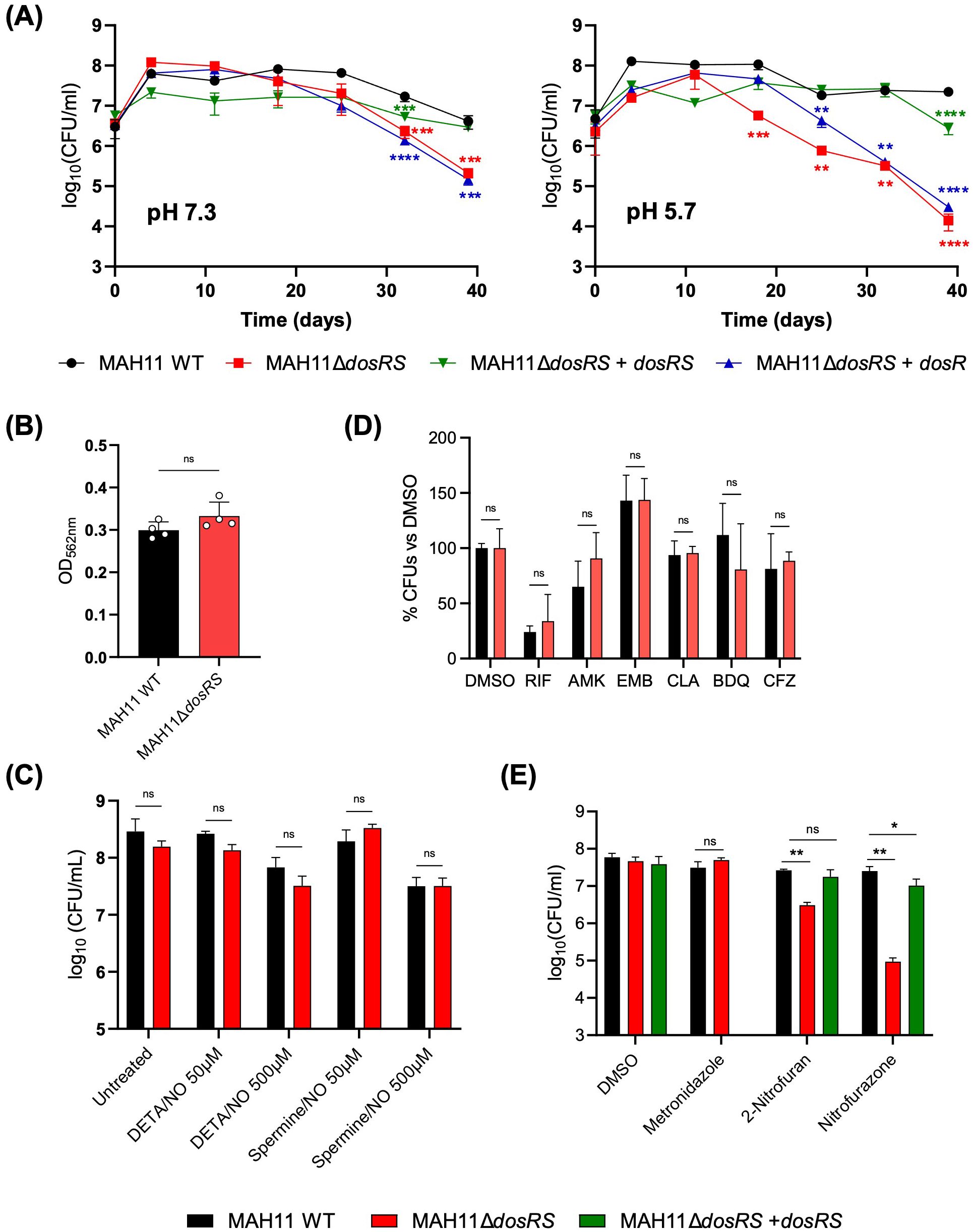
Figure 3. Phenotypic characterization of the M. avium dosRS knock-out mutant. (A) Comparative survival curves of MAH11 WT, MAH11ΔdosRS and MAH11ΔdosRS complemented with dosR or dosRS in the Wayne model at pH 7.3 (left) and 5.7 (right). At the indicated time points, tubes were opened and serial dilutions of the cultures plated for CFU enumeration. The results presented are the means ± SD of technical triplicates. Asterisks denote statistically significant differences relative to WT MAH11 pursuant to Dunnett’s one-way ANOVA, with **p<0.005; ***p<0.0005; and ****p<0.0001. (B) Biofilm formation of MAH11 WT and MAH11ΔdosRS after 14 days of incubation in SCFM in poly-D-lysine-coated microplates was determined by crystal violet staining. Crystal violet absorbed by the biofilm matrix was extracted with 300 μL of 30% acetic acid for 30 min followed by reading of the absorbance of the solution at 562 nm. The values reported on the Y-axis are the means ± SD of four biological replicates. “ns” indicates that the difference between the two strains is not significant pursuant to Dunnett’s one-way ANOVA. (C) Susceptibility of M. avium MAH11 WT and MAH11ΔdosRS to NO. Triplicate cultures of WT MAH11 and MAH11ΔdosRS grown in 7H9-ADS-Tween 80 were either untreated or treated with 50 or 500 μM DETA/NO or Spermine/NO for 24 h and subsequently plated for CFU enumeration. The values reported on the Y-axis are the means ± SD of three technical replicates. “ns” indicates that the difference between the two strains is not significant pursuant to Dunnett’s one-way ANOVA. The susceptibility of MAH11 strains to clinically used antibiotics (RIF, rifampicin; AMK, amikacin; EMB, ethambutol; CLA, clarithromycin; BDQ, bedaquiline; CFZ, clofazimine) (D) and metronidazole and nitrofurans (E) under hypoxic conditions was determined using the Wayne model. Upon onset of anaerobiosis (as determined by decolorization of methylene blue), drugs were added and the cultures incubated for another 7 days. At the end of the incubation time, serial dilutions of the cultures were plated to enumerate CFUs. The values reported on the Y-axis are the means ± SD of technical triplicates. Asterisks denote statistically significant differences relative to WT MAH11 pursuant to Dunnett’s one-way ANOVA, with **p<0.05; ***p<0.0005; and ****p<0.0001; ns, not significant. The results shown in (A) through (E) are representative of three independent experiments.
Consistent with the apparent lack of involvement of MAV_2508 in the hypoxic response of M. avium, an MAH11 mutant harboring a transposon insertion in this gene did not show any survival defects in the same hypoxic model (Supplementary Figure S3).
3.4 Effect of DosRS inactivation on biofilm formationThe impact of DosRS on the ability of M. avium to form biofilms was tested in our recently developed synthetic CF medium (SCFM) model (Belardinelli et al., 2021). A comparative assessment of the ability of the WT, dosRS mutant and complemented mutant strains to form biofilms in SCFM-containing poly-D-lysine-coated microtiter plates revealed no significant differences between strains (Figure 3B).
3.5 Impact of DosRS on the resistance of M. avium to NOTo next determine whether DosRS enhances the resistance of M. avium to NO, WT MAH11 and MAH11ΔdosRS cells were exposed to 50 or 500 μM of DETA/NO and spermine/NO for 24 h and viability was subsequently assessed by CFU plating. CFU counts revealed no difference in susceptibility to NO between the two strains (Figure 3C). Of note, M. avium MAH11 demonstrated a high-level intrinsic resistance to NO. Indeed, exposure to 500 μM of DETA/NO or spermine/NO for 24 h only resulted to in a 0.6 to 1 log10 decrease in viable MAH11 CFUs, whereas a reduction of 2 to 2.8 log10 CFUs was observed with M. abscessus (Supplementary Figure S4).
3.6 Impact of DosRS on drug tolerance in vitroIn M. abscessus, DosRS is important to the development of drug tolerance under early anaerobic dormancy (Belardinelli et al., 2022). To determine if this attribute of the DosRS two-component regulator also applied to M. avium, WT MAH11 and the dosRS knock-out mutant grown under hypoxic conditions were treated for 7 days with either DMSO (negative control) or the clinically relevant antibiotics, rifampicin, amikacin, ethambutol, clarithromycin, bedaquiline and clofazimine. Enumeration of surviving bacteria post-treatment indicated that the WT and mutant strains were both fully tolerant to almost all antibiotics tested (Figure 3D). Only rifampicin showed significant killing in the Wayne model, but WT and mutant strains did not significantly differ in their tolerance to this drug (Figure 3D). We conclude from this experiment that dosRS does not play a significant role in the ability of M. avium to develop tolerance to clinically used antibiotics under hypoxia.
Metronidazole and nitrofurans are classes of drugs that require partial reduction at their nitro groups by dedicated NAD(P)H nitroreductases to generate highly reactive, bactericidal, intermediates. Since as many as three NAD(P)H nitroreductase genes were found to be under control of DosRS in M. avium (Table 1), we set out to determine whether the lack of induction of these genes in the dosRS knock-out mutant under hypoxia enhanced the level of resistance to metronidazole, nitrofurazone and 2-nitrofuran. While the mutant displayed WT susceptibility to metronidazole, we found that it was significantly more susceptible to both nitrofurans (Figure 3E). WT susceptibility to nitrofurans was restored in the dosRS complemented mutant.
3.7 Impact of DosRS on the survival of M. avium inside macrophagesTo investigate potential effects of DosRS on the interactions of M. avium with macrophages, RAW 264.7 cells were infected with WT MAH11, the dosRS mutant and the dosRS complemented mutant, and the different strains compared for their intracellular replication and survival. Macrophages were infected at an MOI of 1. Two DosR regulon genes, MAV_1793 and MAV_2507, were found to be strongly induced in WT M. avium MAH11 (but not in the dosRS knock-out mutant) residing intracellularly, indicating that DosRS is induced upon macrophage infection (Figure 4A). Determination of intracellular CFUs 1, 2, 3 and 5 days post-infection revealed comparable infection kinetics for the three strains (Figure 4B). Thus, despite being induced intracellularly, the two-component regulatory system DosRS is not critical to the survival of M. avium inside macrophages under the conditions of this study.
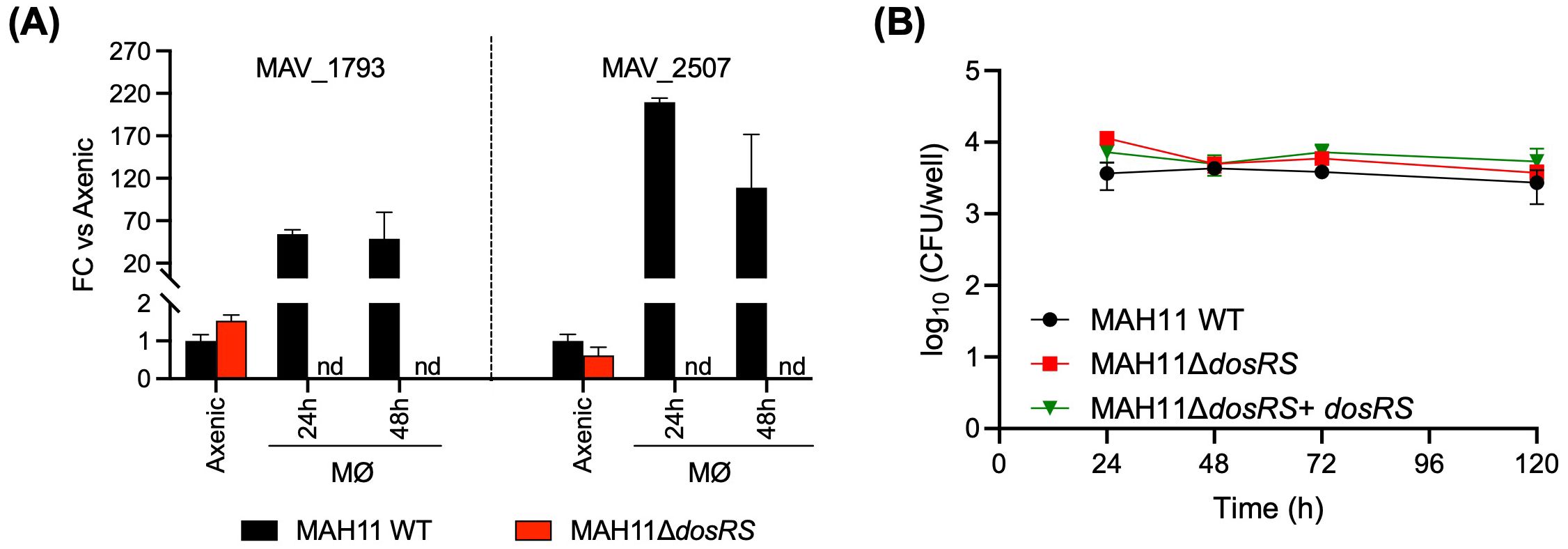
Figure 4. Role of DosRS on the survival of M. avium inside macrophages. (A) Evidence of DosRS regulon induction upon macrophage infection. The expression of DosRS regulon genes, MAV_1793 and MAV_2507, following infection of RAW 264.7 cells with WT MAH11 or MAH11ΔdosRS was monitored by RT-qPCR over time. RAW cells were infected for 2 h at an MOI of 1, followed by a 1 h-treatment with amikacin to eliminate extracellular bacteria. After this treatment (defined as time 0), cells were collected 24 and 48 h post-infection, and RNA extracted and processed for RT-qPCR analyses as described under Materials and Methods. Results are expressed as fold changes over the level of expression of the same genes in WT MAH11 grown to OD600 nm 0.2 in Dubos-Tween 80-albumin under well aerated conditions (labeled as “axenic” on the graph). The results were standardized to sigA expression levels in the same samples and are shown as means ± SD of biological triplicates (n = 3 RNA extractions and RT-qPCR reactions). nd, not detected. (B) RAW 264.7 cells were infected with either WT MAH11, MAH11ΔdosRS or MAH11ΔdosRS complemented with dosRS at an MOI of 1. At the indicated time points, cells were lysed and lysates plated on 7H11-OADC agar for CFU enumeration. Shown are the means ± SD of triplicate wells.
3.8 Virulence of a dosRS knock-out mutant in a murine model of M. avium infectionThe contribution of the DosRS two-component regulator to virulence and pathogenesis was next studied using a BALB/c mouse model of MAC infection. BALB/c mice were infected intratracheally with WT MAH11 and the dosRS knock-out mutant. Over the 56 days of infection, the bacterial burden in the lungs and spleen of WT- and ΔdosRS-infected animals remained comparable, slightly decreasing over time (Figure 5A). Limited disease progression was observed by histopathology regardless of infection with WT or ΔdosRS. Rare areas of inflammation contained a mixture of macrophages and lymphocytes with fewer neutrophils. Across all animals, inflammatory pathology affected less than 2% of the total lung tissue area, indicating a low burden of pulmonary pathology that was comparable between the two strains (Figure 5B).
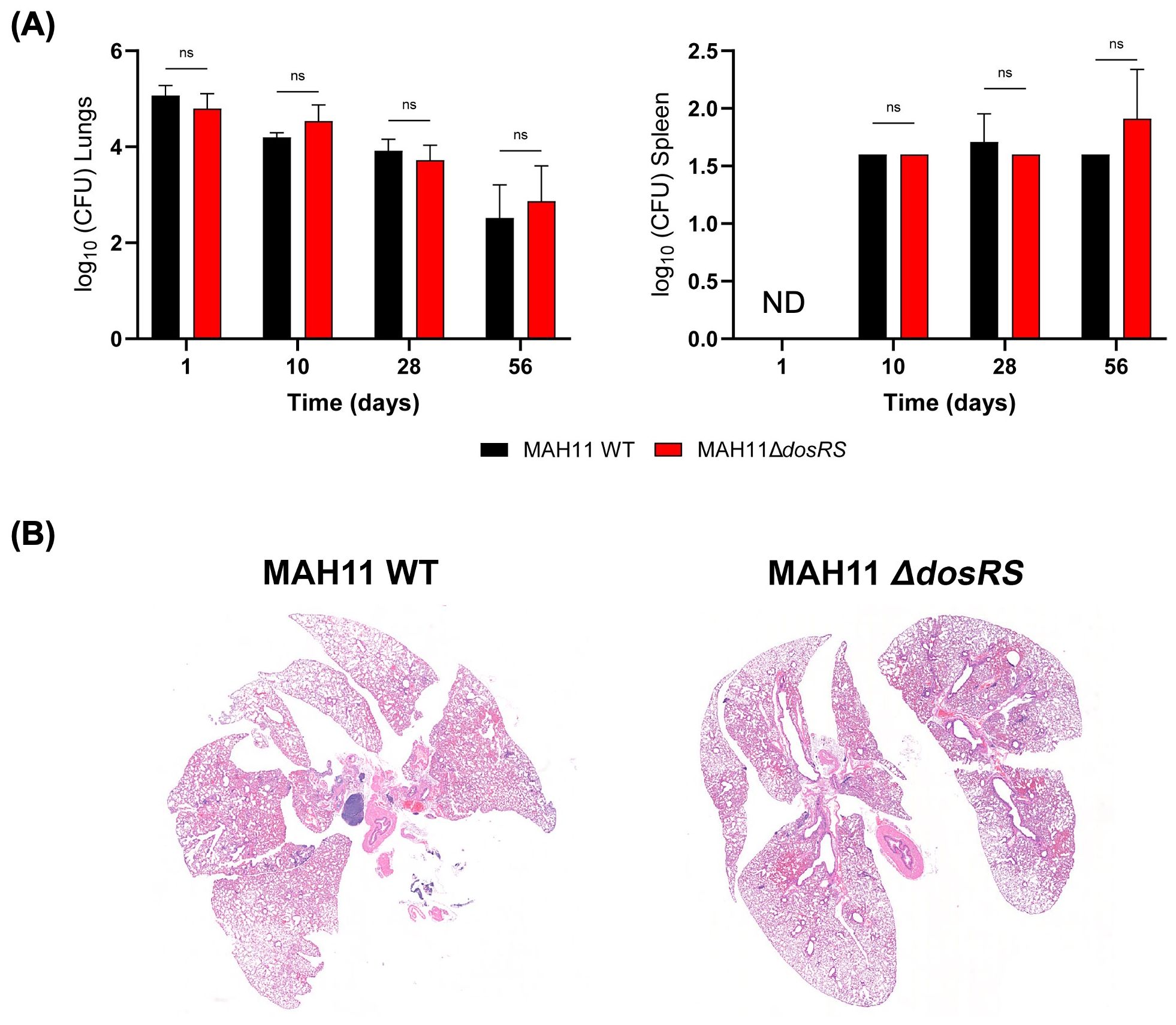
Figure 5. Infection of BALB/c mice with WT M. avium MAH11 and the dosRS knock-out mutant. (A) BALB/c mice were infected intratracheally with 1.0x106 CFU of either MAH11 WT or MAH11ΔdosRS. Groups of mice were euthanized 1, 10, 28 and 56 days post-infection, and lungs and spleens were taken for bacterial enumeration (CFU). For each bacterial strain, the CFU represent the average of 5 mice per time point, and bacterial loads are expressed as log10 CFU ± SD. ND, not determined. “ns” indicates that the difference between the two strains is not significant pursuant to two-way ANOVA. (B) Representative histopathology (on day 56) of mice infected with MAH11 WT or MAH11ΔdosRS is shown. Subgross, low magnification images of total lung tissue are shown. Inflammatory pathology is rare and extent of disease is not different between WT- and ΔdosRS-infected mice.
4 DiscussionHomologs of DosRS regulon genes have been identified not just in the genomes of pathogenic mycobacteria but also in that of environmental mycobacteria as well as other environmental prokaryotes and archaebacteria (Bartek et al., 2009; Gerasimova et al., 2011; Selvaraj et al., 2012). This broad distribution has long suggested that DosR regulons did not primarily evolve for pathogenesis or drug tolerance but rather for adaptation to anaerobic conditions within the environment, and has been adapted by Mtb and other mycobacterial pathogens of the human lung for survival inside activated macrophages and in the avascular necrotic regions of granulomas. Accordingly, the opportunistic NTM, M. avium, responded to a shift from normoxic to microaerophilic conditions by a strong (> 4-fold) upregulation of 16 genes, 13 of which appeared to require DosRS for induction (Figure 2B). The existence of a DosR-binding motif in the promoter region of 10 of these 13 genes indicates that greater than 75% of them are likely under the direct transcriptional control of DosR, thereby implicating the two-component regulatory system DosRS as a major driver of the early response of M. avium to hypoxia. The fact that the upregulation of these 13 genes upon oxygen depletion required both dosR and dosS (complementation of the dosRS mutant with dosR alone failed) establishes DosS as the sole and essential sensor kinase involved in this response (Table 1).
A bioinformatic study predicted a 12-gene DosRS regulon in M. avium 104 and an 8-gene regulon in M. abscessus, the smallest regulons of all mycobacterial species analyzed to date (Gerasimova et al., 2011). Comparatively, the Mtb DosRS regulon comprises 48 genes (Rustad et al., 2008) and that of M. marinum is predicted to contain 66 (Gerasimova et al., 2011). Our RNAseq results confirm the relatively small size of the M. avium DosRS regulon and validate the DosRS-dependent expression of 10 of the 12 genes predicted bioinformatically by Gerasimova et al. Comparing the 13 M. avium DosRS-dependent genes induced under microaerophilic conditions identified in this study (Figure 2B; Table 1) to the DosRS regulons of Mtb and M. abscessus, 6 and 4 homologs were shared between species, respectively, with three genes (MAV_2505 [acg], MAV_2507 and MAV_4010) being found in all three genomes (Table 1). These genes encode an NAD(P)H nitroreductase (Acg) and two putative universal stress proteins, respectively, and are part of the minimal dormancy regulon defined by Gerasimova et al. (2011). The other 10 M. avium DosRS-dependent genes induced under microaerophilic conditions share similar functions with genes reported earlier to be induced under microaerophilic conditions and/or upon exposure to NO in Mtb and M. abscessus (Voskuil et al., 2003; Rustad et al., 2008; Simcox et al., 2023). They encode a putative fatty acid desaturase, a sensor kinase-like protein (MAV_2508), a truncated hemoglobin potentially involved in protection against NO, a pyridoxamine 5’-phosphate oxidase involved in the recycling of enzymatic cofactors, two additional nitroreductases potentially involved in protection against nitrogen stress, two additional universal stress proteins, and two hypothetical proteins of unknown function.
Like Mtb, M. avium has the ability to persist in a non-replicating state inside foamy macrophages that contain abundant lipid bodies (Caire-Brandli et al., 2014). M. avium residing within these cells forms intracytoplasmic triglyceride inclusions that provide a source of nutrients and energy facilitating persistence or bacterial regrowth. In Mtb, triglyceride build-up has been associated with non-replicating persistence and drug tolerance, and is largely controlled by tgs1, a triglyceride synthase gene under control of DosR (Baek et al., 2011). The comparable production of triglycerides by WT MAH11 and MAH11ΔdosRS grown under microaerophilic conditions (Supplementary Figure S2), and apparent lack of regulation of any of the 10 M. avium triglyceride synthase genes by DosR (Figure 2A) point to an alternative mechanism of control of triglyceride synthesis in this species.
In line with the nature of the M. avium DosRS regulon, the ability of MAH11ΔdosRS to survive in the Wayne hypoxic model was significantly reduced (Figure 3A). Lowering the pH from 7.3 to 5.7 further decreased survival, presumably due to the compromised ability of the mutant to maintain cytosolic pH homeostasis (Reichlen et al., 2017). Despite the fitness advantage conferred by DosRS in hypoxia, loss of DosRS had no significant impact on the ability of M. avium to form biofilms in host-relevant SCFM medium. This result is in contrast to observations made in M. abscessus (Belardinelli et al., 2022) and suggests that M. avium has evolved alternative mechanisms to support the growth and survival of bacilli within the hypoxic environment of biofilms. Loss of DosRS also did not significantly impact the tolerance of M. avium MAH11 to clinically used antibiotics in the Wayne model, similar to the situation in Mtb (another slow-growing Mycobacterium) (Bartek et al., 2009) but opposite to that in the fast-growing species, M. abscessus (Belardinelli et al., 2022). A noticeable exception was that of nitrofurans to which the dosRS mutant became significantly more susceptible (Figure 3E). Nitrofurans, like nitroimidazoles such as metronidazole, are prodrugs that require NAD(P)H-dependent nitroreductase activity under anaerobic conditions to reduce their nitro or nitroso groups and release bactericidal intermediates. Some microbial NAD(P)H-dependent nitroreductases, however, inactivate nitro(so) drugs by fully reducing them to non-toxic forms of the antibiotic (Muller et al., 2013; Saghaug et al., 2023). The hypersusceptibility to nitrofurans of the MAH11ΔdosRS mutant (which has lost the ability to upregulate three putative nitroreductases under hypoxia) suggests that one or more of these enzymes have the ability to inactivate nitrofurans. M. avium DosRS-dependent nitroreductases, however, do not appear to confer any protective effect against metronidazole (Figure 3E).
Despite being induced upon macrophage infection, the DosRS regulon is not critical to the survival of M. avium MAH11 inside macrophages under the conditions of this study (Figure 4). It further did not confer any survival or pathogenic advantage in infected BALB/c mice over two months of infection (Figure 5). The fact that only one clinical isolate of M. avium was tested therein combined with the low pathogenicity of MAH11 in murine models of infection and the fact that BALB/c mice do not form necrotic hypoxic granulomas are limitations of this study. It is possible that the MAH11ΔdosRS mutant might only display an attenuation phenotype in animal models of infection that develop hypoxic caseous necrotic granulomas. This situation indeed has precedence with Mtb where dosR mutants only presented attenuation phenotypes in rabbit, guinea pig and nonhuman primate models of infection (Zheng et al., 2020). Whether the same conclusion applies to M. avium will have to await the development of models of infection mimicking the human pathology for this NTM pathogen.
Data availability statementThe data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1191895. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1191895.
Ethics statementThe animal study was approved by Institutional Animal Care and Use Committee (IACUC) at Colorado State University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributionsJB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. CA: Conceptualization, Data curation, Formal Analysis, Investigation, Writing – review & editing. KM: Investigation, Writing – review & editing. HL: Conceptualization, Investigation, Writing – review & editing. MD: Formal Analysis, Investigation, Methodology, Resources, Writing – review & editing. WW: Investigation, Methodology, Writing – review & editing. BP: Formal Analysis, Writing – original draft, Writing – review & editing. MG-J: Conceptualization, Formal Analysis, Writing – review & editing. MJ: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants AI170504 (to MJ and MG-J). The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statementThe author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1545856/full#supplementary-material
ReferencesBartek, I. L., Rutherford, R., Gruppo, V., Morton, R. A., Morris, R. P., Klein, M. R., et al. (2009). The DosR regulon of M. tuberculosis and antibacterial tolerance. Tuberculosis (Edinb) 89, 310–316. doi: 10.1016/j.tube.2009.06.001
PubMed Abstract | Crossref Full Text | Google Scholar
Belardinelli, J. M., Li, W., Avanzi, C., Angala, S. K., Lian, E., Wiersma, C. J., et al. (2021). Unique features of Mycobacterium abscessus biofilms formed in synthetic cystic fibrosis medium. Front. Microbiol. 12, 743126. doi: 10.3389/fmicb.2021.743126
PubMed Abstract | Crossref Full Text | Google Scholar
Belardinelli, J. M., Verma, D., Li, W., Avanzi, C., Wiersma, C. J., Williams, J. T., et al. (2022). Therapeutic efficacy of antimalarial drugs targeting DosRS signaling in Mycobacterium abscessus. Sci. Transl. Med. 14, eabj3860. doi: 10.1126/scitranslmed.abj3860
PubMed Abstract | Crossref Full Text | Google Scholar
Busatto, C., Vianna, J. S., da Silva, L. V. J., Ramis, I. B., da Silva, P. E. A. (2019). Mycobacterium avium: an overview. Tuberculosis (Edinb) 114, 127–134. doi: 10.1016/j.tube.2018.12.004
PubMed Abstract | Crossref Full Text | Google Scholar
Caire-Brandli, I., Papadopoulos, A., Malaga, W., Marais, D., Canaan, S., Thilo, L., et al. (2014). Reversible lipid accumulation and associated division arrest of Mycobacterium avium in lipoprotein-induced foamy macrophages may resemble key events during latency and reactivation of tuberculosis. Infect. Immun. 82, 476–490. doi: 10.1128/IAI.01196-13
留言 (0)